Towards Tabula Gallus
Abstract
1. Introduction
2. Background
2.1. Pasteur, Darwin, and Cajal
2.2. Many Advantages
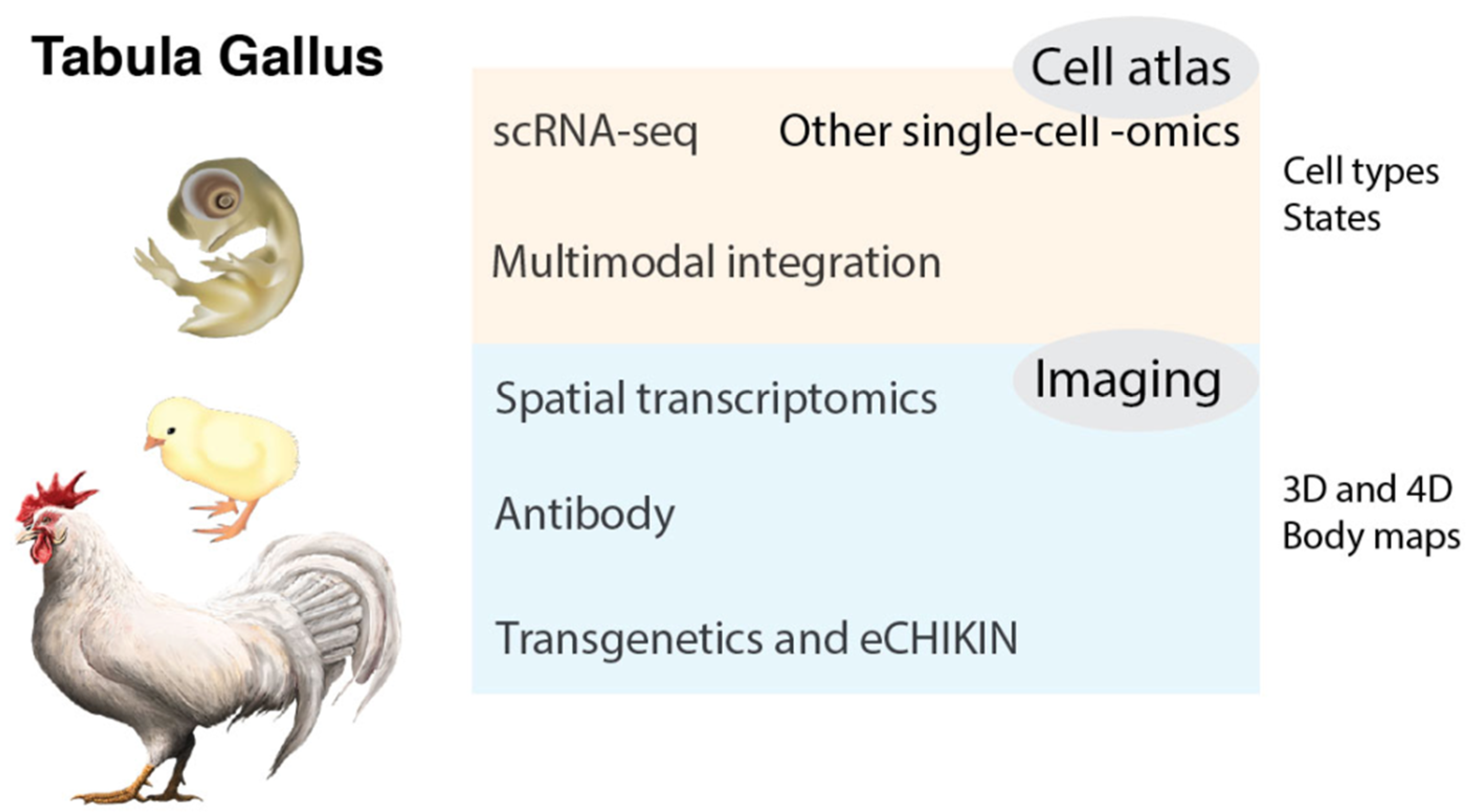
3. Tabula Gallus
3.1. Chicken Cell Atlas
3.2. Cell Types and States
3.3. More Modalities: Epigenome, Protein, Glycan, and Connectome
3.4. Temporal and Spatial Transcriptomics
3.5. Antibodies and Bioimaging
3.6. CRISPR-Mediated and Homology-Instructed Knock-In
3.7. Multimodal Integration and Computational Biology
3.8. Comparative Transcriptomics and Evolution of Cell Types
4. Beyond Tabula Gallus
Funding
Conflicts of Interest
References
- Fumihito, A.; Miyake, T.; Sumi, S.; Takada, M.; Ohno, S.; Kondo, N. One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proc. Natl. Acad. Sci. USA 1994, 91, 12505–12509. [Google Scholar] [CrossRef] [PubMed]
- Lawal, R.A.; Hanotte, O. Domestic chicken diversity: Origin, distribution, and adaptation. Anim. Genet. 2021, 52, 385–394. [Google Scholar] [CrossRef]
- Stern, C.D. The chick: A great model system becomes even greater. Dev. Cell 2005, 8, 9–17. [Google Scholar] [CrossRef]
- Burt, D.W. Emergence of the Chicken as a Model Organism: Implications for Agriculture and Biology. Poult. Sci. 2007, 86, 1460–1471. [Google Scholar] [CrossRef] [PubMed]
- Flores-Santin, J.; Burggren, W.W. Beyond the Chicken: Alternative Avian Models for Developmental Physiological Research. Front. Physiol. 2021, 12, 712633. [Google Scholar] [CrossRef]
- Smith, K.A. Louis Pasteur, the father of immunology? Front. Immunol. 2012, 3, 68. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species; Harvard University Press: Cambridge, MA, USA, 1859. [Google Scholar]
- Ramón y Cajal, S. Histologie du Système Nerveux de l’Homme et des Vertébrés; Maloine: Paris, France, 1911. [Google Scholar] [CrossRef]
- Burkhardt, R.W. Patterns of Behavior: Konrad Lorenz, Niko Tinbergen, and the Founding of Ethology; University of Chicago Press: Chicago, IL, USA, 2005; ISBN 9780226080901. [Google Scholar]
- Pietrzak, K. Christiaan Eijkman (1856–1930). J. Neurol. 2019, 266, 2893–2895. [Google Scholar] [CrossRef] [PubMed]
- Ranek, M.J.; Cotten, S.W.; Willis, M.S. Albert Szent-Györgyi, MD, PhD: Discoverer of Vitamin C and a Pioneer of Cellular Respiration, Muscle Physiology, and Cancer Development. Lab. Med. 2011, 42, 694–698. [Google Scholar] [CrossRef][Green Version]
- Van Epps, H.L. Peyton Rous: Father of the tumor virus. J. Exp. Med. 2005, 201, 320. [Google Scholar] [CrossRef] [PubMed]
- Coffin, J.M.; Fan, H. The discovery of reverse transcriptase. Annu. Rev. Virol. 2016, 3, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Martin, G. The road to Src. Oncogene 2004, 23, 7910–7917. [Google Scholar] [CrossRef]
- Zeliadt, N. Classic Profile of Rita Levi-Montalcini. Proc. Natl. Acad. Sci. USA 2013, 110, 4873–4876. [Google Scholar] [CrossRef]
- Cohen, S. Origins of growth factors: NGF and EGF. J. Biol. Chem. 2008, 283, 33793–33797. [Google Scholar] [CrossRef]
- Bellairs, R.; Osmons, M. The Atlas of Chick Development, 3rd ed.; Academic Press: London, UK, 2014; ISBN 978-0-12-384951-9. [Google Scholar]
- Wolpert, L. Much more from the chicken’s egg than breakfast—A wonderful model system. Mech. Dev. 2004, 121, 1015–1017. [Google Scholar] [CrossRef]
- Mason, I. The Avian Embryo. Methods Mol. Biol. 2008, 461, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Kain, K.H.; Miller, J.W.; Jones-Paris, C.R.; Thomason, R.T.; Lewis, J.D.; Bader, D.M.; Barnett, J.V.; Zijlstra, A. The chick embryo as an expanding experimental model for cancer and cardiovascular research. Dev. Dyn. 2014, 243, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Lillie, F.R. The Development of the Chick; Holt: New York, NY, USA, 1908. [Google Scholar] [CrossRef]
- Schoenwolf, G. Cutting, pasting and painting: Experimental embryology and neural development. Nat. Rev. Neurosci. 2001, 2, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Stern, C.D.; Conrad, H. Waddington’s contributions to avian and mammalian development, 1930–1940. Int. J. Dev. Biol. 2000, 44, 15–23. [Google Scholar]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef] [PubMed]
- Lumsden, A.; Keynes, R. Segmental patterns of neuronal development in the chick hindbrain. Nature 1989, 337, 424–428. [Google Scholar] [CrossRef]
- Jessell, T.M. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat. Rev. Genet. 2000, 1, 20–92. [Google Scholar] [CrossRef]
- Sonoda, E.; Morrison, C.; Yamashita, Y.M.; Takata, M.; Takeda, S. Reverse genetic studies of homologous DNA recombination using the chicken B-lymphocyte line, DT40. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Le Douarin, N. The avian embryo as a model to study the development of the neural crest: A long and still ongoing story. Mech. Dev. 2004, 121, 1089–1102. [Google Scholar] [CrossRef]
- Pourquie, O. The chick embryo: A leading model in somitogenesis studies. Mech. Dev. 2004, 121, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Yokouchi, Y. Establishment of a chick embryo model for analyzing liver development and a search for candidate genes. Dev. Growth Differ. 2005, 47, 357–366. [Google Scholar] [CrossRef]
- Kohonen, P.; Nera, K.P.; Lassila, O. Avian model for B-cell immunology—New genomes and phylotranscriptomics. Scand. J. Immunol. 2007, 66, 113–121. [Google Scholar] [CrossRef]
- Coleman, C.M. Chicken embryo as a model for regenerative medicine. Birth Defects Res. Part C Embryo Today 2008, 84, 245–256. [Google Scholar] [CrossRef]
- Rashidi, H.; Sottile, V. The chick embryo: Hatching a model for contemporary biomedical research. BioEssays 2009, 31, 459–465. [Google Scholar] [CrossRef]
- Swanberg, S.E.; O’Hare, T.H.; Robb, E.A.; Robinson, C.M.; Chang, H.; Delany, M.E. Telomere biology of the chicken: A model for aging research. Exp. Gerontol. 2010, 45, 647–654. [Google Scholar] [CrossRef]
- Datar, S.P.; Bhonde, R.R. Modeling chick to assess diabetes pathogenesis and treatment. Rev. Diabet. Stud. 2011, 8, 245–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vergara, M.N.; Canto-Soler, M.V. Rediscovering the chick embryo as a model to study retinal development. Neural Dev. 2012, 7, 22. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; Segura, T.; Iruela-Arispe, M.L. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis 2014, 17, 779–804. [Google Scholar] [CrossRef] [PubMed]
- Bjørnstad, S.; Austdal, L.P.; Roald, B.; Glover, J.C.; Paulsen, R.E. Cracking the Egg: Potential of the Developing Chicken as a Model System for Nonclinical Safety Studies of Pharmaceuticals. J. Pharmacol. Exp. Ther. 2015, 355, 386–396. [Google Scholar] [CrossRef]
- Cooper, M.D. The early history of B cells. Nat. Rev. Immunol. 2015, 15, 191–197. [Google Scholar] [CrossRef]
- Bronner, M.E.; Simões-Costa, M. The Neural Crest Migrating into the Twenty-First Century. Curr. Top. Dev. Biol. 2016, 116, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Wisely, C.E.; Sayed, J.A.; Tamez, H.; Zelinka, C.; Abdel-Rahman, M.H.; Fischer, A.J.; Cebulla, C.M. The chick eye in vision research: An excellent model for the study of ocular disease. Prog. Retin. Eye Res. 2017, 61, 72–97. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Towers, M.; Vargesson, N.; Tickle, C. The chick limb: Embryology, genetics and teratology. Int. J. Dev. Biol. 2018, 62, 85–95. [Google Scholar] [CrossRef]
- Janesick, A.S.; Heller, S. Stem Cells and the Bird Cochlea-Where Is Everybody? Cold Spring Harb. Perspect. Med. 2019, 9, a033183. [Google Scholar] [CrossRef]
- Vilches-Moure, J.G. Embryonic Chicken (Gallus gallus domesticus) as a Model of Cardiac Biology and Development. Comp. Med. 2019, 69, 184–203. [Google Scholar] [CrossRef]
- Bednarczyk, M.; Dunislawska, A.; Stadnicka, K.; Grochowska, E. Chicken embryo as a model in epigenetic research. Poult. Sci. 2021, 100, 101164. [Google Scholar] [CrossRef] [PubMed]
- Wachholz, G.E.; Rengel, B.D.; Vargesson, N.; Fraga, L.R. From the Farm to the Lab: How Chicken Embryos Contribute to the Field of Teratology. Front. Genet. 2021, 12, 666726. [Google Scholar] [CrossRef]
- Krumlauf, R.; Wilkinson, D.G. Segmentation and patterning of the vertebrate hindbrain. Development 2021, 148, dev186460. [Google Scholar] [CrossRef]
- Tregaskes, C.A.; Kaufman, J. Chickens as a simple system for scientific discovery: The example of the MHC. Mol. Immunol. 2021, 135, 12–20. [Google Scholar] [CrossRef]
- Hammarback, J.A.; Palm, S.L.; Furcht, L.T.; Letourneau, P.C. Guidance of neurite outgrowth by pathways of substratum-adsorbed laminin. J. Neurosci. Res. 1985, 13, 213–220. [Google Scholar] [CrossRef]
- Walter, J.; Kern-Veits, B.; Huf, J.; Stolze, B.; Bonhoeffer, F. Recognition of position-specific properties of tectal cell membranes by retinal axons in vitro. Development 1987, 101, 685–696. [Google Scholar] [CrossRef]
- Luo, Y.; Raible, D.; Raper, J.A. Collapsin: A protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 1993, 75, 217–227. [Google Scholar] [CrossRef]
- Etchevers, H. Primary culture of chick, mouse or human neural crest cells. Nat. Protoc. 2011, 6, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Roycroft, A.; Mayor, R. Michael Abercrombie: Contact inhibition of locomotion and more. Int. J. Dev. Biol. 2018, 62, 5–13. [Google Scholar] [CrossRef]
- Costa, M.L.; Jurberg, A.D.; Mermelstein, C. The Role of Embryonic Chick Muscle Cell Culture in the Study of Skeletal Myogenesis. Front. Physiol. 2021, 12, 668600. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, S.; Sorkin, B.C.; White, P.C.; Brackenbury, R.; Mailhammer, R.; Rutishauser, U.; Cunningham, B.A.; Edelman, G.M. Chemical characterization of a neural cell adhesion molecule purified from embryonic brain membranes. J. Biol. Chem. 1982, 257, 7720–7729. [Google Scholar] [CrossRef]
- Serafini, T.; Kennedy, T.E.; Galko, M.J.; Mirzayan, C.; Jessell, T.M.; Tessier-Lavigne, M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell 1994, 78, 409–424. [Google Scholar] [CrossRef]
- Drescher, U.; Kremoser, C.; Handwerker, C.; Löschinger, J.; Noda, M.; Bonhoeffer, F. In vitro guidance of retinal ganglion cell axons by RAGS, a 25 kDa tectal protein related to ligands for Eph receptor tyrosine kinases. Cell 1995, 82, 359–370. [Google Scholar] [CrossRef]
- Schade, R.; Calzado, E.G.; Sarmiento, R.; Chacana, P.A.; Porankiewicz-Asplund, J.; Terzolo, H.R. Chicken egg yolk antibodies (IgY-technology): A review of progress in production and use in research and human and veterinary medicine. Altern. Lab. Anim. 2005, 33, 129–154. [Google Scholar] [CrossRef]
- Stern, C.D. The chick embryo—Past, present and future as a model system in developmental biology. Mech. Dev. 2004, 121, 1011–1013. [Google Scholar] [CrossRef]
- Bolker, J. Model organisms: There’s more to life than rats and flies. Nature 2012, 491, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Bolker, J.A. Animal Models in Translational Research: Rosetta Stone or Stumbling Block? Bioessays 2017, 39, 1700089. [Google Scholar] [CrossRef]
- Haniffa, M.; Taylor, D.; Linnarsson, S.; Aronow, B.J.; Bader, G.D.; Barker, R.A.; Camara, P.G.; Gray Camp, J.; Chédotal, A.; Copp, A.; et al. A roadmap for the Human Developmental Cell Atlas. Nature 2021, 597, 196–205. [Google Scholar] [CrossRef] [PubMed]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Burt, D.W. Chicken genomics. Int. J. Dev. Biol. 2018, 62, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, E.D. Perspectives from the Avian Phylogenomics Project: Questions that Can Be Answered with Sequencing All Genomes of a Vertebrate Class. Annu. Rev. Anim. Biosci. 2016, 4, 45–59. [Google Scholar] [CrossRef]
- Feng, S.; Stiller, J.; Deng, Y.; Armstrong, J.; Fang, Q.; Hart Reeve, A.; Xie, D.; Chen, G.; Guo, C.; Faircloth, B.C.; et al. Dense sampling of bird diversity increases power of comparative genomics. Nature 2020, 587, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Funahashi, J. Electroporation: Past, present and future. Dev. Growth Differ. 2013, 55, 15–19. [Google Scholar] [CrossRef]
- Farzaneh, M.; Attari, F.; Mozdziak, P.E.; Khoshnam, S.E. The evolution of chicken stem cell culture methods. Br. Poult. Sci. 2017, 58, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; Park, Y.H. Primordial germ cell-mediated transgenesis and genome editing in birds. J. Anim. Sci. Biotechnol. 2018, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Wang, M.; Ling, W.; Xie, D.; Chu, X.; Li, Y.; Huang, Y.; Li, T.; Otieno, E.; Qiu, X.; et al. Advances in Isolation and Culture of Chicken Embryonic Stem Cells in vitro. Cell Reprogram. 2020, 22, 43–54. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.H.; Lee, K. Current Approaches and Applications in Avian Genome Editing. Int. J. Mol. Sci. 2020, 21, 3937. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 2008, 451, 465–469. [Google Scholar] [CrossRef]
- Takahashi, Y.; Watanabe, T.; Nakagawa, S.; Kawakami, K.; Sato, Y. Transposon-mediated stable integration and tetracycline-inducible expression of electroporated transgenes in chicken embryos. Methods Cell Biol. 2008, 87, 271–280. [Google Scholar] [CrossRef]
- Macdonald, J.; Taylor, L.; Sherman, A.; Kawakami, K.; Takahashi, Y.; Sang, H.M.; McGrew, M.J. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc. Natl. Acad. Sci. USA 2012, 109, 1466–1472. [Google Scholar] [CrossRef]
- Morgan, B.A.; Fekete, D.M. Manipulating gene expression with replication-competent retroviruses. Methods Cell Biol. 1996, 51, 185–218. [Google Scholar] [CrossRef]
- Leber, S.M.; Yamagata, M.; Sanes, J.R. Gene transfer using replication-defective retroviral and adenoviral vectors. Methods Cell Biol. 1996, 51, 161–183. [Google Scholar] [CrossRef] [PubMed]
- Ishii, Y.; Reese, D.E.; Mikawa, T. Somatic transgenesis using retroviral vectors in the chicken embryo. Dev. Dyn. 2004, 229, 630–642. [Google Scholar] [CrossRef]
- Hen, G.; Yosefi, S.; Shinder, D.; Or, A.; Mygdal, S.; Condiotti, R.; Galun, E.; Bor, A.; Sela-Donenfeld, D.; Friedman-Einat, M. Gene transfer to chicks using lentiviral vectors administered via the embryonic chorioallantoic membrane. PLoS ONE 2012, 7, e36531. [Google Scholar] [CrossRef]
- Matsui, R.; Tanabe, Y.; Watanabe, D. Avian adeno-associated virus vector efficiently transduces neurons in the embryonic and post-embryonic chicken brain. PLoS ONE 2012, 7, e48730. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ono, M.; Matsui, R.; Watanabe, D.; Ohmori, H. Avian adeno-associated virus as an anterograde transsynaptic vector. J. Neurosci. Methods 2021, 359, 109221. [Google Scholar] [CrossRef]
- Sid, H.; Schusser, B. Applications of Gene Editing in Chickens: A New Era Is on the Horizon. Front. Genet. 2018, 9, 456. [Google Scholar] [CrossRef]
- Chojnacka-Puchta, L.; Sawicka, D. CRISPR/Cas9 gene editing in a chicken model: Current approaches and applications. J. Appl. Genet. 2020, 61, 221–229. [Google Scholar] [CrossRef]
- Park, J.S.; Lee, K.Y.; Han, J.Y. Precise Genome Editing in Poultry and Its Application to Industries. Genes 2020, 11, 1182. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R. CRISPR-mediated Labeling of Cells in Chick Embryos Based on Selectively Expressed Genes. Bio-Protocol 2021, 11, e4105. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Zhuang, X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat. Methods 2021, 18, 18–22. [Google Scholar] [CrossRef]
- Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018, 562, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Regev, A.; Teichmann, S.A.; Lander, E.S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. The Human Cell Atlas. eLife 2017, 6, e27041. [Google Scholar] [CrossRef] [PubMed]
- Osumi-Sutherland, D.; Xu, C.; Keays, M.; Levine, A.P.; Kharchenko, P.V.; Regev, A.; Lein, E.; Teichmann, S.A. Cell type ontologies of the Human Cell Atlas. Nat. Cell Biol. 2021, 23, 1129–1135. [Google Scholar] [CrossRef]
- The Tabula Sapiens Consortium. Tabula Sapiens: An Atlas of Single-Cell Gene Expression. Am. J. Med. Genet. A 2021, 185, 2857–2858. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Ding, J.; Adiconis, X.; Simmons, S.K.; Kowalczyk, M.S.; Hession, C.C.; Marjanovic, N.D.; Hughes, T.K.; Wadsworth, M.H.; Burks, T.; Nguyen, L.T.; et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 2020, 38, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.A.; McLennan, R.; Wolfe, L.A.; Gogol, M.M.; Meier, S.; McKinney, M.C.; Teddy, J.M.; Holmes, L.; Semerad, C.L.; Box, A.C.; et al. Single-cell transcriptome analysis of avian neural crest migration reveals signatures of invasion and molecular transitions. eLife 2017, 6, e28415. [Google Scholar] [CrossRef] [PubMed]
- Vermillion, K.L.; Bacher, R.; Tannenbaum, A.P.; Swanson, S.; Jiang, P.; Chu, L.F.; Stewart, R.; Thomson, J.A.; Vereide, D.T. Spatial patterns of gene expression are unveiled in the chick primitive streak by ordering single-cell transcriptomes. Dev. Biol. 2018, 439, 30–41. [Google Scholar] [CrossRef]
- Feregrino, C.; Sacher, F.; Parnas, O.; Tschopp, P. A single-cell transcriptomic atlas of the developing chicken limb. BMC Genom. 2019, 20, 401. [Google Scholar] [CrossRef]
- Li, J.; Xing, S.; Zhao, G.; Zheng, M.; Yang, X.; Sun, J.; Wen, J.; Liu, R. Identification of diverse cell populations in skeletal muscles and biomarkers for intramuscular fat of chicken by single-cell RNA sequencing. BMC Genom. 2020, 21, 752. [Google Scholar] [CrossRef] [PubMed]
- Tegla, M.G.G.; Buenaventura, D.F.; Kim, D.Y.; Thakurdin, C.; Gonzalez, K.C.; Emerson, M.M. OTX2 represses sister cell fate choices in the developing retina to promote photoreceptor specification. eLife 2020, 9, e54279. [Google Scholar] [CrossRef]
- Hoang, T.; Wang, J.; Boyd, P.; Wang, F.; Santiago, C.; Jiang, L.; Yoo, S.; Lahne, M.; Todd, L.J.; Jia, M.; et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science 2020, 370, eabb8598. [Google Scholar] [CrossRef]
- Gandhi, S.; Hutchins, E.J.; Maruszko, K.; Park, J.H.; Thomson, M.; Bronner, M.E. Bimodal function of chromatin remodeler Hmga1 in neural crest induction and Wnt-dependent emigration. eLife 2020, 9, e57779. [Google Scholar] [CrossRef] [PubMed]
- Estermann, M.A.; Williams, S.; Hirst, C.E.; Roly, Z.Y.; Serralbo, O.; Adhikari, D.; Powell, D.; Major, A.T.; Smith, C.A. Insights into Gonadal Sex Differentiation Provided by Single-Cell Transcriptomics in the Chicken Embryo. Cell Rep. 2020, 31, 107491. [Google Scholar] [CrossRef] [PubMed]
- De Lima, J.E.; Blavet, C.; Bonnin, M.A.; Hirsinger, E.; Comai, G.; Yvernogeau, L.; Delfini, M.C.; Bellenger, L.; Mella, S.; Nassari, S.; et al. Unexpected contribution of fibroblasts to muscle lineage as a mechanism for limb muscle patterning. Nat. Commun. 2021, 12, 3851. [Google Scholar] [CrossRef]
- Guillot, C.; Djeffal, Y.; Michaut, A.; Rabe, B.; Pourquié, O. Dynamics of primitive streak regression controls the fate of neuromesodermal progenitors in the chicken embryo. eLife 2021, 10, e64819. [Google Scholar] [CrossRef]
- Janesick, A.; Scheibinger, M.; Benkafadar, N.; Kirti, S.; Ellwanger, D.C.; Heller, S. Cell-type identity of the avian cochlea. Cell Rep. 2021, 34, 108900. [Google Scholar] [CrossRef]
- Dai, M.; Feng, M.; Li, Z.; Chen, W.; Liao, M. Chicken peripheral blood lymphocyte response to ALV-J infection assessed by single-cell RNA sequencing. bioRxiv 2021. [Google Scholar] [CrossRef]
- Mantri, M.; Scuderi, G.J.; Abedini-Nassab, R.; Wang, M.F.Z.; McKellar, D.; Shi, H.; Grodner, B.; Butcher, J.T.; De Vlaminck, I. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat. Commun. 2021, 12, 1771. [Google Scholar] [CrossRef]
- Yamagata, M.; Yan, W.; Sanes, J.R. A cell atlas of the chick retina based on single-cell transcriptomics. eLife 2021, 10, e63907. [Google Scholar] [CrossRef]
- Zhang, J.; Lv, C.; Mo, C.; Liu, M.; Wan, Y.; Li, J.; Wang, Y. Single-cell RNA sequencing analysis of chicken anterior pituitary: A bird’s-eye view on vertebrate pituitary. Front. Physiol. 2021, 12, 562817. [Google Scholar] [CrossRef]
- Cook, R.G. The comparative psychology of avian visual cognition. Curr. Dir. Psychol. Sci. 2000, 9, 83–89. [Google Scholar] [CrossRef]
- Zhu, C.; Preissl, S.; Ren, B. Single-cell multimodal omics: The power of many. Nat. Methods 2020, 17, 11–14. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- Lareau, C.A.; Duarte, F.M.; Chew, J.G.; Kartha, V.K.; Burkett, Z.D.; Kohlway, A.S.; Pokholok, D.; Aryee, M.J.; Steemers, F.J.; Lebofsky, R.; et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019, 37, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef]
- Peterson, V.M.; Zhang, K.X.; Kumar, N.; Wong, J.; Li, L.; Wilson, D.C.; Moore, R.; McClanahan, T.K.; Sadekova, S.; Klappenbach, J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017, 35, 936–939. [Google Scholar] [CrossRef] [PubMed]
- Stoeckius, M.; Zheng, S.; Houck-Loomis, B.; Hao, S.; Yeung, B.Z.; Mauck, W.M., 3rd; Smibert, P.; Satija, R. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Mair, F.; Erickson, J.R.; Voillet, V.; Simoni, Y.; Bi, T.; Tyznik, A.J.; Martin, J.; Gottardo, R.; Newell, E.W.; Prlic, M. A Targeted Multi-omic Analysis Approach Measures Protein Expression and Low-Abundance Transcripts on the Single-Cell Level. Cell Rep. 2020, 31, 107499. [Google Scholar] [CrossRef]
- Kearney, C.J.; Vervoort, S.J.; Ramsbottom, K.M.; Todorovski, I.; Lelliott, E.J.; Zethoven, M.; Pijpers, L.; Martin, B.P.; Semple, T.; Martelotto, L.; et al. SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Sci. Adv. 2021, 7, eabe3610. [Google Scholar] [CrossRef] [PubMed]
- Lipovsek, M.; Bardy, C.; Cadwell, C.R.; Hadley, K.; Kobak, D.; Tripathy, S.J. Patch-seq: Past, Present, and Future. J. Neurosci. 2021, 41, 937–946. [Google Scholar] [CrossRef]
