Interactions between C8orf37 and FAM161A, Two Ciliary Proteins Essential for Photoreceptor Survival
Abstract
1. Introduction
2. Results
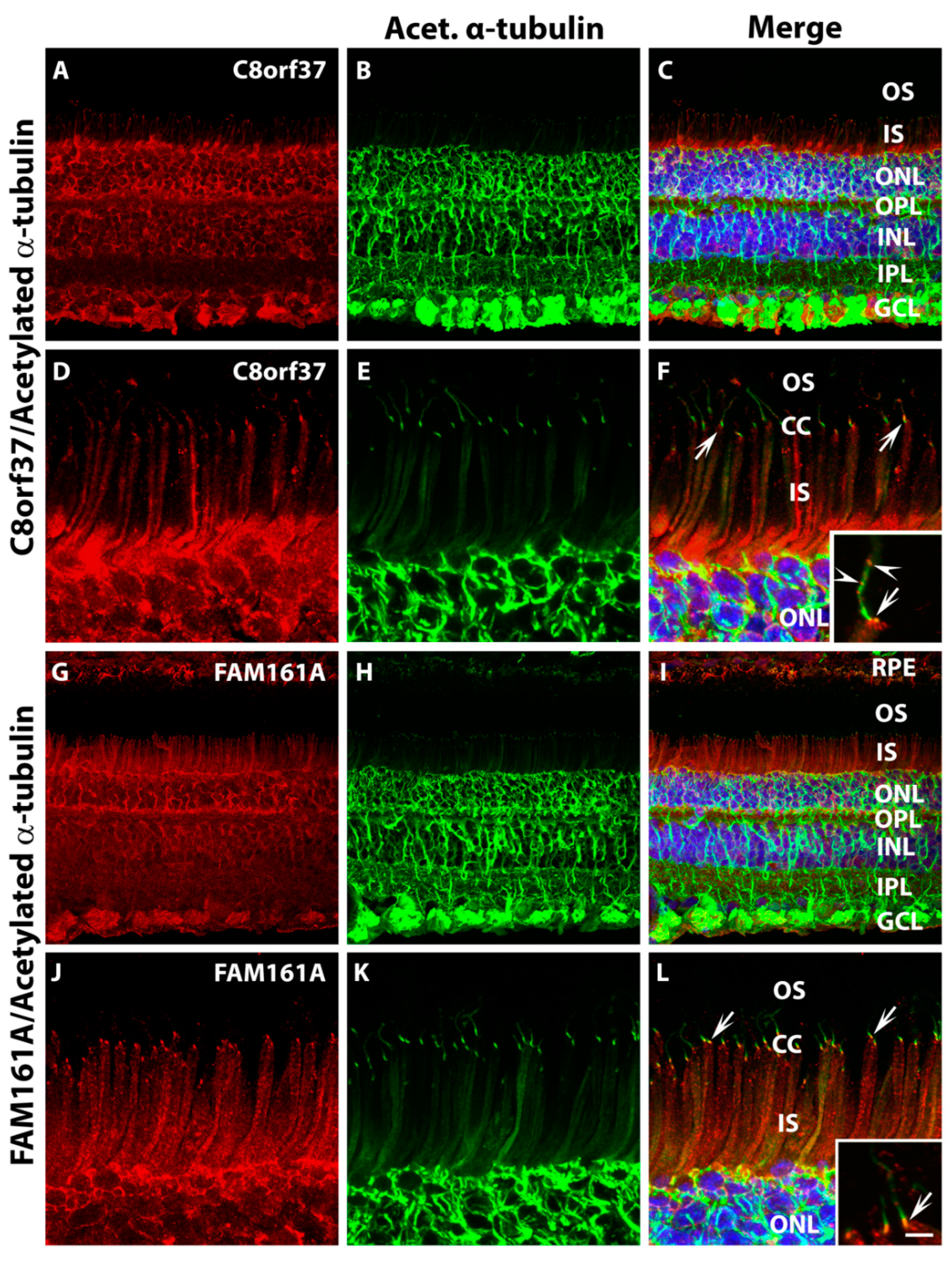
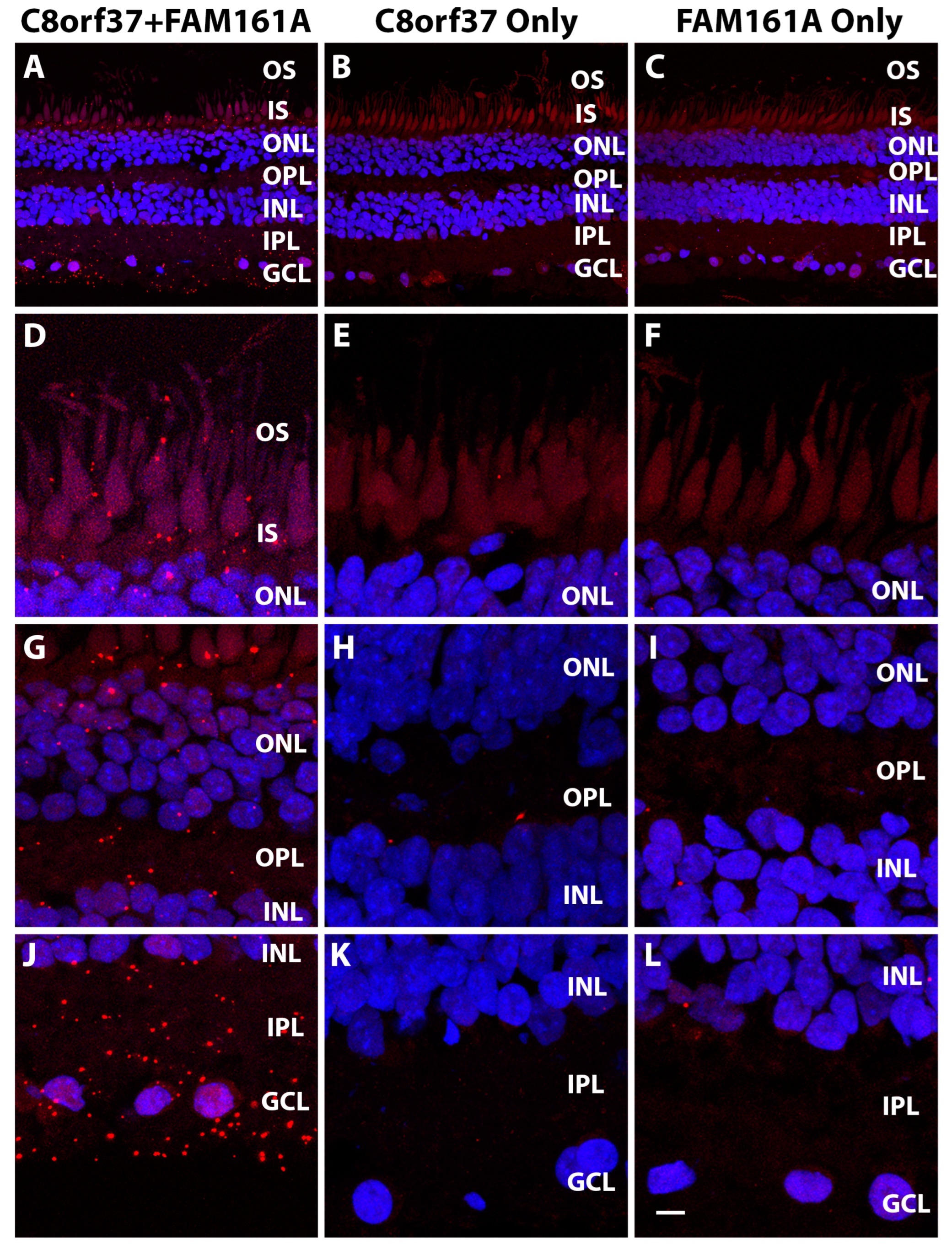
2.1. Co-localization of C8orf37 and FAM161A in the Marmoset Retina
2.2. Co-Localization of Epitope-Tagged C8orf37 and FAM161A in Transfected HEK293 Cells
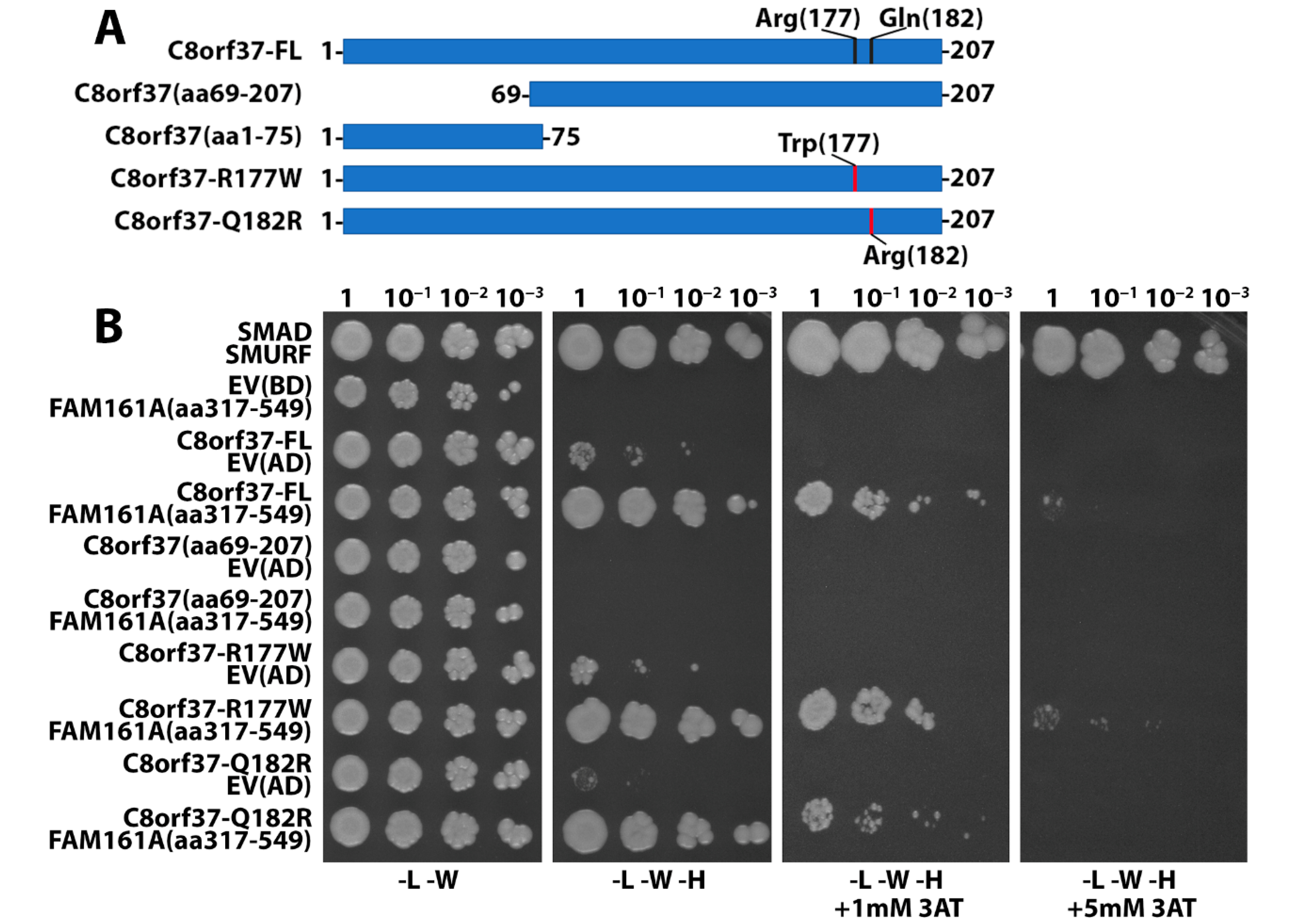
2.3. N-Terminal Region of C8orf37 Interacted with FAM161A
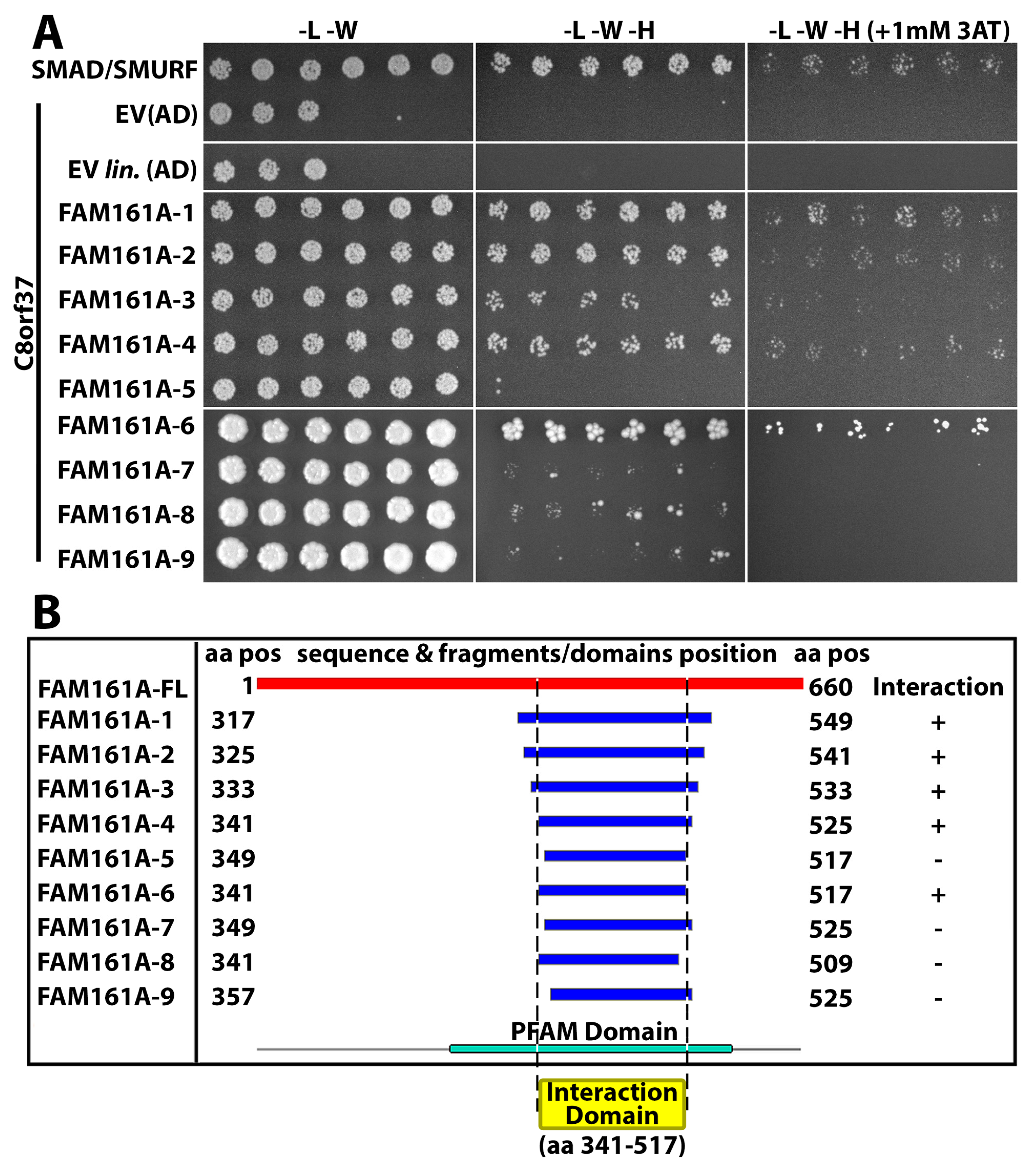
2.4. C8orf37 Interacted with PFAM UPF0564 Domain of FAM161A
3. Discussion
4. Materials and Methods
4.1. Yeast Two-Hybrid
4.2. Transfection of HEK293 Cells
4.3. Immunofluorescence Staining
4.4. Proximity Ligation Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Heon, E.; Kim, G.; Qin, S.; Garrison, J.E.; Tavares, E.; Vincent, A.; Nuangchamnong, N.; Scott, C.A.; Slusarski, D.C.; Sheffield, V.C. Mutations in C8ORF37 cause Bardet Biedl syndrome (BBS21). Hum. Mol. Genet. 2016, 25, 2283–2294. [Google Scholar] [CrossRef]
- Khan, A.O.; Decker, E.; Bachmann, N.; Bolz, H.J.; Bergmann, C. C8orf37 is mutated in Bardet-Biedl syndrome and constitutes a locus allelic to non-syndromic retinal dystrophies. Ophthalmic Genet. 2016, 37, 290–293. [Google Scholar] [CrossRef] [PubMed]
- van Huet, R.A.; Estrada-Cuzcano, A.; Banin, E.; Rotenstreich, Y.; Hipp, S.; Kohl, S.; Hoyng, C.B.; den Hollander, A.I.; Collin, R.W.; Klevering, B.J. Clinical characteristics of rod and cone photoreceptor dystrophies in patients with mutations in the C8orf37 gene. Investig. Ophthalmol. Vis. Sci. 2013, 54, 4683–4690. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, S.; Hayashi, T.; Yoshitake, K.; Akahori, M.; Ikeo, K.; Gekka, T.; Tsuneoka, H.; Iwata, T. Novel C8orf37 Mutations in Patients with Early-onset Retinal Dystrophy, Macular Atrophy, Cataracts, and High Myopia. Ophthalmic Genet. 2016, 37, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.H.; Mutsuddi, M.; Kimchi, A.; Zelinger, L.; Mizrahi-Meissonnier, L.; Marks-Ohana, D.; Boleda, A.; Ratnapriya, R.; Sharon, D.; Swaroop, A.; et al. Whole exome sequencing reveals GUCY2D as a major gene associated with cone and cone-rod dystrophy in Israel. Investig. Ophthalmol. Vis. Sci. 2014, 56, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Cuzcano, A.; Neveling, K.; Kohl, S.; Banin, E.; Rotenstreich, Y.; Sharon, D.; Falik-Zaccai, T.C.; Hipp, S.; Roepman, R.; Wissinger, B.; et al. Mutations in C8orf37, encoding a ciliary protein, are associated with autosomal-recessive retinal dystrophies with early macular involvement. Am. J. Hum. Genet. 2012, 90, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Jinda, W.; Taylor, T.D.; Suzuki, Y.; Thongnoppakhun, W.; Limwongse, C.; Lertrit, P.; Suriyaphol, P.; Trinavarat, A.; Atchaneeyasakul, L.O. Whole exome sequencing in Thai patients with retinitis pigmentosa reveals novel mutations in six genes. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2259–2268. [Google Scholar] [CrossRef]
- Ravesh, Z.; El Asrag, M.E.; Weisschuh, N.; McKibbin, M.; Reuter, P.; Watson, C.M.; Baumann, B.; Poulter, J.A.; Sajid, S.; Panagiotou, E.S.; et al. Novel C8orf37 mutations cause retinitis pigmentosa in consanguineous families of Pakistani origin. Mol. Vis. 2015, 21, 236–243. [Google Scholar]
- Rahner, N.; Nuernberg, G.; Finis, D.; Nuernberg, P.; Royer-Pokora, B. A novel C8orf37 splice mutation and genotype-phenotype correlation for cone-rod dystrophy. Ophthalmic Genet. 2016, 37, 294–300. [Google Scholar] [CrossRef]
- Langmann, T.; Di Gioia, S.A.; Rau, I.; Stohr, H.; Maksimovic, N.S.; Corbo, J.C.; Renner, A.B.; Zrenner, E.; Kumaramanickavel, G.; Karlstetter, M.; et al. Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010, 87, 376–381. [Google Scholar] [CrossRef][Green Version]
- Bandah-Rozenfeld, D.; Mizrahi-Meissonnier, L.; Farhy, C.; Obolensky, A.; Chowers, I.; Pe’er, J.; Merin, S.; Ben-Yosef, T.; Ashery-Padan, R.; Banin, E.; et al. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010, 87, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Di Gioia, S.A.; Letteboer, S.J.; Kostic, C.; Bandah-Rozenfeld, D.; Hetterschijt, L.; Sharon, D.; Arsenijevic, Y.; Roepman, R.; Rivolta, C. FAM161A, associated with retinitis pigmentosa, is a component of the cilia-basal body complex and interacts with proteins involved in ciliopathies. Hum. Mol. Genet. 2012, 21, 5174–5184. [Google Scholar] [CrossRef] [PubMed]
- Zach, F.; Grassmann, F.; Langmann, T.; Sorusch, N.; Wolfrum, U.; Stohr, H. The retinitis pigmentosa 28 protein FAM161A is a novel ciliary protein involved in intermolecular protein interaction and microtubule association. Hum. Mol. Genet. 2012, 21, 4573–4586. [Google Scholar] [CrossRef]
- Formstecher, E.; Aresta, S.; Collura, V.; Hamburger, A.; Meil, A.; Trehin, A.; Reverdy, C.; Betin, V.; Maire, S.; Brun, C.; et al. Protein interaction mapping: A Drosophila case study. Genome Res. 2005, 15, 376–384. [Google Scholar] [CrossRef]
- Sharif, A.S.; Yu, D.; Loertscher, S.; Austin, R.; Nguyen, K.; Mathur, P.D.; Clark, A.M.; Zou, J.; Lobanova, E.S.; Arshavsky, V.Y.; et al. C8ORF37 Is Required for Photoreceptor Outer Segment Disc Morphogenesis by Maintaining Outer Segment Membrane Protein Homeostasis. J. Neurosci. 2018, 38, 3160–3176. [Google Scholar] [CrossRef]
- Hawkins, R.K.; Jansen, H.G.; Sanyal, S. Development and degeneration of retina in rds mutant mice: Photoreceptor abnormalities in the heterozygotes. Exp. Eye Res. 1985, 41, 701–720. [Google Scholar] [CrossRef]
- Sanyal, S.; Hawkins, R.K. Development and degeneration of retina in rds mutant mice: Effects of light on the rate of degeneration in albino and pigmented homozygous and heterozygous mutant and normal mice. Vis. Res. 1986, 26, 1177–1185. [Google Scholar] [CrossRef]
- Zhang, Y.; Molday, L.L.; Molday, R.S.; Sarfare, S.S.; Woodruff, M.L.; Fain, G.L.; Kraft, T.W.; Pittler, S.J. Knockout of GARPs and the beta-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J. Cell Sci. 2009, 122, 1192–1200. [Google Scholar] [CrossRef]
- Rattner, A.; Smallwood, P.M.; Williams, J.; Cooke, C.; Savchenko, A.; Lyubarsky, A.; Pugh, E.N.; Nathans, J. A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron 2001, 32, 775–786. [Google Scholar] [CrossRef]
- Liu, Q.; Lyubarsky, A.; Skalet, J.H.; Pugh, E.N., Jr.; Pierce, E.A. RP1 is required for the correct stacking of outer segment discs. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4171–4183. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.; Hong, D.H.; Pawlyk, B.; Yue, G.; Adamian, M.; Grynberg, M.; Godzik, A.; Li, T. The retinitis pigmentosa GTPase regulator (RPGR)- interacting protein: Subserving RPGR function and participating in disk morphogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 3965–3970. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, Y.; Lillo, C.; Chien, J.; Yu, Z.; Michaelides, M.; Klein, M.; Howes, K.A.; Li, Y.; Kaminoh, Y.; et al. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J. Clin. Investig. 2008, 118, 2908–2916. [Google Scholar] [CrossRef] [PubMed]
- Karlstetter, M.; Sorusch, N.; Caramoy, A.; Dannhausen, K.; Aslanidis, A.; Fauser, S.; Boesl, M.R.; Nagel-Wolfrum, K.; Tamm, E.R.; Jagle, H.; et al. Disruption of the retinitis pigmentosa 28 gene Fam161a in mice affects photoreceptor ciliary structure and leads to progressive retinal degeneration. Hum. Mol. Genet. 2014, 23, 5197–5210. [Google Scholar] [CrossRef] [PubMed]
- Van Schil, K.; Klevering, B.J.; Leroy, B.P.; Pott, J.W.; Bandah-Rozenfeld, D.; Zonneveld-Vrieling, M.N.; Sharon, D.; den Hollander, A.I.; Cremers, F.P.; De Baere, E.; et al. A Nonsense Mutation in FAM161A Is a Recurrent Founder Allele in Dutch and Belgian Individuals With Autosomal Recessive Retinitis Pigmentosa. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7418–7426. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.M.; Sergouniotis, P.; Alfano, G.; Muspratt-Tucker, N.; Barton, S.; Moore, A.T.; Black, G.; Bhattacharya, S.S.; Webster, A.R. Diverse clinical phenotypes associated with a nonsense mutation in FAM161A. Eye 2015, 29, 1226–1232. [Google Scholar] [CrossRef]
