Abstract
The occurrence of stress is unavoidable in the process of livestock production, and prolonged stress will cause the decrease of livestock productivity. The stress response is mainly regulated by the hypothalamic-pituitary-adrenal axis (HPA axis), which produces a large amount of stress hormones, namely glucocorticoids (GCs), and generates a severe impact on the energy metabolism of the animal body. It is reported that m6A modification plays an important role in the regulation of stress response and also participates in the process of muscle growth and development. In this study, we explored the effect of GCs on the protein synthesis procession of porcine skeletal muscle cells (PSCs). We prove that dexamethasone affects the expression of SLC7A7, a main amino acid transporter for protein synthesis by affecting the level of m6A modification in PSCs. In addition, we find that SLC7A7 affects the level of PSC protein synthesis by regulating the conduction of the mTOR signaling pathway, which indicates that the reduction of SLC7A7 expression may alleviate the level of protein synthesis under stress conditions.
1. Introduction
Stress causes significant losses to the pork industry by affecting animal physiology and reducing their production performance. A previous paper has shown that compared with a control group, pigs showed a markedly reduced final body weight, average daily gain, average daily feed intake and the ratios of the feed intake to weight gain under chronic oxidative stress condition [1]. Under a stress condition, the body secretes a cascade of glucocorticoids (GCs) through the HPA axis, which is involved in many metabolic processes, such as immune system, reproduction, behavior, and cognitive function [2]. As a primary target organ, skeletal muscle is vulnerable to the effects of GCs, resulting in reduced synthesis and increased degradation of muscle proteins, leading to skeletal muscle atrophy and impaired meat quality [3,4,5].
The synthesis of muscle proteins needs free amino acids as substrates [6], and amino acid transporters are required in this process [7]. y+L+ amino acid transporter-1 (y+LAT1) is encoded by the SLC7A7 gene, and the substrates of this system are arginine, lysine, leucine, glutamine and methionine [8]. The expression level of SLC7A7 is high in intestine and kidney but low in skeletal muscle, and the mutation of SLC7A7 gene usually induces many complications, such as persistent hypotonia, birth retardation, osteoporosis, coma and mental development disorder [9,10]. It has been demonstrated that SLC7A7−/− mice exhibit intrauterine growth restriction and persistent growth retardation that generates fetal growth retardation through downregulation of IGF1, thus resulting in mouse death [11]. Similarly, SLC7A7−/− mice had significantly lower weights of liver and skeletal muscle compared with control littermates [12].
The regulation of m6A is particularly critical under stress conditions such as extreme temperature, hypoxia nutrient deprivation and toxin exposure due to short- or long-term effects brought about by stress [13]. m6A plays an important role in body when subjected to various stresses, and researches have shown that m6A levels in the liver and abdominal fat of offspring piglets were higher than those in heat stress conditions, and heat stress also increased the expression of genes related to adipogenesis [14]. It is also possible that METTL3 targets HSP90 and HSP70 to increase their mRNA levels in response to heat stress [15]. m6A is mainly involved with three proteases: methyltransferases, demethylase and readers. m6A methyltransferases mainly include METTL3, METTL14 and WTAP. Among them, METTL3 serves as a methyl donor, METTL14 forms a stable heterodimer with METTL3 and WTAP is responsible for recruiting METTL3 and METTL14 to form a stable complex [16,17]. Besides, m6A can be removed by ALKBH5 and FTO demethylases. mRNAs with m6A modification can be recognized by specific recognition proteins, such as YTHDF1, YTHDF2, YTHDC1, etc. Recognition proteins bind to methylated mRNA transcripts and regulate their stability, degradation and translation of mRNA [18].
In this study, we further analyzed the effects of stress on m6A modification and the expression of the amino acid transporter SLC7A7 and studied the molecular mechanisms regulated by SLC7A7 in protein synthesis in porcine skeletal muscle.
2. Results
2.1. Effects of Glucocorticoid (GC) and RU486 on Protein Synthesis in Porcine Skeletal Muscle Cells
We found that the mRNA expression levels of genes such as Atrogin-1, MuRF, FoxO1 and MSTN, which are related to protein breakdown and synthesis, showed upregulation when treated with 0.1μΜ DEX in PSCs for 48 h. Among them, the protein expression levels of Atrogin-1, FoxO1 and MSTN were significantly upregulated according to the RT-PCR and Western blot results (Figure 1A–C). To further explore the effect of GCs on protein synthesis, we used a sunset non-radioactive (SUnSET) method to examine the protein synthesis of cells after treatment of DEX and its antagonist RU486, which shows that the protein synthesis rate decreased after adding DEX but then increased with the addition of RU486 (Figure 1D,F). The synthesis of muscle proteins requires activation of the mTOR signaling pathway. We then extracted proteins after treatment with DEX for 48 h in PSCs and found that the phosphorylation levels of mTOR as well as two key functional downstream proteins, S6K1 and 4EBP1, were significantly downregulated after treated with DEX. These results showed an increase in the expression levels of protein breakdown genes and a decrease in the expression levels of protein synthesis genes under stress condition in PSCs (Figure 1E,G).

Figure 1.
Glucocorticoids (GCs) increase protein breakdown and decrease protein synthesis in porcine skeletal muscle cells. (A) The mRNA expression levels of Atrogin-1, MuRF1, FOXO1 and MSTN treated with DEX; (B) the protein expression levels of Atrogin-1, FOXO1 and MSTN treated with DEX; (C) quantification of Western blotting under the treatment of DEX; (D) effect of glucocorticoid and its antagonist on the protein synthesis rate; (E) the phosphorylation level of mTOR, S6K1 and 4EBP1 in mTOR signaling pathway after adding DEX; (F) quantification of protein synthesis rate under the treatment of DEX and RU486; (G) quantification of Western blotting under the treatment of DEX. *, ** indicate significant difference at p < 0.05 and p < 0.01, respectively.
2.2. Effects of GCs and RU486 on Intracellular m6A Levels in Porcine Skeletal Muscle Cells
To confirm whether GCs and their antagonist RU486 are involved in the regulation of intracellular m6A levels affecting the expression of intracellular genes after treatment with DEX in PSCs, we used dot blot hybridization and high-performance liquid chromatography-mass spectrometry methods and found a reduction of overall m6A modification level in cells after treatment with DEX for 48 h while then restored to original level after adding with RU486 (Figure 2A), and mass spectrometry assay showed the same result (Figure 2B). To gain further insight into the involvement of DEX in the regulation of m6A modification, we treated the cells with DEX for 48 h and then examined the expression levels of m6A modification enzymes, and found that the mRNA and protein expression levels of METTL3 and METTL14 were significantly downregulated while the expression of the recognition protein YTHDF2 was significantly upregulated (Figure 2C–E). The results implied that m6A levels were downregulated in porcine skeletal muscle cells under stress conditions likely through METTL3, METTL14 as well as the YTHDF2 protein.
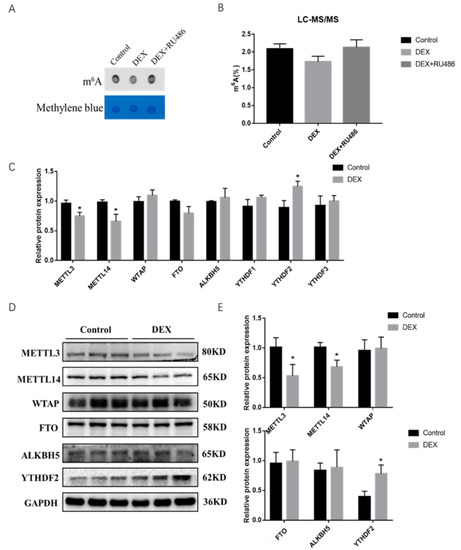
Figure 2.
m6A modification level changes after DEX and RU486 treatment in porcine skeletal muscle cells. (A) Dot blot confirmed the m6A modification after DEX and RU486 treatment; (B) the m6A ratio in RNA was measured using LC-MS/MS; (C) the mRNA expression levels of METL3, METL14, WTAP, FTO, ALKBH5 and YTHDF1-3 after treatment with DEX; (D) protein expression levels of METTL3, METTL14, WTAP, FTO and YTHDF2 after treatment with DEX; (E) quantification of Western blotting under the treatment of DEX. * Indicates a significant difference at p < 0.05.
2.3. Effect of GCs on the Amino Acid Transporter SLC7A7
We extracted RNA after DEX treatment for 48h in porcine skeletal muscle cells and used QPCR to determine the mRNA expression levels of relevant amino acid transporters, which revealed a highly significant downregulation of the mRNA expression levels of the amino acid transporters SLC1A3, SLC7A7, and SLC7A10 (Figure S1). The expression level of SLC7A7 mRNA was found to be greatly decreased using cortisol-fed samples from piglets (Figure 3A). SLC7A7 mRNA expression levels as well as the protein expression levels were found to be significantly downregulated after treated with DEX for 24 h and 48 h in PSCs (Figure 3B–D). To further explore whether SLC7A7 regulated protein synthesis by means of m6A modification, we predicted m6A single base sites of SLC7A7 gene using the online database SRAMP, and the results showed a high confidence of three m6A modification sites in the SLC7A7 gene cDNA sequence (Figure S2). Then we examined the m6A modification levels of SLC7A7 after treating DEX in PSCs and found a significant decrease of SLC7A7 compared with the control group, demonstrating that its expression was subjected to m6A post transcriptional modification mediated by DEX (Figure 3E). The stability of SLC7A7 mRNA was later examined by treating PSCs with the pan-transcriptional inhibitor ActD. After treating cells with DEX for 48 h and then adding ActD for 0, 2, and 4 h respectively, we found that the expression of SLC7A7 was significantly downregulated using QPCR, demonstrating that DEX treatment to the cells for 48 h accelerated the decay of SLC7A7 mRNA (p < 0.05) (Figure 3F). We used dot blot hybridization to detect the m6A methyl modification level after treating PSCs with 3-deazaadenosine (DAA) for 48 h, an inhibitor of m6A methylation (Figure 3G). QPCR and Western blotting were performed to detect the expression levels of SLC7A7 and treating PSCs with DAAs reduced the expression levels of SLC7A7 mRNA and protein, which was consistent with previous results (Figure 3H–J). Overall, above results all suggest that stress is able to reduce the expression of the amino acid transporter SLC7A7 and DEX can modulate the level of m6A modification of SLC7A7 mRNA to affect its expression.
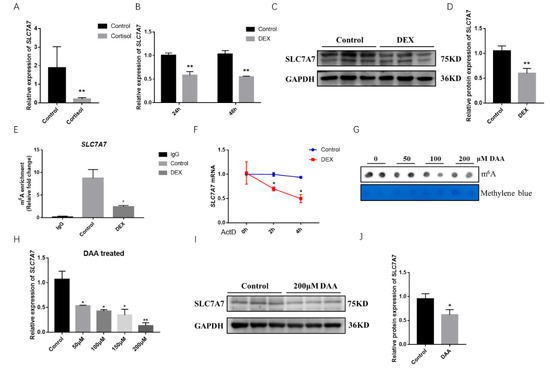
Figure 3.
Effect of GCs on the amino acid transporter SLC7A7. (A) The mRNA expression levels of SLC7A7 in longissimus dorsi muscle of the cortisol group and the control group, respectively; (B) the mRNA expression levels of SLC7A7 after DEX treatment for 24 h and 48 h; (C) the protein expression levels of SLC7A7 after DEX treatment; (D): quantification of Western blotting under the treatment of DEX; (E) validation of m6A modification in SLC7A7 mRNA by MeRIP-QPCR; (F) the mRNA expression of SLC7A7 was detected at 0, 2 and 4 h after ActD treatment; (G) Dot blot detection of m6A modification levels after treatment with 0, 50, 100 and 200 μM DAA; (H) the mRNA expression level of SLC7A7 after treatment with 0, 50, 100 and 200 μM DAA; (I) the protein expression levels of SLC7A7 after treatment with 200 μM DAA; (J) quantification of Western blotting under the treatment of DAA. *, ** indicate significant difference at p < 0.05 and p < 0.01, respectively.
2.4. Effect of SLC7A7 on Protein Synthesis in Porcine Skeletal Muscle Cells
Previous results have shown that DEX affects the expression of protein synthesis-related genes in PSCs, so we further investigated the effect of SLC7A7 on protein synthesis in PSCs by overexpression and interference of SLC7A7. The overexpression and interference of SLC7A7 has been constructed successfully in our research work (Figure 4). After transfection of SLC7A7 overexpression vector and interference fragment into PSCs, we found that the concentration of the protein was significantly increased after overexpression of SLC7A7 compared with the control PCMV-HA group and the protein concentration was dramatically downregulated after interference of SLC7A7 compared with the control NC group (Figure 5A,D). We also used a SUnSET approach and found that the rate of cellular protein synthesis upregulated upon overexpression of SLC7A7 to PSCs but dramatically downregulated after perturbing SLC7A7 in cells (Figure 5B,C,E,F). The above results demonstrate that the amino acid transporter SLC7A7 is able to promote protein synthesis in PSCs.
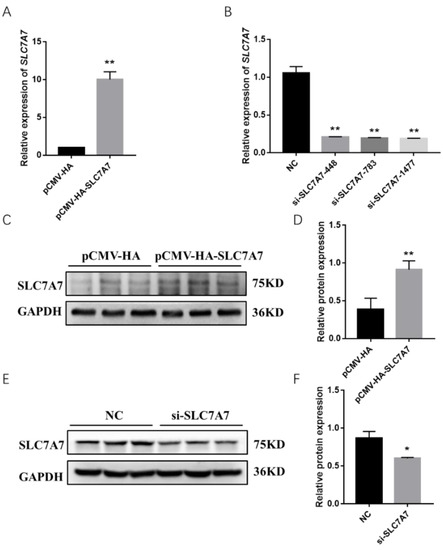
Figure 4.
Expression level changes following overexpression and interference of SLC7A7 in PSCs. (A) The mRNA expression level of SLC7A7 after overexpression of SLC7A7; (B) the mRNA expression level of SLC7A7 after interference of SLC7A7; (C) protein expression level of SLC7A7 after overexpression of SLC7A7; (D) Quantification of Western blotting after overexpression of SLC7A7; (E) the protein expression level of SLC7A7 after interference of SLC7A7; (F) quantification of Western blotting after interference of SLC7A7. *, ** indicate significant difference at p < 0.05 and p < 0.01, respectively.
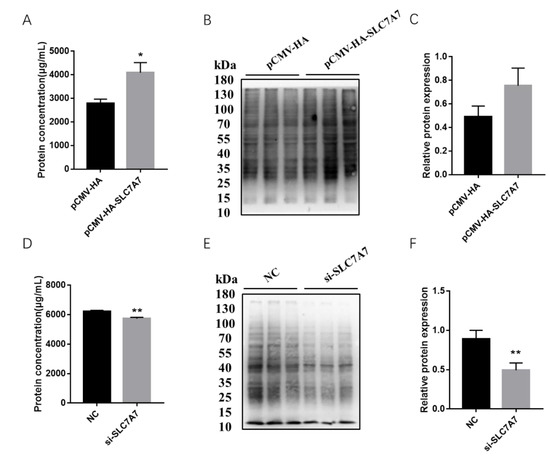
Figure 5.
Overexpression and interference of SLC7A7 affect cell protein synthesis. (A) Total cell protein after overexpression of SLC7A7; (B) the effect of overexpression of SLC7A7 on the rate of protein synthesis; (C) protein quantification of puromycin; (D) total cell protein after interfering of SLC7A7; (E) the effect of interfering of SLC7A7 on the rate of protein synthesis; (F) protein quantification of puromycin. *, ** indicate significant difference at p < 0.05 and p < 0.01, respectively.
To further explore the role of SLC7A7 in PSCs, we examined the expression levels of genes affecting protein synthesis using QPCR after transfection of a SLC7A7 overexpression vector into PSCs, and the results showed that MSTN, Atrogin-1, and FoxO1, genes which reduce the protein synthesis, were decreased after overexpression of SLC7A7. However, the expression of IGF1, a gene promoting protein synthesis, was significantly upregulated (Figure 6A). Similarly, we transfected interfering fragments of SLC7A7 into PSCs and found an elevated expression of MSTN, a gene that promotes proteolysis, but a decrease of IGF1, a gene that promotes protein synthesis (Figure 6B). mTOR signaling pathway is a key regulator in the process of protein synthesis, and it promotes total protein synthesis by phosphorylating two key downstream proteins S6K1 and 4EBP1. After overexpression of SLC7A7, it was found that the phosphorylation levels of mTOR and S6K1 were significantly upregulated SLC7A7(Figure 6C,D) while significantly downregulated after transfected with SLC7A7 interference fragment, and the phosphorylation level of 4EBP1 was also significantly downregulated (Figure 6E,F). These results suggest that amino acid transporter SLC7A7 can increase cellular protein synthesis through the mTOR signaling pathway.
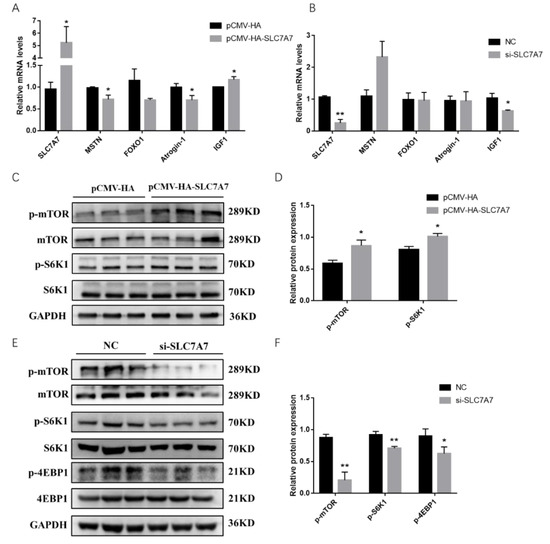
Figure 6.
Overexpression and interference of SLC7A7 in PSCs affect expression of genes involved in protein synthesis and mTOR signaling pathway. (A) The mRNA expression level of SLC7A7, MSTN, FOXO1, Atrogin-1 and IFG1 after overexpression of SLC7A7; (B) the mRNA expression level of SLC7A7, MSTN, FOXO1, Atrogin-1 and IFG1 after interference with SLC7A7; (C) the phosphorylation level of mTOR and S6K1 after overexpression of SLC7A7; (D) quantification of Western blotting after overexpression of SLC7A7; (E) the phosphorylation level of mTOR, S6K1 and 4EBP1 after interference with SLC7A7; (F) quantification of Western blotting after interference with SLC7A7. *, ** indicate significant difference at p < 0.05 and p < 0.01, respectively.
3. Discussion
Stress is an inevitable physiological response in the animal body in the face of detrimental environmental changes. Once the stress stimulus is detected during the development of the animals, a large cascade of GCs will be released through the HPA axis, which will promote the catabolism of protein and fat and affect the growth and development of skeletal muscle, making the production performance of livestock decline sharply [19,20,21]. Our research treated 0.1 µmol/L DEX to PSCs in 24 and 48 h to imitate the effect of GCs emerged in the animal body when they are confronting stress conditions, thus to find a universal indication about how GCs affects protein degradation and synthesis. Skeletal muscle serves as the largest protein repository and its quality is dependent on protein and cell turnover [22]. In the event of stress, GCs decrease the level of muscle protein synthesis and increase the level of protein degradation, ultimately leading to muscle atrophy [23]. GCs affect muscle protein synthesis through a variety of genes and pathways, among which GCs mainly through the UPS system, resulting in proteolysis [24]. DEX treatment of C2C12 cells, L6 myotubes or direct application of DEX to mice have all been reported to increase the expression of genes associated with muscle atrophy like Atrogin-1 and MuRF-1, and activate the ubiquitin proteasome pathway, which promotes the breakdown of skeletal muscle proteins [25,26]. Previous research has found that puromycin binding efficiency was significantly reduced after treatment of C2C12 cells with DEX by using a non-set non-radioactive technique that detects protein synthesis rates in skeletal muscle cells, showing the anti-synthesis effect of DEX on muscle proteins [27]. In our work, it demonstrated similarly that stress evoked highly significant upregulation of atrophy related genes Atrogin-1 and MuRF1 in PSCs as well as significant decreases in the protein synthesis efficiency (Figure 1A–D), which indicated that stress brought about increased degradation and decreased synthesis rates of skeletal muscle proteins. mTOR is critical for maintaining muscle mass and function during skeletal muscle growth and preventing muscle wasting in mice [28,29]. Excessive glucocorticoid treatment is responsible for the reduction of protein synthesis levels in PSCs by inhibiting mTOR to S6K1 signaling pathway [30,31,32]. GC-induced leucine zipper (GILZ), a GC-target protein to mediate several actions of GCs, has been found to promote apoptosis by inhibition of the Akt/mTOR signaling pathway in myeloma cells [33]. Since the mTOR signaling pathway is mainly studied in our research and essential in protein synthesis process, GILZ may also bind to mTORC2 to inhibit phosphorylation of AKT (at Ser473) and activate FoxO3a-mediated transcription of the pro-apoptotic protein BIMIN to inhabit protein synthesis procedure under stress condition. In L6 myoblasts, GCs inhibited IGF1 secretion and the phosphorylation levels of 4EBP1 and P70S6K1, which play key roles in protein synthesis, thereby reducing skeletal muscle protein synthesis [34]. In our study, the phosphorylation levels of mTOR, P70S6K, and 4EBP1 were all significantly decreased after DEX treatment in PSCs, indicating that GCs reduce the protein synthesis levels in skeletal muscle by downregulating mTOR signaling pathway. (Figure 1E,F).
Some epigenetic mechanisms, such as DNA methylation and chromatin modifications, are frequently involved in the regulation of gene expression in response to stimuli in an organism or under pathological conditions [35]. Among them, m6A modification is a part of the RNA epigenetics field, with evidence from studies that changes were found in m6A modification levels in vivo upon organismal stress [13]. As the body develops heat stress, the m6A modification of RNA is significantly higher in sheep than a control group, and ALKBH5 promotes RNA demethylation in mammary cells when subjected to hypoxic stress [36,37]. Treatment of PSCs with DEX in our assay resulted in a decrease in overall intracellular m6A levels (Figure 2A,B). As a dynamic and reversible internal modification, m6A mainly plays a dynamic regulatory role through three enzymes: methylase, demethylase and reader protein. Previous research work found that METTL3 and METTL14 affect the process of cell differentiation by regulating the expression level of MyoD during mouse myoblast differentiation process [38,39]. A role of GR-mediated transcriptional regulation of m6A metabolic genes on m6A-depenent post-transcriptional activation of lipogenic genes in the chicken has been described in the literature [40], indicating that m6A/methylation genes were involved in GR pathways. The above studies both pointed out that m6A/methylation genes were affected by GR and the expression of subsequent GR-related genes in stress events [41]. As for our results, the mRNA and protein expression levels of METT3 and METTL14 were significantly downregulated in PSCs after DEX treatment (Figure 2C–E). It is tentatively concluded here that stress may affect the process of cell growth and development by affecting the expression levels of METTL3 and METTL14 in PSCs. However, the specific mechanism of how DEX affects the level of m6A modification in the cells needs further study.
GCs suppress protein synthesis to induced muscle atrophy through a variety of mechanisms, among which the catabolism of GCs inhibits the transport of amino acids into muscle cells, thereby reducing protein synthesis [34,42]. Cells will have an increasing demand for amino acids to satisfy the body’s needs in special events like cancer or stress which are detrimental to cell growth or development [43,44]. It has been reported that heat stress and exercise-induced stress responses can alter amino acid digestibility and the expression of amino acid transporters, export amino acids from muscle cells by increasing the expression of amino acid transporters [44,45,46]. In our results, the expression of SLC7A7, an amino acid transporter, was significantly downregulated in samples from cortisol-fed piglets and in PSCs treated with DEX (Figure 3A–D). The results above suggest that stress can affect the expression of the corresponding amino acid transporters in skeletal muscle, leading to reduced protein synthesis and loss of muscle mass. DEX causes significant changes in m6A modification levels in PSCs (Figure 2), so are there any such modifications at the SLC7A7 gene as well? The m6A post transcriptional modification mainly occurs in response to stress by regulating gene expression through a series of RNA metabolic ways that affect genes mRNA translation and degradation [47]. It has been shown that amino acid transporters can be regulated by m6A modification, as the main glutamine transporter SLC1A5 in cancer cells can be regulated by the m6A modified demethylase FTO and SLC7A11 is promoted to decay by preferentially bound to YTHDC2 [48]. We detected a highly significant downregulation of SLC7A7 mRNA m6A modification compared with the control (Figure 3E,F), and the expression level of SLC7A7 was significantly downregulated after treatment with DAA, an inhibitor of m6A methylation. These results confirm that SLC7A7 is subject to m6A posttranscriptional modification after glucocorticoid treatment (Figure 3G–J). Also, the m6A modified reader protease YTHDF2 was significantly upregulated in cells with DEX treatment (Figure 2). Thus, we hypothesized that the degradation of SLC7A7 might be controlled by YTHDF2 in an m6A-dependent manner.
Amino acid transporters can act the upstream of the mTOR signaling pathway, enabling cells to sense the availability of amino acids and initiate anabolic responses [49]. Studies have shown that leucine is required for activation of the mTOR signaling pathway, which is mainly transported by members of SLC7 family of cationic amino acid transporter solute carriers, among which SLC7A7 can transport leucine intracellularly, and it has also been shown that SLC7A7 can regulate the mTOR signaling pathway and affect the phosphorylation levels of the downstream effector S6K1 [50]. In our study, after overexpression of SLC7A7, the synthesis levels of cellular proteins were found to be upregulated, and the phosphorylation levels of mTOR signaling pathway and its two downstream S6K1 and 4EBP1 were also significantly upregulated; in addition, the opposite results were obtained after the interference with SLC7A7 in cells (Figure 4, Figure 5 and Figure 6), implying that SLC7A7 is required for PSCs protein synthesis by regulating the signaling of mTOR pathway.
In summary, when animals fall into a stress situation, the body’s HPA axis will be activated, and the level of GCs increases, making mTOR signaling significantly downregulate. Furthermore, when stress occurs, the overall m6A modification level of the skeletal muscle cell would significantly downregulate, resulting in a significant decrease of mRNA and protein expression of SLC7A7 and mTOR signaling pathway, and finally decrease total protein synthesis in porcine skeletal muscle (Figure 7).
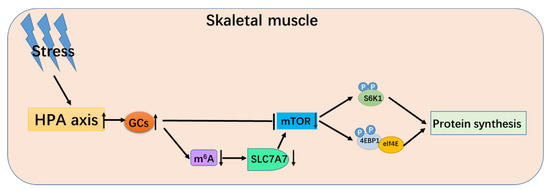
Figure 7.
The schematic diagram of glucocorticoid regulating protein deposition process.
4. Materials and Methods
4.1. Animals and Samples
The Yorkshire piglets of 1–2 days were provided by the Farm of Huazhong Agricultural University (Wuhan, China). After the piglets were slaughtered, the longissimus dorsi and leg muscles were taken to separate for the primary skeletal muscle cells. Finally, the skeletal muscle cells were grown in incubators at 37 °C and 5% CO2, and proliferating cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (DMEM, Gibco, Grand Island, NY, USA). The 28-day-old Yorkshire pigs were fed with cortisol in 120 mg/kg diet for 7 days to mimic stress, then the longissimus dorsi (LD) muscle was sampled and analyzed for gene expression, and more detailed information can be found in our previous work [50].
Animals: all experimental animal procedures in this study were performed according to the guidelines of Good Laboratory Practice, and the animals were supplied with nutritional food and sufficient water. Animal feeding and tests were conducted based on the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University.
Isolation and culture of primary skeletal muscle cells: primary skeletal muscle cells were isolated from the longissimus dorsi skeletal muscles of 2–3 day old piglets, minced and digested in a mixture of type I collagenase and DMEM (DMEM, Gibco, Grand Island, NY, USA).
4.2. Total RNA Preparation and cDNA Synthesis
Total RNA was isolated at 48 h after cell transfection, using total RNA extraction kit (Omega bio-tek, Norcross, GA, USA) according to the manufacturer’s protocol. The RNA integrity was checked using denaturing gel electrophoresis and the RNA concentration was measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Total RNA was reverse transcribed using a RevertAidTM First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA).
4.3. Quantitative Real-Time Polymerase Chain Reaction (PCR)
The specific fluorescent quantitative PCR primers were designed using cDNA as a template; the mRNA level was quantified using β-actin gene as an internal reference; the real-time quantitative PCR experiments were performed on the CFX384 TouchTM fluorescence quantitative PCR instrument using SYBR Green qPCR Master Mix (Bio-Rad). Finally, the 2−ΔΔCt method was used for data analysis, and one-way analysis of variance was performed to determine the significance at p < 0.05 (significant) and p < 0.01 (extremely significant). Primer sets are listed in Table 1.

Table 1.
Primer sequences used for quantitative real-time polymerase chain reaction (qPCR).
4.4. BCA Method for Detecting Protein Concentration
We used the BCA Protein Concentration Test Kit (Thermo Fisher America) to perform detection according to the procedure.
4.5. Plasmid Construction, siRNA Synthesis and Cell Transfection
Briefly, For the SLC7A7 overexpression plasmids, full-length sequences were cloned into the pCMV-HA plasmid. Full-length GRα and SLC2A4 sequences were amplified with full-length-F/R primers (Table 2). LC7A7 siRNA was synthesized by Shanghai GenePharm Company, using the following SLC7A7 siRNA sequences: SLC7A7 siRNA (sense): CCUACAUCCUCGAGGCCUUTT; SLC7A7 siRNA (anti-sense):

Table 2.
SLC7A7 CDS region amplification primer sequence.
AAGGCCUCGAGGAUGUAGGTT. For cell transfection, primary cells were transfected with 4 μg of expression vectors or 3.3 μg/μL of siRNA oligo using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) in each well of a 6-well plate.
4.6. Western Blot Analysis
Briefly, PSC was washed with PBS and lysed in RIPA lysis buffer (Beyotime Bio-technology Company, Shanghai, China). Next, 20 μg of total protein was resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electro-transferred onto a poly-vinylidene fluoride (PVDF) (Millipore, Burlington, MA, USA) membrane. The PVDF membrane was blocked in 5% skim milk powder dissolved in TBST for 90 min at room temperature. Primary antibodies were applied in sealing fluid at 4 °C overnight. Subsequently, the PVDF membrane was washed with TBST and stained with the appropriate HPR-labeled secondary antibodies (goat anti-rabbit or mouse) for 1 h at room temperature. After washing with TBST, the membrane was analyzed using the ECL Reagent (Beyotime Biotechnology). The antibodies used included: GAPDH (GB11002) antibody purchased from Servicebio, Wuhan, China; β-Actin (AC037), MSTN (A6913), FOXO1 (A7637), Atrogin-1 (MAFbx) (A3193) and secondary antibody purchased from Abclonal Technology, Wuhan, China, SLC7A7 (NBP1-59856, Novusbio); m6A(202003,SynapticSystems); p70S6K1 (sc-230, Santa); P-S6K1 (AF3228, Affinity Biosciences), mTOR (2971S, Cell Signaling Technology); P-mTOR(S2448, Cell Signaling Technology); 4EBP1 (RT1002,HUABIO); p-4EBP1 (RT1004, HUABIO) and mouse puromycin antibody 12D10 (320591, Millipore).
4.7. Surface Sensing of Translation (SUnSET) Non-Radioactive Method for Detecting Protein Synthesis Rate
In this study, the surface sensing of translation (SUnSET) non-radioactive method was used to measure the protein synthesis rate of porcine skeletal muscle cells. After transfection with the SLC7A7 overexpression vector and SLC7A7 interference fragment, the cells were treated separately with DEX and RU486 for 48 h, followed by the addition of 1 μg/mL puromycin (Beyotime Biotechnology, Shanghai, China) and treatment of 30 min. Finally, the change of puromycin was detected using a Western blot.
4.8. m6A Immunoprecipitation (MeRIP) qPCR
1 μg m6A antibody (Abcam, ab151230) with 20 μL beads A/G (Bio-rad, 161-4013; 161-4023) Incubate overnight at 4 °C. This was followed by 1 × IP buffer containing 40 U RNase inhibitors (50 mM Tris · HCl, 750 mM NaCl, and 0.5% NP40) Wash the above beads, add 100 μg of fragmented mRNA, respectively, and continue the incubation overnight. Then, we discarded the solution and added the eluent containing 8.4 μg proteinase K (5 mM Tris · HCl, 1 mM EDTA, and 0.05% SDS) then incubated it at 50 °C for 1.5 h. Finally, phenol chloroform was used to extract RNA and combine qPCR to detect the expression of related genes.
4.9. Statistical Analysis
The results are presented as means ± standard deviation (SD). Statistical analysis of the groups was performed using Student’s t-test or one-way ANOVA with the LSD post hoc test. Statistical significance was set at * p < 0.05 and ** p < 0.01.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23020661/s1.
Author Contributions
Conceptualization, J.C., W.-J.X. and K.G.; methodology., W.-J.X., K.G., J.-L.S., C.-T.G., J.-L.X., R.Z. and S.-W.J.; validation, W.-J.X. and K.G.; writing—original draft preparation, W.-J.X.; writing—review and editing, W.-J.X. and K.G.; supervision, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China (2021YFD1301200, 2017YFD0502000), National Natural Science Foundation of China (31301946, 31402051), The Fundamental Research Funds for the Central Universities (2662020DKPY012), Research Funds for Hubei Key Laboratory of Animal Embryo Engineering and Molecular Breeding (KLAEMB-2019-01) and Key R & D pro-jects of Hubei Province (2021BBA082, 2020BBB069, 2020ABA016).
Institutional Review Board Statement
The study was conducted according to the guidelines of Good Laboratory Practice, and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University. (protocol code HZAUSW-2017-001, approved 26 January 2017).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, Y.; Wang, P.; Yin, J.; Jin, S.; Su, W.; Tian, J.; Li, T.; Yao, K. Effects of ornithine alpha-ketoglutarate on growth performance and gut microbiota in a chronic oxidative stress pig model induced by d-galactose. Food Funct. 2020, 11, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Gjerstad, J.K.; Lightman, S.L.; Spiga, F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress 2018, 21, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reser, J.E. Chronic stress, cortical plasticity and neuroecology. Behav. Processes 2016, 129, 105–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyola, M.G.; Handa, R.J. Hypothalamic-pituitary-adrenal and hypothalamic-pituitary-gonadal axes: Sex differences in regulation of stress responsivity. Stress 2017, 20, 476–494. [Google Scholar] [CrossRef]
- Löfberg, E.; Gutierrez, A.; Wernerman, J.; Anderstam, B.; Mitch, W.E.; Price, S.R.; Bergström, J.; Alvestrand, A. Effects of high doses of GCs on free amino acids, ribosomes and protein turnover in human muscle. Eur. J. Clin. Invest. 2002, 32, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Tipton, K.D.; Gurkin, B.E.; Matin, S.; Wolfe, R.R. Nonessential amino acids are not necessary to stimulate net muscle protein synthesis in healthy volunteers. J. Nutr. Biochem. 1999, 10, 89–95. [Google Scholar] [CrossRef]
- Javed, K.; Fairweather, S.J. Amino acid transporters in the regulation of insulin secretion and signalling. Biochem. Soc. Trans. 2019, 47, 571–590. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.; Yang, X.; Wang, N.; Kang, M.; Wang, Y.; Rong, L.; Fang, Y.; Xue, Y. Function of SLC7A7 in T-Cell Acute Lymphoblastic Leukemia. Cell. Physiol. Biochem. 2018, 48, 731–740. [Google Scholar] [CrossRef]
- Ishida, A.; Ashihara, A.; Nakashima, K.; Katsumata, M. Expression of cationic amino acid transporters in pig skeletal muscles during postnatal development. Amino Acids 2017, 49, 1805–1814. [Google Scholar] [CrossRef]
- de Baulny, H.O.; Schiff, M.; Dionisi-Vici, C. Lysinuric protein intolerance (LPI): A multi organ disease by far more complex than a classic urea cycle disorder. Mol. Genet. Metab. 2012, 106, 12–17. [Google Scholar] [CrossRef]
- Sperandeo, M.P.; Annunziata, P.; Bozzato, A.; Piccolo, P.; Maiuri, L.; D’Armiento, M.; Ballabio, A.; Corso, G.; Andria, G.; Borsani, G.; et al. Slc7a7 disruption causes fetal growth retardation by downregulating Igf1 in the mouse model of lysinuric protein intolerance. Am. J. Physiol.-Cell Physiol. 2007, 293, C191–C198. [Google Scholar] [CrossRef] [Green Version]
- Bodoy, S.; Sotillo, F.; Espino-Guarch, M.; Sperandeo, M.P.; Ormazabal, A.; Zorzano, A.; Sebastio, G.; Artuch, R.; Palacín, M. Inducible Slc7a7 Knockout Mouse Model Recapitulates Lysinuric Protein Intolerance Disease. Int. J. Mol. Sci. 2019, 20, 5294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Roh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403. [Google Scholar] [CrossRef]
- Heng, J.H.; Tian, M.; Zhang, W.F.; Chen, F.; Guan, W.T.; Zhang, S.H. Maternal heat stress regulates the early fat deposition partly through modification of m(6)A RNA methylation in neonatal piglets. Cell Stress Chaperones 2019, 24, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, Y.; Wang, T.; Zhong, X. Modification of N6-methyladenosine RNA methylation on heat shock protein expression. PLoS ONE 2018, 13, e0198604. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Yue, Y.N.; Han, D.L.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.F.; Yu, M.; Lu, Z.K.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N-6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Tang, H.F.; Kai, Y. N6-methyladenine RNA Modification (m(6)A): An Emerging Regulator of Metabolic Diseases. Curr. Drug Targets 2020, 21, 1056–1067. [Google Scholar] [CrossRef]
- Dallman, M.F.; Akana, S.F.; Strack, A.M.; Hanson, E.S.; Sebastian, R.J. The neural network that regulates energy balance is responsive to GCs and insulin and also regulates HPA axis responsivity at a site proximal to CRF neurons. Ann. N. Y. Acad. Sci. 1995, 29, 730–742. [Google Scholar] [CrossRef]
- Chang, W.H.; Li, J.J.; Zhang, S.; Zheng, A.J.; Yuan, J.L.; Cai, H.Y.; Liu, G.H. Effects of glucocorticoid-induced stress on absorption of glycylsarcosine in jejunum of broilers. Poult. Sci. 2015, 94, 700–705. [Google Scholar] [CrossRef]
- Zhao, L.D.; McMillan, R.P.; Xie, G.H.; Giridhar, S.; Raumgard, L.H.; El-Kadi, S.; Selsby, J.; Ross, J.; Gabler, N.; Hulver, M.W.; et al. Heat stress decreases metabolic flexibility in skeletal muscle of growing pigs. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2018, 315, R1096–R1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartorelli, V.; Fulco, M. Molecular and cellular determinants of skeletal muscle atrophy and hypertrophy. Sci. STKE Signal Transduct. Knowl. Environ. 2004, 2004, re11. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, P.; Sandri, M. Cellular and molecular mechanisms of muscle atrophy. Dis. Models Mech. 2013, 6, 25–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schakman, O.; Kalista, S.; Barbe, C.; Loumaye, A.; Thissen, J.P. Glucocorticoid-induced skeletal muscle atrophy. Int. J. Biochem. Cell Biol. 2013, 45, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Reznick, A.Z. The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free. Radic. Biol. Med. 2016, 98, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B.A.; Drujan, D.; Willis, M.S.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef] [Green Version]
- Kuci, O.; Archambault, E.; Dodacki, A.; Nubret, E.; De Bandt, J.P.; Cynober, L. Effect of citrulline on muscle protein turnover in an in vitro model of muscle catabolism. Nutrition 2020, 71, 110597. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Kim, J.; Park, M.Y.; Kim, H.K.; Park, Y.; Whang, K.Y. Cortisone and dexamethasone inhibit myogenesis by modulating the AKT/mTOR signaling pathway in C2C12. Biosci. Biotechnol. Biochem. 2016, 80, 2093–2099. [Google Scholar] [CrossRef] [Green Version]
- Shah, O.J.; Kimball, S.R.; Jefferson, L.S. Acute attenuation of translation initiation and protein synthesis by GCs in skeletal muscle. Am. J. Physiol.-Endocrinol. Metab. 2000, 278, E76–E82. [Google Scholar] [CrossRef]
- Shah, O.J.; Kimball, S.R.; Jefferson, L.S. Among translational effecters, p70(S6k) is uniquely sensitive to inhibition by GCs. Biochem. J. 2000, 347, 389–397. [Google Scholar] [CrossRef]
- Jellyman, J.K.; Martin-Gronert, M.S.; Cripps, R.L.; Giussani, D.A.; Ozanne, S.E.; Shen, Q.W.W.; Du, M.; Fowden, A.L.; Forhead, A.J. Effects of Cortisol and Dexamethasone on Insulin Signalling Pathways in Skeletal Muscle of the Ovine Fetus during Late Gestation. PLoS ONE 2012, 7, e52363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joha, S.; Nugues, A.L.; Hetuin, D.; Berthon, C.; Dezitter, X.; Dauphin, V.; Mahon, F.X.; Roche-Lestienne, C.; Preudhomme, C.; Quesnel, B.; et al. GILZ inhibits the mTORC2/AKT pathway in BCR-ABL(+) cells. Oncogene 2012, 31, 1419–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.K.; Choi, J.W.; Choi, Y.H.; Nam, T.J. Protective Effect of Pyropia yezoensis Peptide on Dexamethasone-Induced Myotube Atrophy in C2C12 Myotubes. Mar. Drugs 2019, 17, 284. [Google Scholar] [CrossRef] [Green Version]
- de Kloet, E.R.; Joels, M.; Holsboer, F. Stress and the brain: From adaptation to disease. Nat. Rev. Neurosci. 2005, 6, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.K.; Ma, Y.J.; Li, Q.; Liu, E.M.; Jin, M.L.; Zhang, L.P.; Wei, C.H. The role of N-6-methyladenosine RNA methylation in the heat stress response of sheep (Ovis aries). Cell Stress Chaperones 2019, 24, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Samanta, D.; Lu, H.Q.; Bullen, J.W.; Zhang, H.M.; Chen, I.; He, X.S.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [Green Version]
- Kudou, K.; Komatsu, T.; Nogami, J.; Maehara, K.; Harada, A.; Saeki, H.; Oki, E.; Maehara, Y.; Ohkawa, Y. The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol. 2017, 7, 170119. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.X.; Yao, Y.L.; Han, J.H.; Yang, Y.L.; Chen, Y.; Tang, Z.L.; Gao, F. Longitudinal epitranscriptome profiling reveals the crucial role of N-6-methyladenosine methylation in porcine prenatal skeletal muscle development. J. Genet. Genom. 2020, 47, 466–476. [Google Scholar] [CrossRef]
- Feng, Y.; Li, Y.; Jiang, W.; Hu, Y.; Jia, Y.; Zhao, R. GR-mediated transcriptional regulation of m(6)A metabolic genes contributes to diet-induced fatty liver in hens. J. Anim. Sci. Biotechnol. 2021, 12, 117. [Google Scholar] [CrossRef]
- Kostyo, J.L.; Redmond, A.F. Role of protein synthesis in the inhibitory action of adrenal steroid hormones on amino acid transport by muscle. Endocrinology 1966, 79, 531–540. [Google Scholar] [CrossRef]
- Li, Z.Y.; Zhang, H.F. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016, 73, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Poncet, N.; Halley, P.A.; Lipina, C.; Gierlinski, M.; Dady, A.; Singer, G.A.; Febrer, M.; Shi, Y.B.; Yamaguchi, T.P.; Taylor, P.M.; et al. Wnt regulates amino acid transporter Slc7a5 and so constrains the integrated stress response in mouse embryos. EMBO Rep. 2020, 21, e48469. [Google Scholar] [CrossRef] [PubMed]
- Habashy, W.S.; Milfort, M.C.; Adomako, K.; Attia, Y.A.; Rekaya, R.; Aggrey, S.E. Effect of heat stress on amino acid digestibility and transporters in meat-type chickens. Poult. Sci. 2017, 96, 2312–2319. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.J.; Fry, C.S.; Glynn, E.L.; Timmerman, K.L.; Dickinson, J.M.; Walker, D.K.; Gundermann, D.M.; Volpi, E.; Rasmussen, B.B. Skeletal muscle amino acid transporter expression is increased in young and older adults following resistance exercise. J. Appl. Physiol. 2011, 111, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Dorn, L.E.; Lasman, L.; Chen, J.; Xu, X.Y.; Hund, T.J.; Medvedovic, M.; Hanna, J.H.; van Berlo, J.H.; Accornero, F. The N-6-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019, 139, 533–545. [Google Scholar] [CrossRef]
- Ma, L.F.; Chen, T.X.; Zhang, X.; Miao, Y.Y.; Tian, X.T.; Yu, K.K.; Xu, X.; Niu, Y.J.; Guo, S.S.; Zhang, C.C.; et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021, 38, 101801. [Google Scholar] [CrossRef]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C.; et al. Bidirectional Transport of Amino Acids Regulates mTOR and Autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef] [Green Version]
- Asaoka, Y.; Nagai, Y.; Namae, M.; Furutani-Seiki, M.; Nishina, H. SLC7 family transporters control the establishment of left-right asymmetry during organogenesis in medaka by activating mTOR signaling. Biochem. Biophys. Res. Commun. 2016, 474, 146–153. [Google Scholar] [CrossRef]
- Wan, X.; Wang, D.; Xiong, Q.; Xiang, H.; Li, H.; Wang, H.; Liu, Z.; Niu, H.; Peng, J.; Jiang, S.; et al. Elucidating a molecular mechanism that the deterioration of porcine meat quality responds to increased cortisol based on transcriptome sequencing. Sci. Rep. 2016, 6, 36589. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).