- Sun, Y.C.; Chen, X.; Fischer, S.; Lu, S.; Zhan, H.; Gillis, J.; Zador, A.M. Integrating barcoded neuroanatomy with spatial transcriptional profiling enables identification of gene correlates of projections. Nat. Neurosci. 2021, 24, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Frisén, J.; Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 2021, 18, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Eichhorn, S.W.; Zingg, B.; Yao, Z.; Cotter, K.; Zeng, H.; Dong, H.; Zhuang, X. Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 2021, 598, 137–143. [Google Scholar] [CrossRef]
- Eng, C.L.; Lawson, M.; Zhu, Q.; Dries, R.; Koulena, N.; Takei, Y.; Yun, J.; Cronin, C.; Karp, C.; Yuan, G.C.; et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 2019, 568, 235–239. [Google Scholar] [CrossRef]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef]
- Rood, J.E.; Stuart, T.; Ghazanfar, S.; Biancalani, T.; Fisher, E.; Butler, A.; Hupalowska, A.; Gaffney, L.; Mauck, W.; Eraslan, G.; et al. Toward a Common Coordinate Framework for the Human Body. Cell 2019, 179, 1455–1467. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, S.L.; Li, Y.; Royall, J.; Feng, D.; Lesnar, P.; Graddis, N.; Naeemi, M.; Facer, B.; Ho, A.; et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 2020, 181, 936–953. [Google Scholar] [CrossRef]
- Cherf, G.M.; Cochran, J.R. Applications of Yeast Surface Display for Protein Engineering. Methods Mol. Biol. 2015, 1319, 155–175. [Google Scholar] [CrossRef]
- Almagro, J.C.; Pedraza-Escalona, M.; Arrieta, H.I.; Pérez-Tapia, S.M. Phage Display Libraries for Antibody Therapeutic Discovery and Development. Antibodies 2019, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Tombácz, I.; Weissman, D.; Pardi, N. Vaccination with Messenger RNA: A Promising Alternative to DNA Vaccination. Methods Mol. Biol. 2021, 2197, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Vieites-Prado, A.; Renier, N. Tissue clearing and 3D imaging in developmental biology. Development 2021, 148, dev199369. [Google Scholar] [CrossRef]
- Hallou, A.; Yevick, H.G.; Dumitrascu, B.; Uhlmann, V. Deep learning for bioimage analysis in developmental biology. Development 2021, 148, dev199616. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Callaway, E.M.; Svoboda, K. Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 2018, 98, 256–281. [Google Scholar] [CrossRef]
- Davey, M.G.; Balic, A.; Rainger, J.; Sang, H.M.; McGrew, M.J. Illuminating the chicken model through genetic modification. Int. J. Dev. Biol. 2018, 62, 257–264. [Google Scholar] [CrossRef]
- Joesch, M.; Mankus, D.; Yamagata, M.; Shahbazi, A.; Schalek, R.; Suissa-Peleg, A.; Meister, M.; Lichtman, J.W.; Scheirer, W.J.; Sanes, J.R. Reconstruction of genetically identified neurons imaged by serial-section electron microscopy. eLife 2016, 5, e15015. [Google Scholar] [CrossRef]
- Wolf, S.; Wan, Y.; McDole, K. Current approaches to fate mapping and lineage tracing using image data. Development 2021, 148, dev198994. [Google Scholar] [CrossRef]
- Gray, G.E.; Leber, S.M.; Sanes, J.R. Migratory patterns of clonally related cells in the developing central nervous system. Experientia 1990, 46, 929–940. [Google Scholar] [CrossRef]
- Woodworth, M.B.; Girskis, K.M.; Walsh, C.A. Building a lineage from single cells: Genetic techniques for cell lineage tracking. Nat. Rev. Genet. 2017, 18, 230–244. [Google Scholar] [CrossRef]
- Kester, L.; van Oudenaarden, A. Single-Cell Transcriptomics Meets Lineage Tracing. Cell Stem Cell 2018, 23, 166–179. [Google Scholar] [CrossRef]
- Wagner, D.E.; Klein, A.M. Lineage tracing meets single-cell omics: Opportunities and challenges. Nat. Rev. Genet. 2020, 21, 410–427. [Google Scholar] [CrossRef]
- VanHorn, S.; Morris, S.A. Next-Generation Lineage Tracing and Fate Mapping to Interrogate Development. Dev. Cell 2021, 56, 7–21. [Google Scholar] [CrossRef] [PubMed]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Alemany, A.; Florescu, M.; Baron, C.S.; Peterson-Maduro, J.; van Oudenaarden, A. Whole-organism clone tracing using single-cell sequencing. Nature 2018, 556, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Raj, B.; Wagner, D.E.; McKenna, A.; Pandey, S.; Klein, A.M.; Shendure, J.; Gagnon, J.A.; Schier, A.F. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 2018, 36, 442–450. [Google Scholar] [CrossRef]
- Chow, K.K.; Budde, M.W.; Granados, A.A.; Cabrera, M.; Yoon, S.; Cho, S.; Huang, T.H.; Koulena, N.; Frieda, K.L.; Cai, L.; et al. Imaging cell lineage with a synthetic digital recording system. Science 2021, 372, eabb3099. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Arendt, D.; Bertucci, P.Y.; Achim, K.; Musser, J.M. Evolution of neuronal types and families. Curr. Opin. Neurobiol. 2019, 56, 144–152. [Google Scholar] [CrossRef]
- Yuste, R.; Hawrylycz, M.; Aalling, N.; Aguilar-Valles, A.; Arendt, D.; Armañanzas, R.; Ascoli, G.A.; Bielza, C.; Bokharaie, V.; Bergmann, T.B.; et al. A community-based transcriptomics classification and nomenclature of neocortical cell types. Nat. Neurosci. 2020, 23, 1456–1468. [Google Scholar] [CrossRef]
- Tosches, M.A. From Cell Types to an Integrated Understanding of Brain Evolution: The Case of the Cerebral Cortex. Annu. Rev. Cell Dev. Biol. 2021, 37, 495–517. [Google Scholar] [CrossRef]
- BRAIN Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 2021, 598, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Arendt, D.; Musser, J.M.; Baker, C.V.H.; Bergman, A.; Cepko, C.; Erwin, D.H.; Pavlicev, M.; Schlosser, G.; Widder, S.; Laubichler, M.D.; et al. The origin and evolution of cell types. Nat. Rev. Genet. 2016, 17, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Shafer, M.E.R. Cross-Species Analysis of Single-Cell Transcriptomic Data. Front. Cell Dev. Biol. 2019, 7, 175. [Google Scholar] [CrossRef]
- Tanay, A.; Sebé-Pedrós, A. Evolutionary cell type mapping with single-cell genomics. Trends Genet. 2021, 37, 919–932. [Google Scholar] [CrossRef]
- Tarashansky, A.J.; Musser, J.M.; Khariton, M.; Li, P.; Arendt, D.; Quake, S.R.; Wang, B. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 2021, 10, e66747. [Google Scholar] [CrossRef]
- Levine, M.; Davidson, E.H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 2005, 102, 4936–4942. [Google Scholar] [CrossRef]
- Arendt, D. The evolution of cell types in animals: Emerging principles from molecular studies. Nat. Rev. Genet. 2008, 9, 868–882. [Google Scholar] [CrossRef] [PubMed]
- Tosches, M.A.; Yamawaki, T.M.; Naumann, R.K.; Jacobi, A.A.; Tushev, G.; Laurent, G. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 2018, 360, 881–888. [Google Scholar] [CrossRef]
- Emery, N.J. Cognitive ornithology: The evolution of avian intelligence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Mooney, R. Neural mechanisms for learned birdsong. Learn. Mem. 2009, 16, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Güntürkün, O.; Bugnyar, T. Cognition without Cortex. Trends Cogn. Sci. 2016, 20, 291–303. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamagata, M. Towards Tabula Gallus. Int. J. Mol. Sci. 2022, 23, 613. https://doi.org/10.3390/ijms23020613
Yamagata M. Towards Tabula Gallus. International Journal of Molecular Sciences. 2022; 23(2):613. https://doi.org/10.3390/ijms23020613
Chicago/Turabian StyleYamagata, Masahito. 2022. "Towards Tabula Gallus" International Journal of Molecular Sciences 23, no. 2: 613. https://doi.org/10.3390/ijms23020613
APA StyleYamagata, M. (2022). Towards Tabula Gallus. International Journal of Molecular Sciences, 23(2), 613. https://doi.org/10.3390/ijms23020613