- Ellingford, J.M.; Barton, S.; Bhaskar, S.; O’Sullivan, J.; Williams, S.G.; Lamb, J.A.; Panda, B.; Sergouniotis, P.I.; Gillespie, R.L.; Daiger, S.P.; et al. Molecular findings from 537 individuals with inherited retinal disease. J. Med. Genet. 2016, 53, 761–767. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, V.W.; Feng, Y.; Tian, X.; Li, F.Y.; Truong, C.; Wang, G.; Chiang, P.W.; Lewis, R.A.; Wong, L.J. Dependable and efficient clinical utility of target capture-based deep sequencing in molecular diagnosis of retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6213–6223. [Google Scholar] [CrossRef]
- Jespersgaard, C.; Fang, M.; Bertelsen, M.; Dang, X.; Jensen, H.; Chen, Y.; Bech, N.; Dai, L.; Rosenberg, T.; Zhang, J.; et al. Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Sci. Rep. 2019, 9, 1219. [Google Scholar] [CrossRef]
- Venturini, G.; Di Gioia, S.A.; Harper, S.; Weigel-DiFranco, C.; Rivolta, C.; Berson, E.L. Molecular genetics of FAM161A in North American patients with early-onset retinitis pigmentosa. PLoS ONE 2014, 9, e92479. [Google Scholar] [CrossRef][Green Version]
- Beryozkin, A.; Shevah, E.; Kimchi, A.; Mizrahi-Meissonnier, L.; Khateb, S.; Ratnapriya, R.; Lazar, C.H.; Blumenfeld, A.; Ben-Yosef, T.; Hemo, Y.; et al. Whole Exome Sequencing Reveals Mutations in Known Retinal Disease Genes in 33 out of 68 Israeli Families with Inherited Retinopathies. Sci. Rep. 2015, 5, 13187. [Google Scholar] [CrossRef]
- Glockle, N.; Kohl, S.; Mohr, J.; Scheurenbrand, T.; Sprecher, A.; Weisschuh, N.; Bernd, A.; Rudolph, G.; Schubach, M.; Poloschek, C.; et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur. J. Hum. Genet. 2014, 22, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Carss, K.J.; Arno, G.; Erwood, M.; Stephens, J.; Sanchis-Juan, A.; Hull, S.; Megy, K.; Grozeva, D.; Dewhurst, E.; Malka, S.; et al. Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am. J. Hum. Genet. 2017, 100, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.S.; Song, H.; Li, Y.; Xiao, Z.Y.; Li, T. Whole-exome sequencing identifies novel mutations in genes responsible for retinitis pigmentosa in 2 nonconsanguineous Chinese families. Int. J. Ophthalmol. 2019, 12, 915–923. [Google Scholar] [CrossRef]
- Duncan, J.L.; Biswas, P.; Kozak, I.; Navani, M.; Syed, R.; Soudry, S.; Menghini, M.; Caruso, R.C.; Jeffrey, B.G.; Heckenlively, J.R.; et al. Ocular Phenotype of a Family with FAM161A-associated Retinal Degeneration. Ophthalmic Genet. 2016, 37, 44–52. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zobor, D.; Balousha, G.; Baumann, B.; Wissinger, B. Homozygosity mapping reveals new nonsense mutation in the FAM161A gene causing autosomal recessive retinitis pigmentosa in a Palestinian family. Mol. Vis. 2014, 20, 178–182. [Google Scholar]
- Barbelanne, M.; Hossain, D.; Chan, D.P.; Peranen, J.; Tsang, W.Y. Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum. Mol. Genet. 2015, 24, 2185–2200. [Google Scholar] [CrossRef]
- Stowe, T.R.; Wilkinson, C.J.; Iqbal, A.; Stearns, T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell 2012, 23, 3322–3335. [Google Scholar] [CrossRef]
- Singla, V.; Romaguera-Ros, M.; Garcia-Verdugo, J.M.; Reiter, J.F. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell 2010, 18, 410–424. [Google Scholar] [CrossRef]
- Boldt, K.; Mans, D.A.; Won, J.; van Reeuwijk, J.; Vogt, A.; Kinkl, N.; Letteboer, S.J.; Hicks, W.L.; Hurd, R.E.; Naggert, J.K.; et al. