Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Development and Tissue-Dependent Flower Color Formation in Cymbidium lowianum
Abstract
:1. Introduction
2. Results
2.1. Selection of Candidate Reference Genes Based on Transcriptome Data
2.2. Primer Specificity and Amplification Efficiency Test of Candidate Reference Genes
2.3. Expression Profiles of Candidate Reference Genes
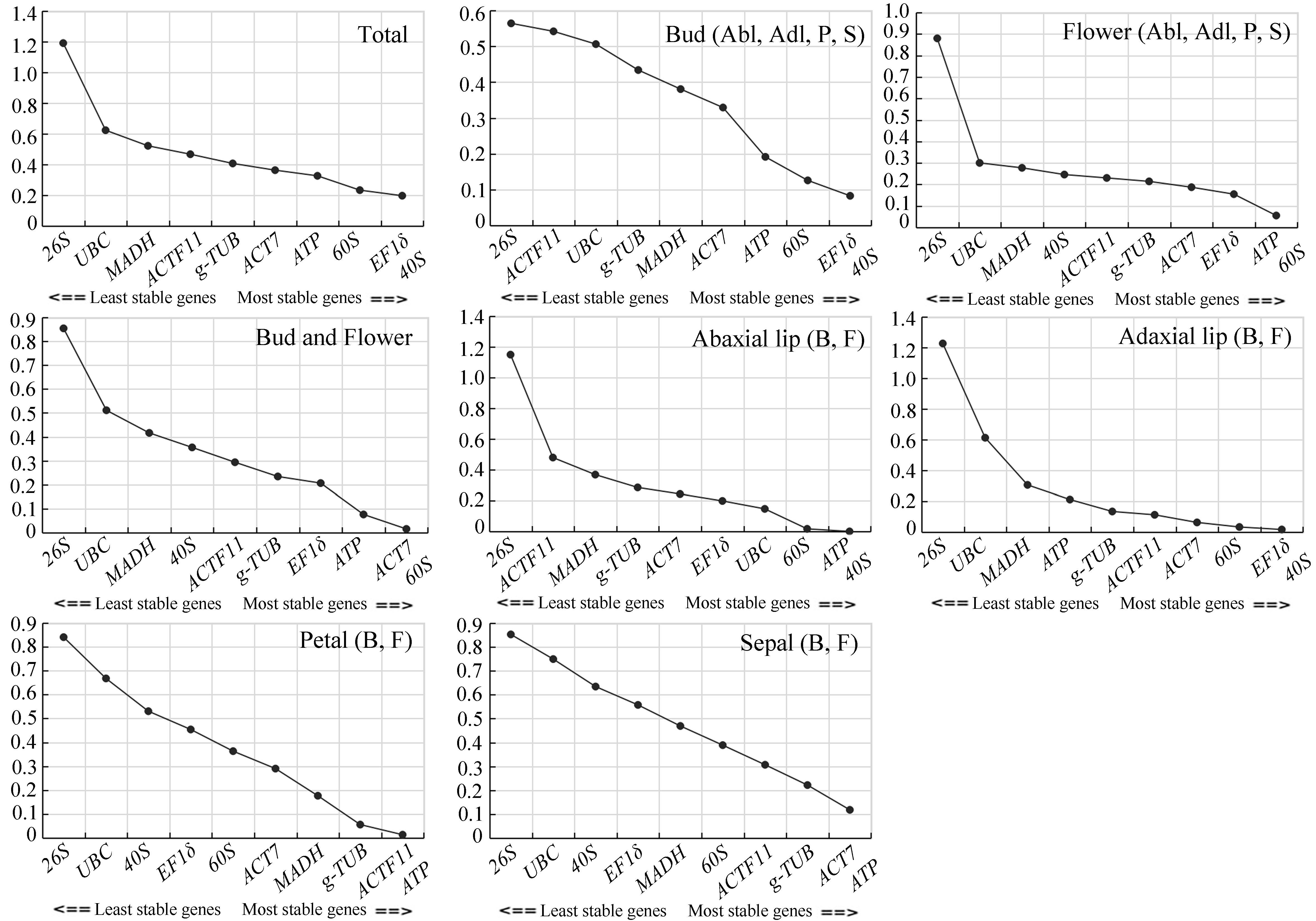
2.4. Expression Stability of Candidate Reference Genes
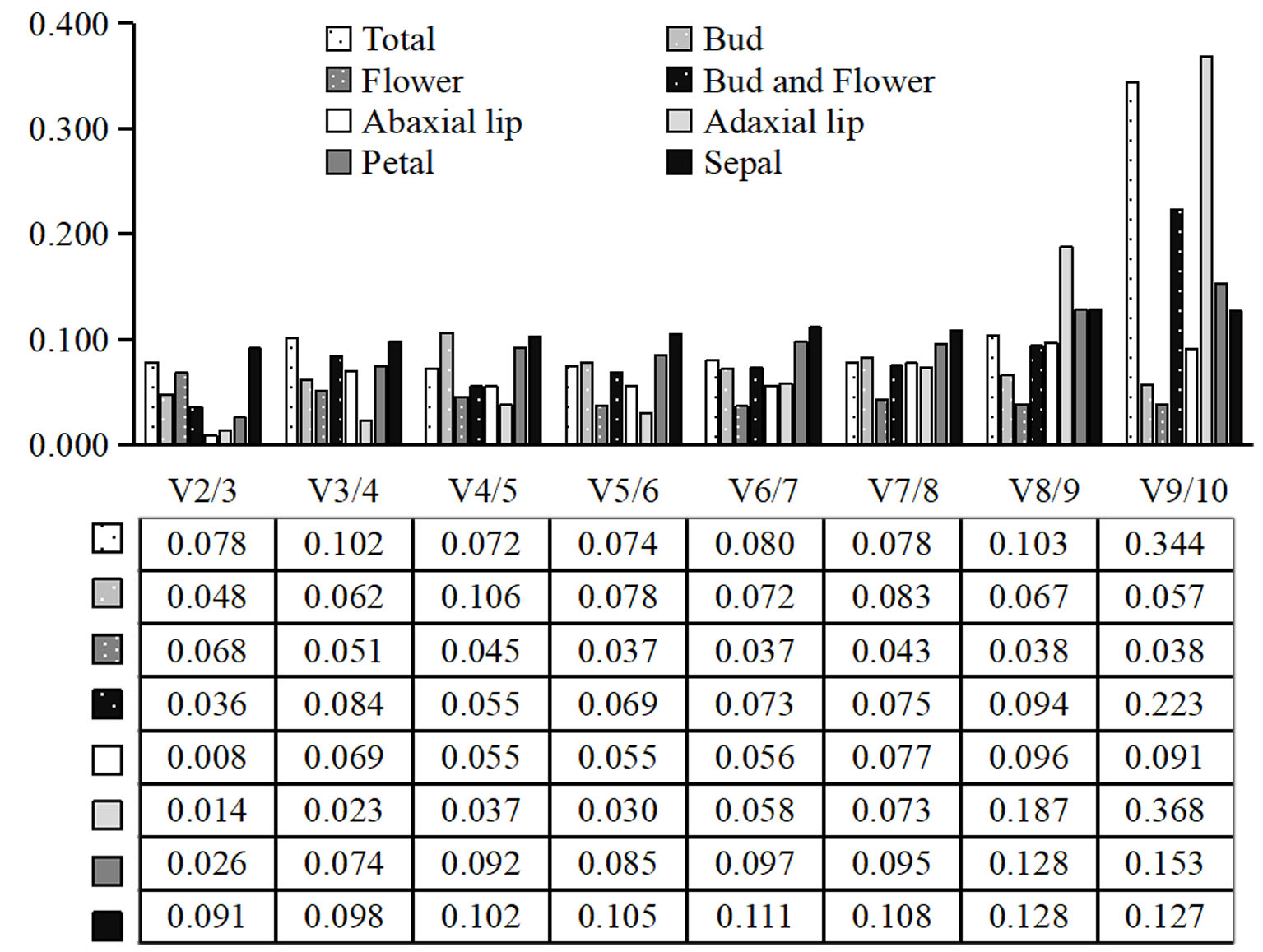
2.4.1. geNorm Analysis
2.4.2. NormFinder Analysis
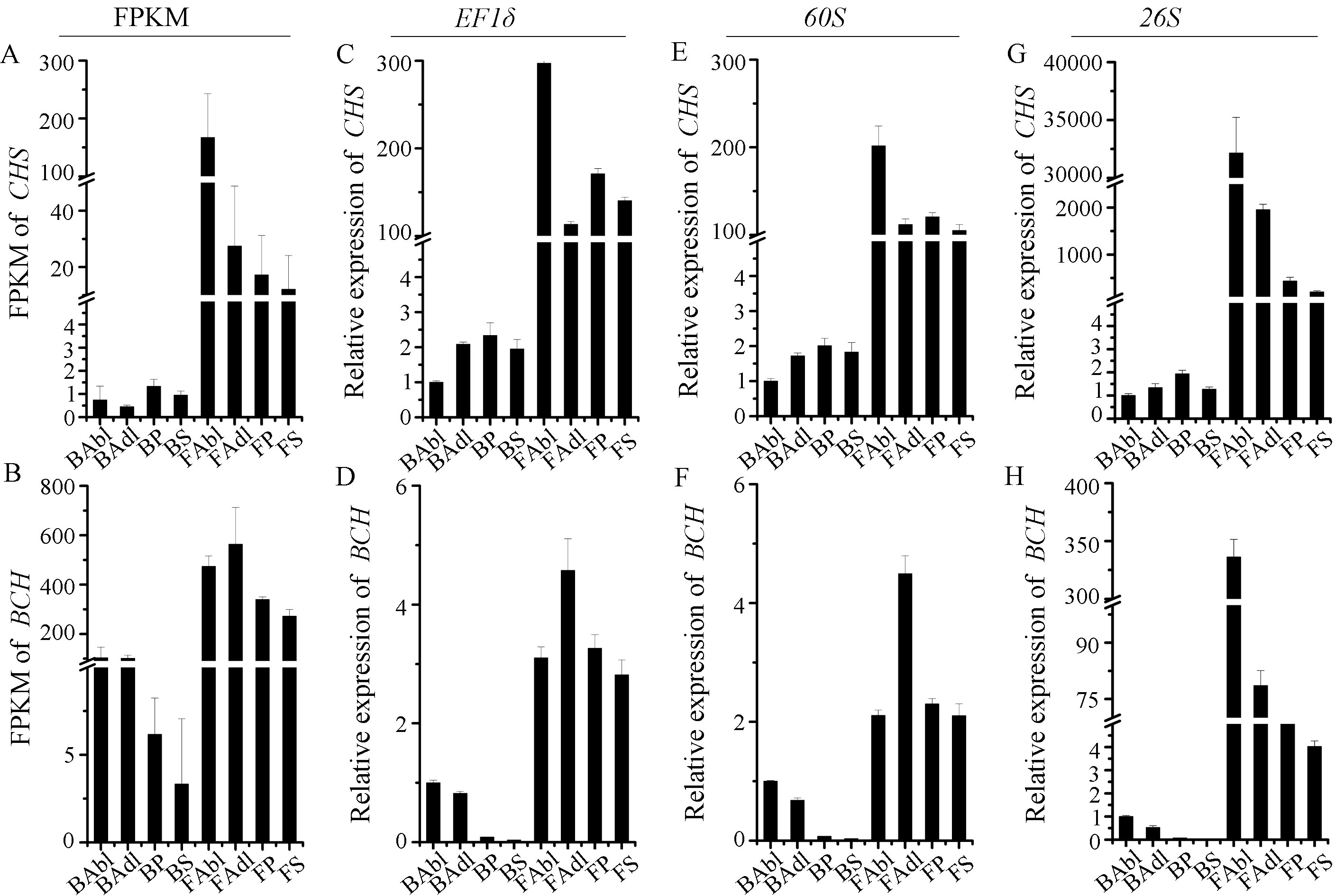
2.5. The Expression Profiles of CHS and BCH Validated by the Most Stable/unstable Reference Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Preparation, Reverse Transcription and qRT-PCR
4.3. Screening of Candidate Reference Genes
4.4. Primer Design and qRT-PCR Analysis of Candidate Reference Genes
4.5. Selection of Reference Genes with Stable Expression
4.6. Validation of the Candidate Reference Genes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| Cq | Cycle threshold |
| ACT7 | Actin-7-like |
| ACTF11 | Actin-depolymerizing factor 11-like |
| ATP | ATP synthase subunit O |
| EF1 δ | Elongation factor 1-delta |
| MADH | Malate dehydrogenase |
| g-TUB | gamma-tubulin complex component 2 |
| UBC | Ubiquitin-conjugating enzyme |
| 26S | 26S proteasome non-ATPase regulatory subunit 2 |
| 40S | 40S ribosomal protein |
| 60S | 60S ribosomal protein |
| ABP | Anthocyanins biosynthetic pathways |
| CBP | Carotenoids biosynthetic pathways |
| CHS | Chalcone synthase |
| BCH | β-carotene hydroxylase |
| FPKM | Fragments per Kilobase of transcript per millions base pairs sequenced |
References
- Gigord, L.D.B.; Macnair, M.R.; Smithson, A. Negative frequency-dependent selection maintains a dramatic flower color polymorphism in the rewardless orchid Dactylorhiza sambucina (L.) Soo. Proc. Natl. Acad. Sci. USA 2001, 98, 6253–6255. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, R.; Rausher, M.D. Pollinator-Mediated Selection on Flower Color Allele Drives Reinforcement. Science 2012, 335, 1090–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazzonelli, C.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Aqeel, M.; Deng, J.; Khalid, N.; Sanaullah, T.; Shuilin, H. Biotechnological Advancements for Improving Floral Attributes in Ornamental Plants. Front. Plant Sci. 2017, 8, 530. [Google Scholar] [CrossRef] [Green Version]
- Li, X.J.; Li, Z.L.; Huang, L.P. Induction and identification of wild polyploid C. lowianum. Chin. Agric. Sci. Bull. 2010, 26, 261–266. [Google Scholar]
- Cheng, L.; Wang, Y.; Zhao, Y.; Li, Y.; He, F.; Li, Z. Characterization of the complete chloroplast genome of the endangered Chinese species Cymbidium lowianum (Rchb.f.) Rchb.f. Mitochondrial DNA Part B 2020, 5, 3318–3319. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Ahmed, R.; Haque, M.S.; Alam, M.M.; Islam, M.S. Identification and validation of reference genes for real-time quantitative RT-PCR analysis in jute. BMC Mol. Biol. 2019, 20, 13. [Google Scholar] [CrossRef]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef] [Green Version]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PC.R. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Chi, C.; Shen, Y.; Yin, L.; Ke, X.; Han, D.; Zuo, Y. Selection and validation of reference genes for gene expression analysis in vigna angularis using quantitative real-time RT-PC.R. PLoS ONE 2016, 11, e0168479. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Dai, S. Selection of Reference Genes for Real-time Polymerase Chain Reaction Analysis of Light-dependent Anthocyanin Biosynthesis in Chrysanthemum. J. Am. Soc. Hortic. Sci. 2015, 140, 68–77. [Google Scholar] [CrossRef]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference Gene Selection for RT-qPCR Analysis of Flower Development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 7, 287. [Google Scholar] [CrossRef] [Green Version]
- Hou, F.; Li, S.; Wang, J.; Kang, X.; Weng, Y.; Xing, G. Identification and validation of reference genes for quantitative real-time PCR studies in long yellow daylily, Hemerocallis citrina Borani. PLoS ONE 2017, 12, e0174933. [Google Scholar]
- Lu, Y.P.; Liao, J.H.; Guo, Z.J.; Zheng, H.Q.; Lu, L.F.; Cai, Z.X.; Zeng, Z.H.; Ying, Z.H.; Chen, M.Y. Selection and Evaluation of Potential Reference Genes for Quantitative Real-Time PCR in Agaricus blazei Based on Transcriptome Sequencing Data. BioMed Res. Int. 2021, 2021. [Google Scholar] [CrossRef]
- Wang, M.; Ren, T.; Marowa, P.; Du, H.; Xu, Z. Identification and selection of reference genes for gene expression analysis by quantitative real-time PCR in Suaeda glauca’s response to salinity. Sci. Rep. 2021, 11, 8569. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Zou, X.; Carballar-Lejarazú, R.; Wu, L.; Sun, W.; Yuan, X.; Wu, S.; Li, P.; Ding, H.; Ni, L.; et al. Selection and evaluation of reference genes for qRT-PCR analysis in Euscaphis konishii Hayata based on transcriptome data. Plant Methods 2018, 14, 42. [Google Scholar] [CrossRef]
- He, Y.; Yan, H.; Hua, W.; Huang, Y.; Wang, Z. Selection and validation of reference genes for quantitative real-time PCR in Gentiana macrophylla. Front. Plant Sci. 2016, 7, 945. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, Y.; Zhou, P. Validation of reference genes for quantitative real-time PCR during Chinese wolfberry fruit development. Plant Physiol. Biochem. 2013, 70, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Gantasala, N.P.; Papolu, P.K.; Thakur, P.K.; Kamaraju, D.; Sreevathsa, R.; Rao, U. Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Res. Notes 2013, 6, 312. [Google Scholar] [CrossRef] [Green Version]
- Galli, V.; da Silva Messias, R.; dos Anjos e Silva, S.D.; Rombaldi, C.V. Selection of reliable reference genes for quantitative real-time polymerase chain reaction studies in maize grains. Plant Cell. Rep. 2013, 32, 1869–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.S.; Yang, H.L.; Zhang, D.Y.; Zhang, Y.M.; Wood, A.J. Reference gene selection in the desert plant Eremosparton songoricum. Int. J. Mol. Sci. 2012, 13, 6944–6963. [Google Scholar] [CrossRef] [PubMed]
- Alves Oliveira, D.; Tang, J.D.; Warburton, M.L. Reference Gene Selection for RT-qPCR Analysis in Maize Kernels Inoculated with Aspergillus flavus. Toxins 2021, 13, 386. [Google Scholar] [CrossRef] [PubMed]
- Qu, R.; Miao, Y.; Cui, Y.; Cao, Y.; Zhou, Y.; Tang, X.; Yang, J.; Wang, F. Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune. BMC Mol. Biol. 2019, 20, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Qi, X.; Yan, H.; Huang, L.; Nie, G.; Zhang, X. Reference gene selection for quantitative real-time Reverse-transcriptase PCR in annual ryegrass (Lolium multiflorum) subjected to various abiotic stresses. Molecules 2018, 23, 172. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Y.; Fang, H.; Shi, H.; Chen, K.; Zhang, Z.; Tan, X. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genom. 2014, 289, 1023–1035. [Google Scholar] [CrossRef]
- Borowski, J.M.; Galli, V.; Messias Rda, S.; Perin, E.C.; Buss, J.H.; dos Anjos e Silva, S.D.; Rombaldi, C.V. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 2014, 239, 1187–1200. [Google Scholar] [CrossRef]
- Liu, X.; Guan, H.; Song, M.; Fu, Y.; Han, X.; Lei, M.; Ren, J.; Guo, B.; He, W.; Wei, Y. Reference gene selection for qRT-PCR assays in Stellera chamaejasme subjected to abiotic stresses and hormone treatments based on transcriptome datasets. PeerJ 2018, 6, e4535. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Roche, J.R.; Donaghy, D.J.; Thrush, A.; Sathish, P. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol. Biol. 2010, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coker, J.S.; Davies, E. Selection of candidate housekeeping controls in tomato plants using EST data. Biotechniques 2003, 35, 740–746. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.J.; Lee, H.S.; Kwak, S.S. Down-regulation of β-carotene hydroxylase increases β-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweetpotato. Phytochemistry 2012, 74, 69–78. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Z.; Ding, Z.; Meng, H.; Shen, R.; Tang, H.; Liu, Y.G.; Chen, L. Public-transcriptome-database-assisted selection and validation of reliable reference genes for qRT-PCR in rice. Sci. China Life Sci. 2020, 63, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Tao, J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Chen, G.-Z.; Huang, J.; Liu, D.-K.; Xue, F.; Chen, X.-L.; Chen, S.-Q.; Liu, C.-G.; Liu, H.; Ma, H.; et al. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 2021, 1, 10. [Google Scholar] [CrossRef]
- Yang, F.; Gao, J.; Wei, Y.; Ren, R.; Zhang, G.; Lu, C.; Jin, J.; Ai, Y.; Wang, Y.; Chen, L.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Gao, M.; Liu, Y.; Ma, X.; Shuai, Q.; Gai, J.; Li, Y. Evaluation of Reference Genes for Normalization of Gene Expression Using Quantitative RT-PCR under Aluminum, Cadmium, and Heat Stresses in Soybean. PLoS ONE 2017, 12, e0168965. [Google Scholar] [CrossRef]
- Le, D.T.; Aldrich, D.L.; Valliyodan, B.; Watanabe, Y.; Ha, C.V.; Nishiyama, R.; Guttikonda, S.K.; Quach, T.N.; Gutierrez-Gonzalez, J.J.; Tran, L.S.; et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE 2012, 7, e46487. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Q.; Sun, M.; Zhu, L.; Yang, M.; Zhao, Y. Selection of reference genes for quantitative real-time PCR normalization in Panax ginseng at different stages of growth and in different organs. PLoS ONE 2014, 9, e112177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, X.; Qi, J.; Zhang, G.; Xu, J.; Tao, A.; Fang, P.; Su, J. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 2015, 6, 848. [Google Scholar] [CrossRef] [Green Version]
- Chang, E.; Shi, S.; Liu, J.; Cheng, T.; Xue, L.; Yang, X.; Yang, W.; Lan, Q.; Jiang, Z. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using real-time PC.R. PLoS ONE 2012, 7, e33278. [Google Scholar]
- Li, L.; Li, N.; Fang, H.; Qi, X.; Zhou, Y. Selection and Validation of Reference Genes for Normalisation of Gene Expression in Glehnia littoralis. Sci. Rep. 2020, 10, 7374. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wang, X.; Qi, Y.; Ma, Y. Selection and validation of reference genes for qRT-PCR analysis during fruit ripening of red pitaya (Hylocereus polyrhizus). FEBS Open Bio. 2021, 11, 3142–3152. [Google Scholar] [CrossRef]
- Huis, R.; Hawkins, S.; Neutelings, G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Liu, X.; Chen, S.; Li, B.; Wang, X.; Fan, C.; Wang, G.; Ni, H. Selection of relatively exact reference genes for gene expression studies in flixweed (Descurainia sophia) by quantitative real-time polymerase chain reaction. Pestic Biochem Physiol. 2016, 127, 59–66. [Google Scholar] [CrossRef]
- Ray, D.L.; Johnson, J.C. Validation of reference genes for gene expression analysis in olive (Olea europaea) mesocarp tissue by quantitative real-time RT-PC.R. BMC Res. Notes 2014, 7, 304. [Google Scholar] [CrossRef] [Green Version]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzber, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA–Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-time PCR, Methods and Application; Meuer, S., Wittwer, C., Nakagawara, K., Eds.; Springer Press: Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PC.R. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

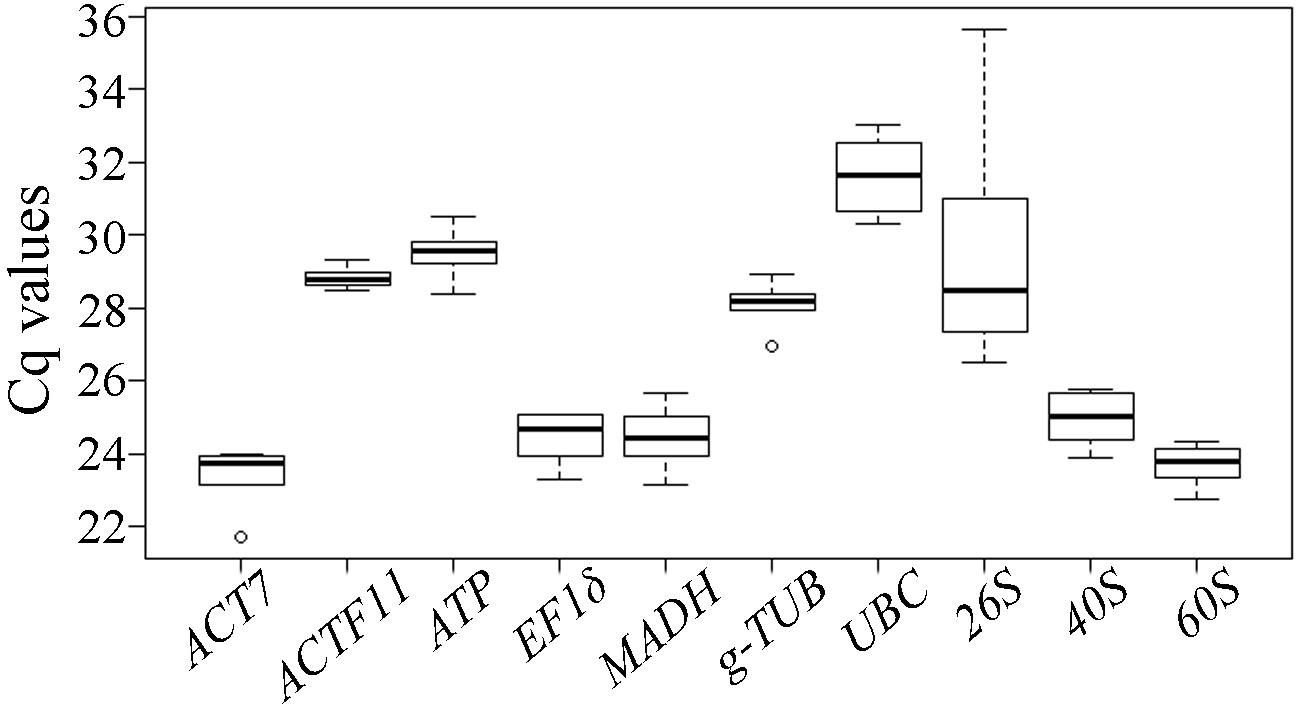
| Gene Name | Description | Primer Sequence (5′-3′) | Amplicon Length (bp) | RT-qPCR Efficiency (%) | R2 |
|---|---|---|---|---|---|
| ACT7 | actin-7-like | F:AACTGGTATTGTGCTGGATTC R:TCATCAGTGAATCTGTAAGGTC | 128 | 88.58 | 0.9970 |
| ACTF11 | actin-depolymerizing factor 11-like | F:ATGTTCATCAACAGTTGCAG R:GGCAGTGATCATCAACTC | 130 | 158.30 | 0.8844 |
| ATP | ATP synthase subunit O | F:TTCACTGATCAATTACGGC R:GATGCGTAGTTTCCAGTAC | 141 | 102.44 | 0.9496 |
| EF1 δ | elongation factor 1-delta | F:CTACCAAGCTTCAAAGGATG R:CTCAGATACAGTAGTAGACC | 143 | 98.32 | 0.9984 |
| MADH | malate dehydrogenase | F:CTACGATATCGCTGGTACTC R:ACGAGTTCTGATCCCTCC | 123 | 105.62 | 0.9948 |
| g-TUB | gamma-tubulin complex component 2 | F:ATCCATTGTGATTGAGAAGGC R:ACTGTAGTATCACCTGCCATG | 102 | 93.05 | 0.9996 |
| UBC | Ubiquitin-conjugating enzyme | F:ATCTCTCAGGCAAGCATTAC R:GTAGAGGTATGGCACTAATC | 125 | 130.87 | 0.9856 |
| 26S | 26S proteasome non-ATPase regulatory subunit 2 | F:CAGAAGCTCGCACTAGAG R:TATGGGCAGATCATCATACTG | 162 | 102.60 | 0.9953 |
| 40S | 40S ribosomal protein | F:GAAGATGGTATTCCTGCAG R:TAGCCTTGGCTGCTTCATG | 139 | 93.18 | 0.9999 |
| 60S | 60S ribosomal protein | F:GTCCAAGTCGAATCAGTATG R:ATAGTGCGTGCCATTCTTC | 139 | 96.68 | 0.9960 |
| Sample | Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Gene | EF1δ | 60S | 40S | ACT7 | ATP | g-TUB | ACTF11 | UBC | MADH | 26S |
| stability | 0.068 | 0.074 | 0.124 | 0.188 | 0.255 | 0.332 | 0.427 | 0.494 | 0.645 | 2.383 | |
| Bud | Gene | 60S | EF1δ | ATP | 40S | g-TUB | ACT7 | ACTF11 | UBC | 26S | MADH |
| stability | 0.143 | 0.166 | 0.193 | 0.227 | 0.287 | 0.333 | 0.360 | 0.368 | 0.385 | 0.388 | |
| Flower | Gene | ATP | 60S | EF1δ | ACTF11 | ACT7 | g-TUB | 40S | MADH | UBC | 26S |
| stability | 0.020 | 0.020 | 0.052 | 0.084 | 0.104 | 0.156 | 0.279 | 0.341 | 0.437 | 2.615 | |
| Bud and Flower | Gene | ACT7 | 60S | EF1δ | 40S | ATP | g-TUB | UBC | ACTF11 | MADH | 26S |
| stability | 0.004 | 0.004 | 0.034 | 0.081 | 0.280 | 0.303 | 0.364 | 0.465 | 0.623 | 1.519 | |
| Abaxial lip | Gene | ATP | 40S | 60S | EF1δ | UBC | ACT7 | g-TUB | MADH | ACTF11 | 26S |
| stability | 0.004 | 0.004 | 0.011 | 0.026 | 0.026 | 0.043 | 0.356 | 0.658 | 0.962 | 4.283 | |
| Adaxial lip | Gene | ACTF11 | g-TUB | 60S | EF1δ | 40S | ACT7 | ATP | MADH | UBC | 26S |
| stability | 0.012 | 0.012 | 0.124 | 0.165 | 0.192 | 0.289 | 0.558 | 0.766 | 0.822 | 2.476 | |
| Petal | Gene | ACT7 | 60S | EF1δ | 40S | ATP | ACTF11 | g-TUB | UBC | MADH | 26S |
| stability | 0.049 | 0.049 | 0.062 | 0.245 | 0.383 | 0.393 | 0.454 | 0.645 | 0.654 | 1.053 | |
| Sepal | Gene | ACT7 | 60S | EF1δ | ATP | 40S | g-TUB | ACTF11 | UBC | MADH | 26S |
| stability | 0.041 | 0.087 | 0.163 | 0.191 | 0.343 | 0.389 | 0.561 | 0.676 | 0.756 | 0.844 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.-M.; Zhang, W.; Zhang, S.-B. Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Development and Tissue-Dependent Flower Color Formation in Cymbidium lowianum. Int. J. Mol. Sci. 2022, 23, 738. https://doi.org/10.3390/ijms23020738
Dong X-M, Zhang W, Zhang S-B. Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Development and Tissue-Dependent Flower Color Formation in Cymbidium lowianum. International Journal of Molecular Sciences. 2022; 23(2):738. https://doi.org/10.3390/ijms23020738
Chicago/Turabian StyleDong, Xiu-Mei, Wei Zhang, and Shi-Bao Zhang. 2022. "Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Development and Tissue-Dependent Flower Color Formation in Cymbidium lowianum" International Journal of Molecular Sciences 23, no. 2: 738. https://doi.org/10.3390/ijms23020738
APA StyleDong, X.-M., Zhang, W., & Zhang, S.-B. (2022). Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Development and Tissue-Dependent Flower Color Formation in Cymbidium lowianum. International Journal of Molecular Sciences, 23(2), 738. https://doi.org/10.3390/ijms23020738





