Transcriptome Analysis Reveals Multiple Genes and Complex Hormonal-Mediated Interactions with PEG during Adventitious Root Formation in Apple
Abstract
:1. Introduction
2. Results
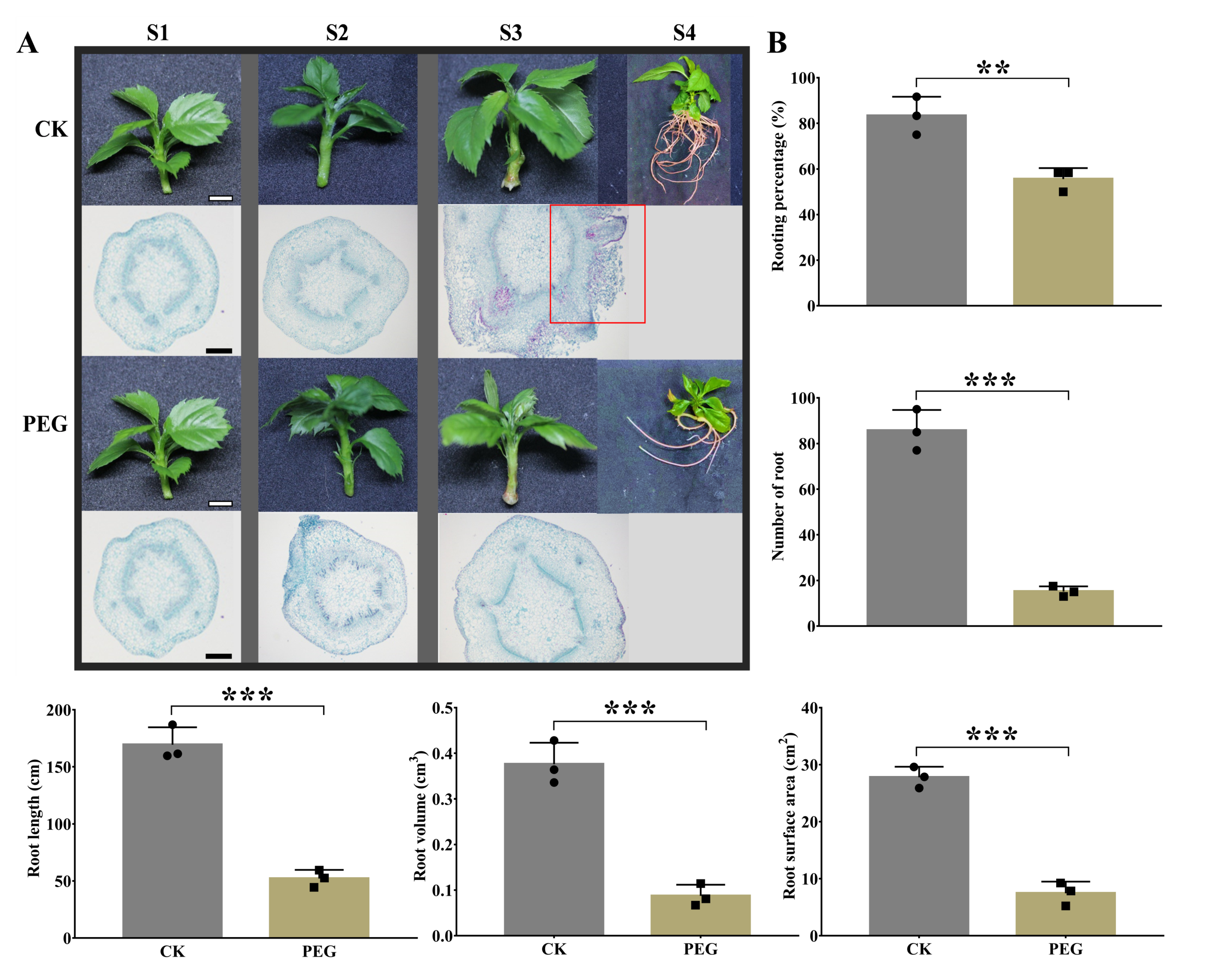
2.1. Morphological and Anatomical Changes by PEG Treatment during AR Formation
2.2. RNA-Seq and Analysis of Differentially Expressed Genes
2.3. Identification and Functional Annotation of DEGs Associated with AR Formation
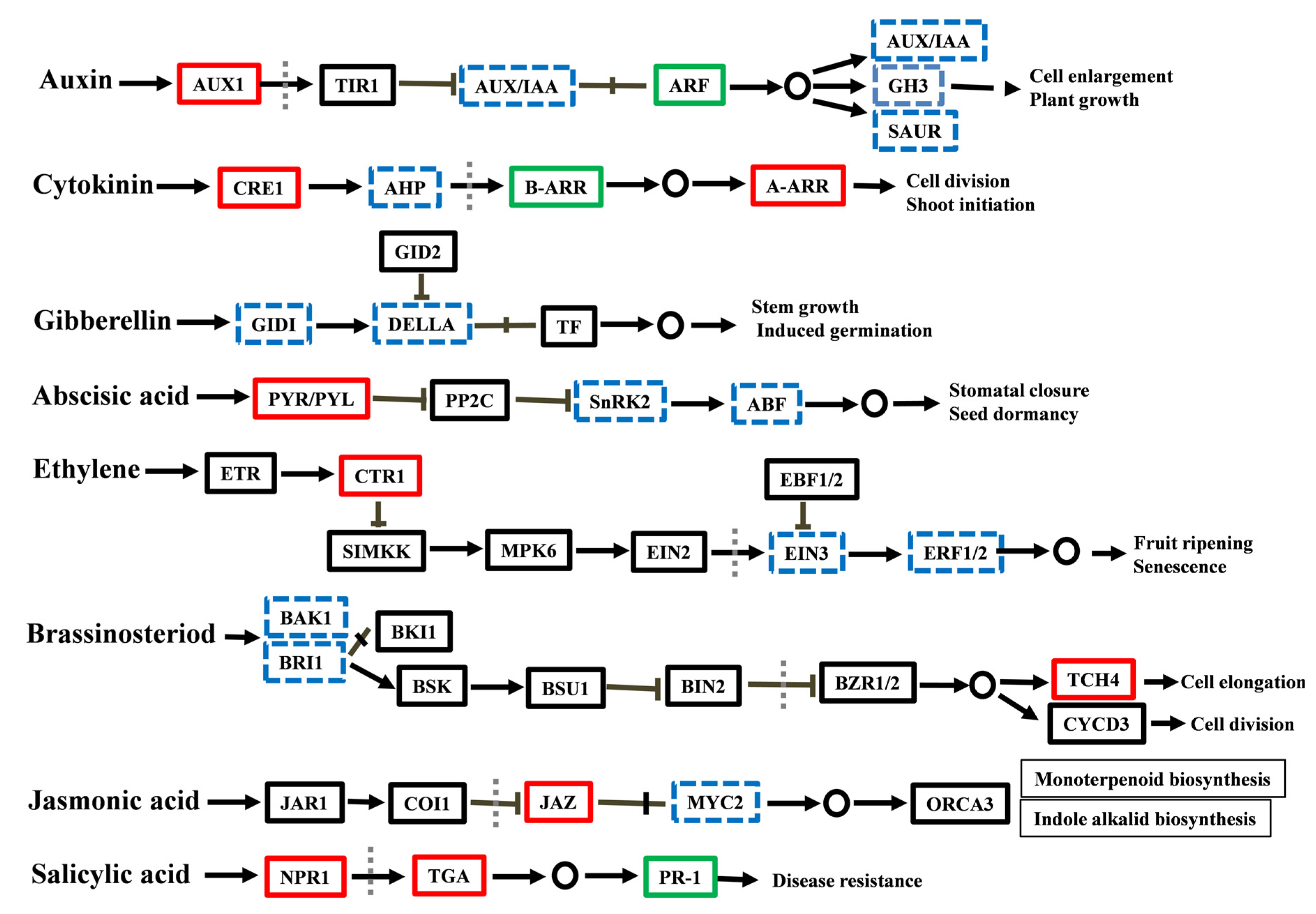
2.4. Endogenous Hormone Measurement and Gene Expression of Hormone Signaling-Related Genes
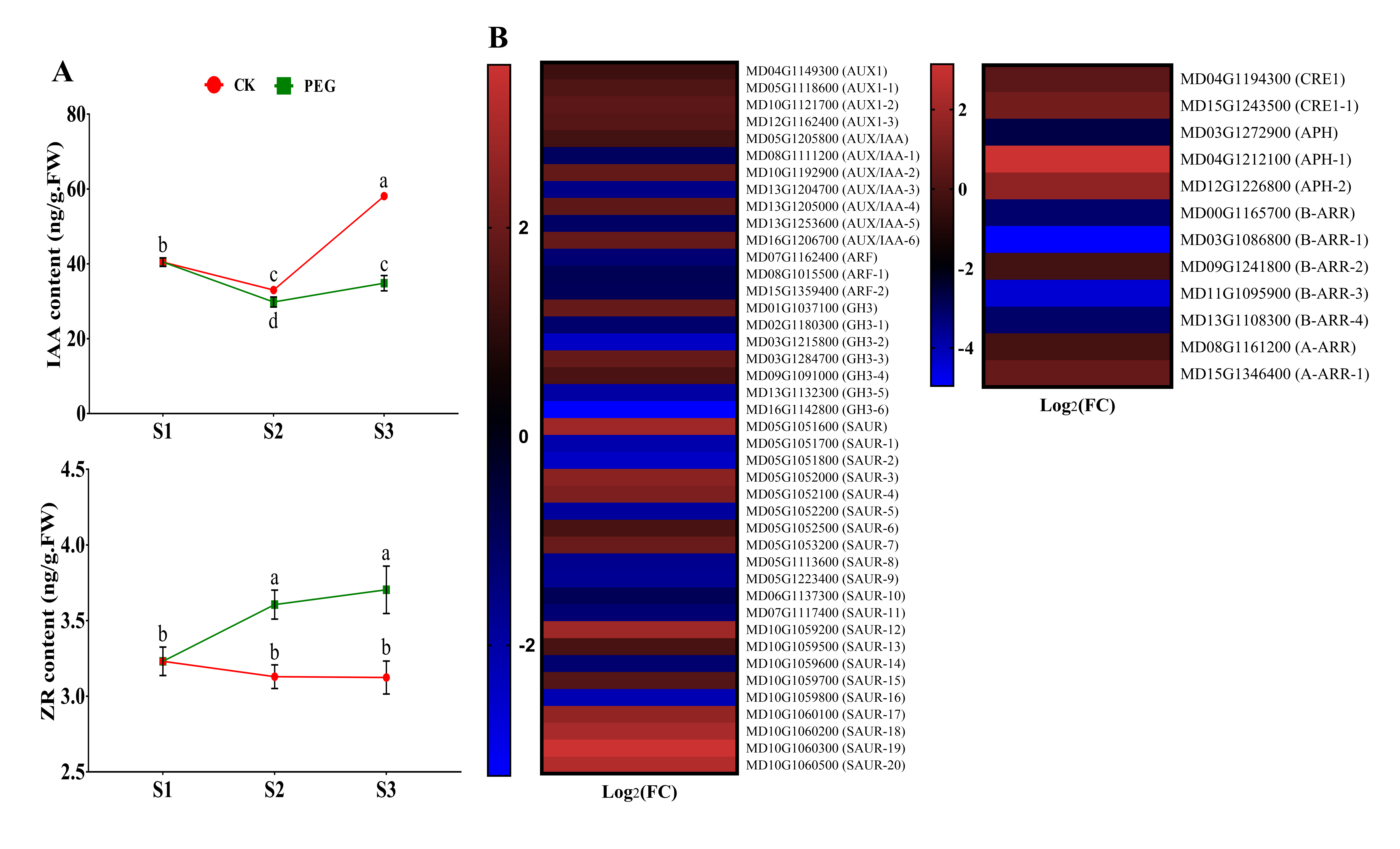
2.4.1. Measurement of IAA and ZR and Analysis of Related DEGs
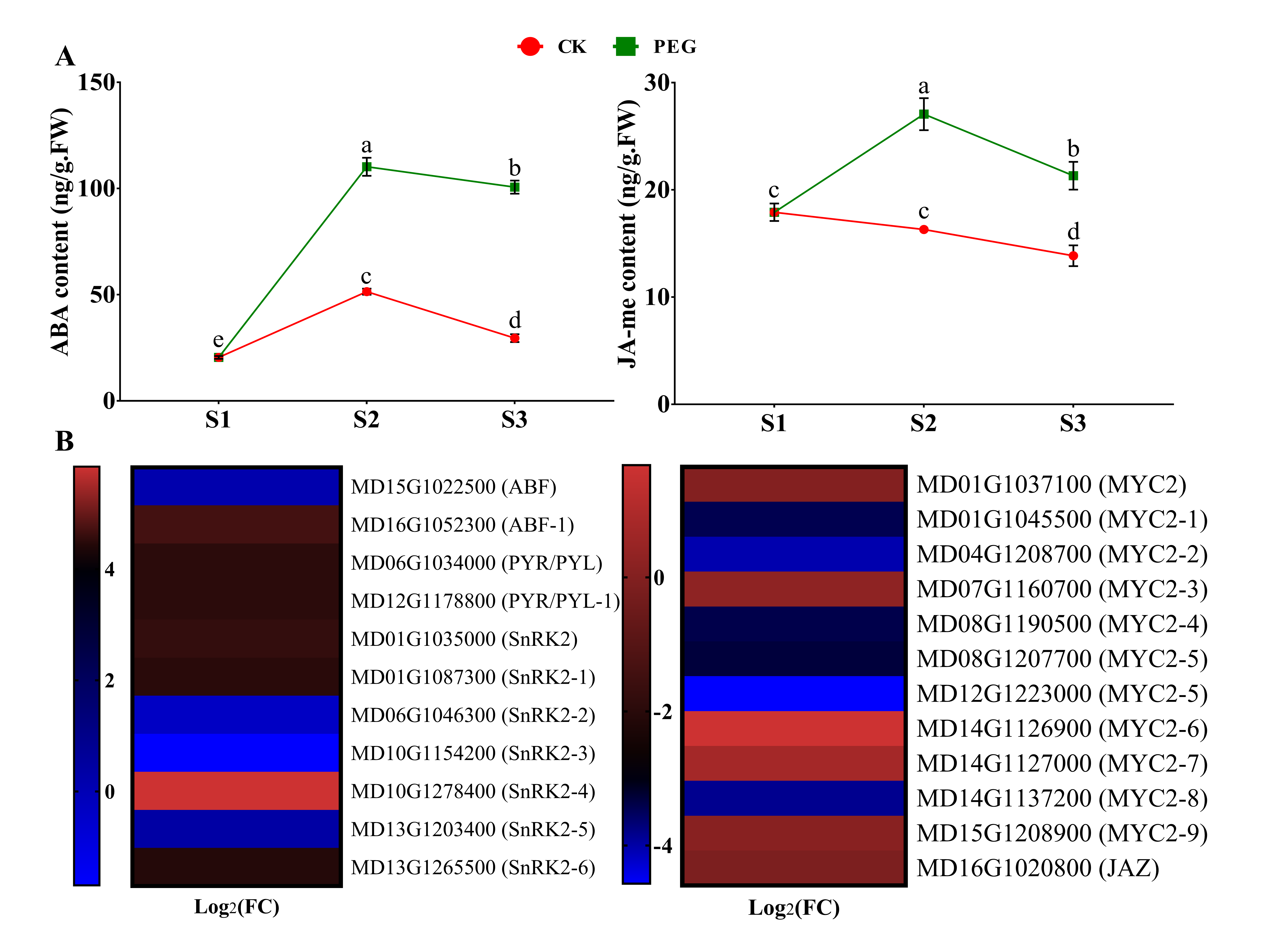
2.4.2. Measurement of ABA and JA-me and Analysis of Related DEGs
2.4.3. Measurement of BR and GA and Analysis of Related DEGs
2.4.4. Expression Analysis of ETH- and SA-Related DEGs
2.5. Expression Analysis of Sugar Metabolism-Related DEGs
2.6. Expression Analysis of Root Development and Cell Cycle-Related Genes
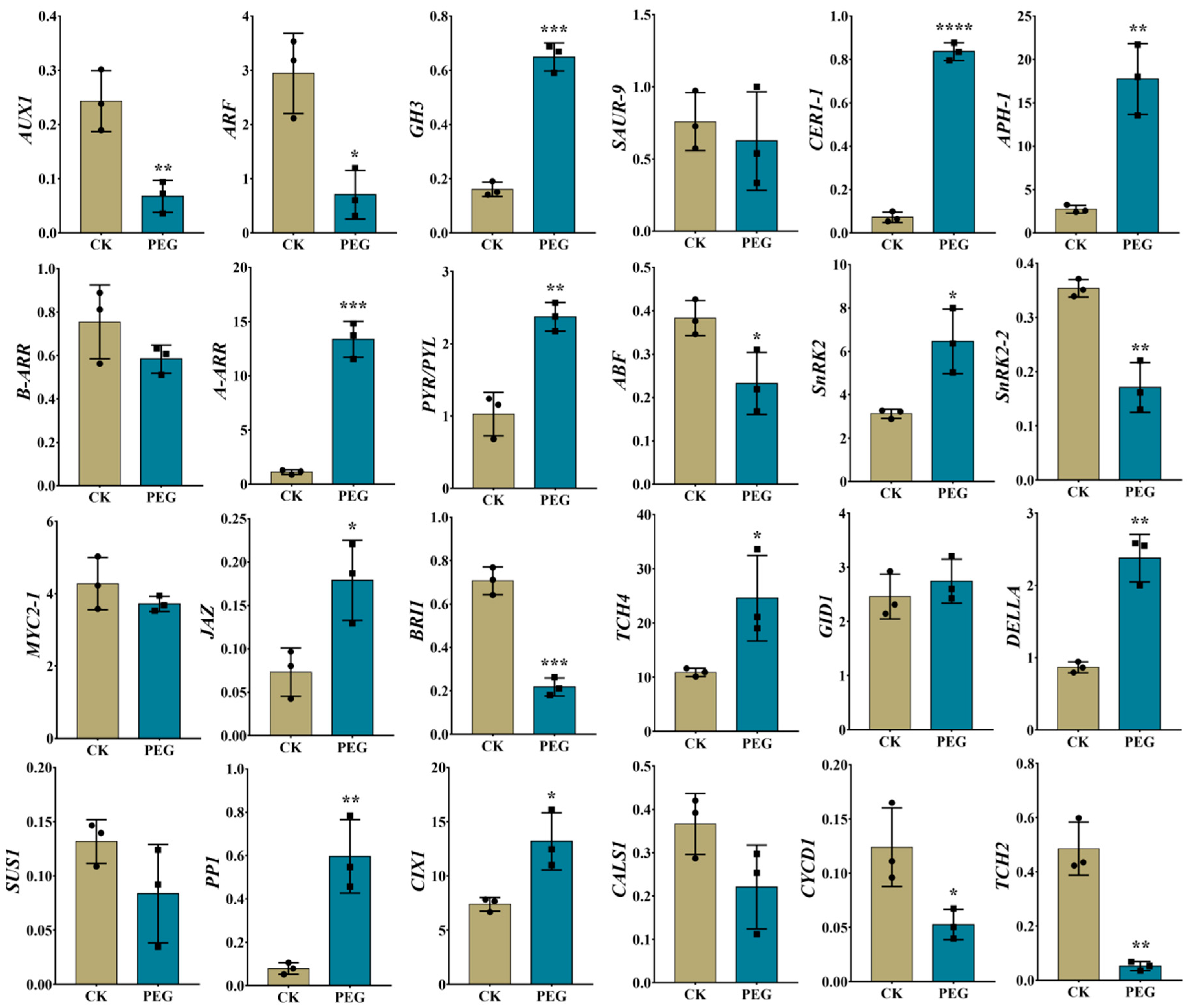
2.7. Validation of RNA-Seq Data Using RT-qPCR
3. Discussion
3.1. PEG Treatment Limits AR Formation in Apple GL-3 Cuttings
3.2. The Role of Hormonal Contents and Related DEGs in PEG-Induced AR Inhibition
3.3. PEG Inhibited ARs by Repressing Sugar Metabolism-Related DEGs
3.4. PEG Inhibited ARs by Repressing Root Development and Cell Cycle-Related Genes
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Anatomical and Morphological Observations
4.3. Measurement of Endogenous Hormones
4.4. RNA Extraction, cDNA Synthesis, and Library Preparation
4.5. Identification of DEGs and Bioinformatics Analysis
4.6. RT-qPCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Druege, U.; Hilo, A.; Pérez-Pérez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef] [Green Version]
- Naija, S.; Elloumi, N.; Jbir, N.; Ammar, S.; Kevers, C. Anatomical and biochemical changes during adventitious rooting of apple rootstocks MM 106 cultured in vitro. Comptes Rendus Biol. 2008, 331, 518–525. [Google Scholar] [CrossRef]
- Meng, Y.; Xing, L.; Li, K.; Wei, Y.; Wang, H.; Mao, J.; Dong, F.; Ma, D.; Zhang, Z.; Han, M. Genome-wide identification, characterization and expression analysis of novel long non-coding RNAs that mediate IBA-induced adventitious root formation in apple rootstocks. Plant Growth Regul. 2019, 87, 287–302. [Google Scholar] [CrossRef]
- Tahir, M.M.; Lu, Z.; Wang, C.; Shah, K.; Li, S.; Zhang, X.; Mao, J.; Liu, Y.; Shalmani, A.; Li, K. Nitrate Application Induces Adventitious Root Growth by Regulating Gene Expression Patterns in Apple Rootstocks. J. Plant Growth Regul. 2021, 1–12. [Google Scholar] [CrossRef]
- Zobel, R.W.; Kinraide, T.B.; Baligar, V.C. Fine root diameters can change in response to changes in nutrient concentrations. Plant Soil 2007, 297, 243–254. [Google Scholar] [CrossRef]
- Ge, Y.; Fang, X.; Liu, W.; Sheng, L.; Xu, L. Adventitious lateral rooting: The plasticity of root system architecture. Physiol. Plant. 2019, 165, 39–43. [Google Scholar] [CrossRef]
- Al-Mayahi, A.M.W. Effect of silicon (Si) application on Phoenix dactylifera L. growth under drought stress induced by polyethylene glycol (PEG) in vitro. Am. J. Plant Sci. 2016, 7, 1711–1728. [Google Scholar] [CrossRef] [Green Version]
- Rauf, M.; Munir, M.; ul Hassan, M.; Ahmad, M.; Afzal, M. Performance of wheat genotypes under osmotic stress at germination and early seedling growth stage. Afr. J. Biotechnol. 2007, 6, 971–975. [Google Scholar]
- Shi, G.; Xia, S.; Ye, J.; Huang, Y.; Liu, C.; Zhang, Z. PEG-simulated drought stress decreases cadmium accumulation in castor bean by altering root morphology. Environ. Exp. Bot. 2015, 111, 127–134. [Google Scholar] [CrossRef]
- Cajero-Sanchez, W.; Aceves-Garcia, P.; Fernández-Marcos, M.; Gutiérrez, C.; Rosas, U.; García-Ponce, B.; Álvarez-Buylla, E.R.; Sánchez, M.d.l.P.; Garay-Arroyo, A. Natural root cellular variation in responses to osmotic stress in arabidopsis thaliana accessions. Genes 2019, 10, 983. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Liang, Y.; Xing, L.; Mao, J.; Liu, Z.; Dong, F.; Meng, Y.; Han, M.; Zhao, C.; Bao, L. Transcriptome analysis reveals multiple hormones, wounding and sugar signaling pathways mediate adventitious root formation in apple rootstock. Int. J. Mol. Sci. 2018, 19, 2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plant. 2019, 166, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Werr, W. Cytokinin–auxin crosstalk in cell type specification. Trends Plant Sci. 2015, 20, 291–300. [Google Scholar] [CrossRef]
- Marhavý, P.; Bielach, A.; Abas, L.; Abuzeineh, A.; Duclercq, J.; Tanaka, H.; Pařezová, M.; Petrášek, J.; Friml, J.; Kleine-Vehn, J. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 2011, 21, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 17, 3007–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Tahir, M.M.; Li, S.; Mao, J.; Nawaz, M.A.; Liu, Y.; Li, K.; Xing, L.; Niu, J.; Zhang, D. Transcriptome analysis reveals the inhibitory nature of high nitrate during adventitious roots formation in the apple rootstock. Physiol. Plant. 2021, 173, 867–882. [Google Scholar] [CrossRef]
- Zhang, X.; Tahir, M.M.; Li, S.; Tang, T.; Mao, J.; Li, K.; Shao, Y.; Yang, W.; Niu, J.; Zhang, D. Effect of exogenous abscisic acid (ABA) on the morphology, phytohormones, and related gene expression of developing lateral roots in ‘Qingzhen 1’apple plants. Plant Cell Tissue Organ Cult. (PCTOC) 2021, 148, 23–34. [Google Scholar] [CrossRef]
- Vanneste, S.; De Rybel, B.; Beemster, G.T.; Ljung, K.; De Smet, I.; Van Isterdael, G.; Naudts, M.; Iida, R.; Gruissem, W.; Tasaka, M. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 2005, 17, 3035–3050. [Google Scholar] [CrossRef] [Green Version]
- Tahir, M.M.; Li, S.; Mao, J.; Liu, Y.; Li, K.; Zhang, X.; Lu, X.; Ma, X.; Zhao, C.; Zhang, D. High nitrate inhibited adventitious roots formation in apple rootstock by altering hormonal contents and miRNAs expression profiles. Sci. Hortic. 2021, 286, 110230. [Google Scholar] [CrossRef]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009, 181, 613–625. [Google Scholar] [CrossRef]
- Tahir, M.M.; Chen, S.; Ma, X.; Li, S.; Zhang, X.; Shao, Y.; Shalmani, A.; Zhao, C.; Bao, L.; Zhang, D. Transcriptome analysis reveals the promotive effect of potassium by hormones and sugar signaling pathways during adventitious roots formation in the apple rootstock. Plant Physiol. Biochem. 2021, 165, 123–136. [Google Scholar] [CrossRef]
- Li, G.; Ma, J.; Tan, M.; Mao, J.; An, N.; Sha, G.; Zhang, D.; Zhao, C.; Han, M. Transcriptome analysis reveals the effects of sugar metabolism and auxin and cytokinin signaling pathways on root growth and development of grafted apple. BMC Genom. 2016, 17, 150. [Google Scholar] [CrossRef] [Green Version]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; Van Der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2. 4 and SnRK2. 10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four A rabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Muscolo, A.; Sidari, M.; Anastasi, U.; Santonoceto, C.; Maggio, A. Effect of PEG-induced drought stress on seed germination of four lentil genotypes. J. Plant Interact. 2014, 9, 354–363. [Google Scholar] [CrossRef]
- Nadir, M.; Ansyar, I.; Khaerani, P. Effect of Various Polyethylene Glycol Concentrations on the Growth of Seedlings of Indigofera Zollingeriana; IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2019; p. 12040. [Google Scholar]
- Kumari, A.; Sairam, R.; Singh, S.K.; Krishna, G. Early growth response: An indicator of subsequent growth and yield of wheat genotypes grown under simulated water stress condition. Indian J. Plant Physiol. 2014, 19, 94–100. [Google Scholar] [CrossRef]
- Ljung, K.; Nemhauser, J.L.; Perata, P. New mechanistic links between sugar and hormone signalling networks. Curr. Opin. Plant Biol. 2015, 25, 130–137. [Google Scholar] [CrossRef]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’h, S.; Lucas, M.; De Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, J.; Xu, J.; Seifertová, D.; Brewer, P.B.; Růžička, K.; Blilou, I.; Rouquié, D.; Benková, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinohara, N.; Taylor, C.; Leyser, O. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef] [Green Version]
- Ioio, R.D.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [Green Version]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [Green Version]
- Aimar, D.; Calafat, M.; Andrade, A.M.; Carassay, L.; Bouteau, F.; Abdala, G.; Molas, M.L. Drought effects on the early development stages of Panicum virgatum L.: Cultivar differences. Biomass Bioenergy 2014, 66, 49–59. [Google Scholar] [CrossRef]
- Liting, W.; Lina, W.; Yang, Y.; Pengfei, W.; Tiancai, G.; Guozhang, K. Abscisic acid enhances tolerance of wheat seedlings to drought and regulates transcript levels of genes encoding ascorbate-glutathione biosynthesis. Front. Plant Sci. 2015, 6, 458. [Google Scholar]
- Kirkham, M. Effect of ABA on the water relations of winter-wheat cultivars varying in drought resistance. Physiol. Plant. 1983, 59, 153–157. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Păcurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Păcurar, M.; Demailly, H.; Geiss, G. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [Green Version]
- Mussig, C.; Shin, G.-H.; Altmann, T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003, 133, 1261–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pop, T.I.; Pamfil, D.; Bellini, C. Auxin control in the formation of adventitious roots. Not. Bot. Horti Agrobot. Cluj-Napoca 2011, 39, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.; Chen, L.-J.; Yu, S.-M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Conway, S.R.; Poethig, R.S. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 2011, 138, 245–249. [Google Scholar] [CrossRef] [Green Version]
- Pawlicki, N.; Welander, M. Influence of carbohydrate source, auxin concentration and time of exposure on adventitious rooting of the apple rootstock Jork 9. Plant Sci. 1995, 106, 167–176. [Google Scholar] [CrossRef]
- Appeldoorn, N.J.; Sergeeva, L.; Vreugdenhil, D.; Van Der Plas, L.H.; Visser, R.G. In situ analysis of enzymes involved in sucrose to hexose-phosphate conversion during stolon-to-tuber transition of potato. Physiol. Plant. 2002, 115, 303–310. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voß, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching out in roots: Uncovering form, function, and regulation. Plant Physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioio, R.D.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef] [Green Version]
- Müller, D.; Leyser, O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011, 107, 1203–1212. [Google Scholar] [CrossRef] [Green Version]
- Dai, H.; Li, W.; Han, G.; Yang, Y.; Ma, Y.; Li, H.; Zhang, Z. Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 2013, 164, 202–208. [Google Scholar] [CrossRef]
- Li, K.; Wei, Y.-H.; Wang, R.-H.; Mao, J.-P.; Tian, H.-Y.; Chen, S.-Y.; Li, S.-H.; Tahir, M.-M.; Zhang, D. Mdm-MIR393b-mediated adventitious root formation by targeted regulation of MdTIR1A expression and weakened sensitivity to auxin in apple rootstock. Plant Sci. 2021, 308, 110909. [Google Scholar] [CrossRef]
- Tahir, M.M.; Wang, H.; Ahmad, B.; Liu, Y.; Fan, S.; Li, K.; Lei, C.; Shah, K.; Li, S.; Zhang, D. Identification and characterization of NRT gene family reveals their critical response to nitrate regulation during adventitious root formation and development in apple rootstock. Sci. Hortic. 2021, 275, 109642. [Google Scholar] [CrossRef]
- Mao, J.; Niu, C.; Li, K.; Chen, S.; Tahir, M.M.; Han, M.; Zhang, D. Melatonin promotes adventitious root formation in apple by promoting the function of MdWOX11. BMC Plant Biol. 2020, 20, 536. [Google Scholar] [CrossRef]
- Meng, Y.; Mao, J.; Tahir, M.M.; Wang, H.; Wei, Y.; Zhao, C.; Li, K.; Ma, D.; Zhao, C.; Zhang, D. Mdm-miR160 participates in auxin-induced adventitious root formation of apple rootstock. Sci. Hortic. 2020, 270, 109442. [Google Scholar] [CrossRef]
- Xu, Q.; Chai, F.; An, X.; Han, S. Production method for paraffin section of invasive species of Bemisia tabaci. Plant Dis. Pests 2012, 3, 46. [Google Scholar]
- Dobrev, P.I.; Kamınek, M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 2002, 950, 21–29. [Google Scholar] [CrossRef]
- Wang, H.; Tahir, M.M.; Nawaz, M.A.; Mao, J.; Li, K.; Wei, Y.; Ma, D.; Lu, X.; Zhao, C.; Zhang, D. Spermidine application affects the adventitious root formation and root morphology of apple rootstock by altering the hormonal profile and regulating the gene expression pattern. Sci. Hortic. 2020, 266, 109310. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Muhammad, I.; Sharif, R.; Zhao, C.; Ullah, U.; Zhang, D.; Jing, X.-Q.; Amin, B.; Jia, P.; Mobeen Tahir, M. Zinc finger-homeodomain genes: Evolution, functional differentiation, and expression profiling under flowering-related treatments and abiotic stresses in plants. Evol. Bioinform. 2019, 15, 1176934319867930. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Ullah, U.; Muhammad, I.; Zhang, D.; Sharif, R.; Jia, P.; Saleem, N.; Gul, N.; Rakhmanova, A.; Tahir, M.M. The TAZ domain-containing proteins play important role in the heavy metals stress biology in plants. Environ. Res. 2021, 197, 111030. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Niu, C.; Li, K.; Mobeen Tahir, M.; Khan, A.; Wang, H.; Li, S.; Liang, Y.; Li, G.; Yang, Z. Exogenous 6-benzyladenine application affects root morphology by altering hormone status and gene expression of developing lateral roots in Malus hupehensis. Plant Biol. 2020, 22, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.M.; Zhang, X.; Shah, K.; Hayat, F.; Li, S.; Mao, J.; Liu, Y.; Shao, Y.; Zhang, D. Nitrate application affects root morphology by altering hormonal status and gene expression patterns in B9 apple rootstock nursery plants. Fruit Res. 2021, 1, 14. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

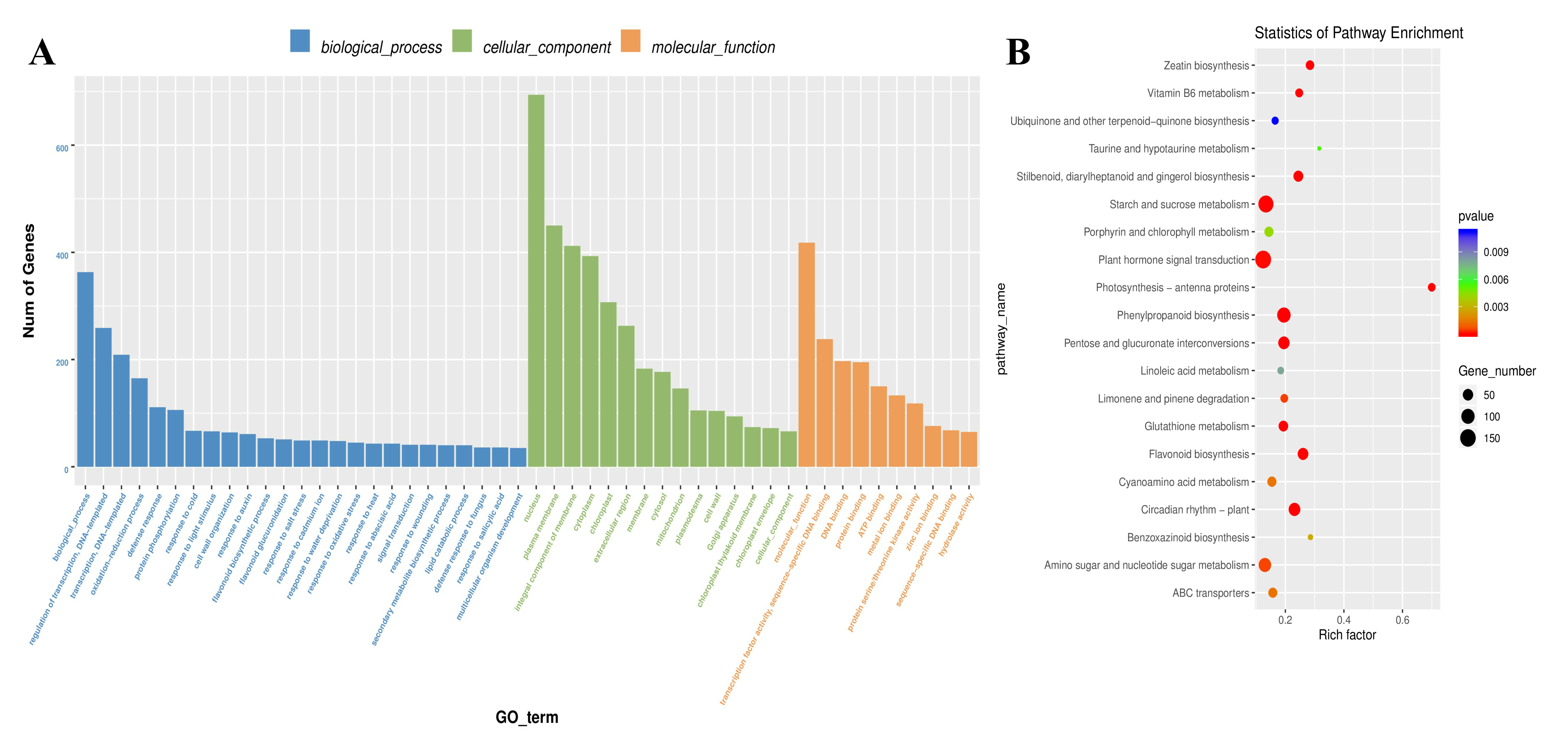

| Pathway Name | Pathway ID | Gene Number | Upregulated | Downregulated |
|---|---|---|---|---|
| Plant hormone signal transduction | ko04075 | 142 | 78 | 64 |
| Starch and sucrose metabolism | ko00500 | 138 | 49 | 89 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 84 | 23 | 61 |
| Circadian rhythm-plant | ko04712 | 71 | 41 | 30 |
| Carbon metabolism | ko01200 | 58 | 21 | 37 |
| Flavonoid biosynthesis | ko00941 | 56 | 27 | 29 |
| Biosynthesis of amino acids | ko01230 | 51 | 16 | 35 |
| Glycerolipid metabolism | ko00561 | 40 | 30 | 10 |
| Porphyrin and chlorophyll metabolism | ko00860 | 35 | 22 | 13 |
| ABC transporters | ko02010 | 33 | 15 | 18 |
| RNA transport | ko03013 | 30 | 14 | 16 |
| Zeatin biosynthesis | ko00908 | 29 | 11 | 18 |
| Glycolysis/Gluconeogenesis | ko00010 | 27 | 6 | 21 |
| Fructose and mannose metabolism | ko00051 | 25 | 8 | 17 |
| alpha-Linolenic acid metabolism | ko00592 | 24 | 7 | 17 |
| Vitamin B6 metabolism | ko00750 | 23 | 12 | 11 |
| Limonene and pinene degradation | ko00903 | 22 | 16 | 6 |
| Photosynthesis-antenna proteins | ko00196 | 21 | 21 | 0 |
| Pentose phosphate pathway | ko00030 | 20 | 6 | 14 |
| Terpenoid backbone biosynthesis | ko00900 | 19 | 10 | 9 |
| Carotenoid biosynthesis | ko00906 | 18 | 11 | 7 |
| mRNA surveillance pathway | ko03015 | 17 | 9 | 8 |
| beta-Alanine metabolism | ko00410 | 16 | 6 | 10 |
| Phenylalanine metabolism | ko00360 | 15 | 5 | 10 |
| Nitrogen metabolism | ko00910 | 14 | 7 | 7 |
| Sulfur metabolism | ko00920 | 13 | 8 | 5 |
| Cutin, suberine and wax biosynthesis | ko00073 | 12 | 8 | 4 |
| Diterpenoid biosynthesis | ko00904 | 11 | 7 | 4 |
| Photosynthesis | ko00195 | 10 | 9 | 1 |
| Base excision repair | ko03410 | 9 | 6 | 3 |
| Brassinosteroid biosynthesis | ko00905 | 8 | 3 | 5 |
| Citrate cycle (TCA cycle) | ko00020 | 7 | 3 | 4 |
| RNA polymerase | ko03020 | 6 | 1 | 5 |
| Indole alkaloid biosynthesis | ko00901 | 5 | 2 | 3 |
| Phagosome | ko04145 | 4 | 3 | 1 |
| Flavone and flavonol biosynthesis | ko00944 | 3 | 2 | 1 |
| Basal transcription factors | ko03022 | 2 | 0 | 2 |
| Histidine metabolism | ko00340 | 1 | 1 | 0 |
| Total | 1119 (100%) | 524 (46.83%) | 595 (53.17%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Tahir, M.M.; Wu, T.; Xie, L.; Zhang, X.; Mao, J.; Ayyoub, A.; Xing, L.; Zhang, D.; Shao, Y. Transcriptome Analysis Reveals Multiple Genes and Complex Hormonal-Mediated Interactions with PEG during Adventitious Root Formation in Apple. Int. J. Mol. Sci. 2022, 23, 976. https://doi.org/10.3390/ijms23020976
Li S, Tahir MM, Wu T, Xie L, Zhang X, Mao J, Ayyoub A, Xing L, Zhang D, Shao Y. Transcriptome Analysis Reveals Multiple Genes and Complex Hormonal-Mediated Interactions with PEG during Adventitious Root Formation in Apple. International Journal of Molecular Sciences. 2022; 23(2):976. https://doi.org/10.3390/ijms23020976
Chicago/Turabian StyleLi, Shaohuan, Muhammad Mobeen Tahir, Tong Wu, Lingling Xie, Xiaoyun Zhang, Jiangping Mao, Anam Ayyoub, Libo Xing, Dong Zhang, and Yun Shao. 2022. "Transcriptome Analysis Reveals Multiple Genes and Complex Hormonal-Mediated Interactions with PEG during Adventitious Root Formation in Apple" International Journal of Molecular Sciences 23, no. 2: 976. https://doi.org/10.3390/ijms23020976
APA StyleLi, S., Tahir, M. M., Wu, T., Xie, L., Zhang, X., Mao, J., Ayyoub, A., Xing, L., Zhang, D., & Shao, Y. (2022). Transcriptome Analysis Reveals Multiple Genes and Complex Hormonal-Mediated Interactions with PEG during Adventitious Root Formation in Apple. International Journal of Molecular Sciences, 23(2), 976. https://doi.org/10.3390/ijms23020976





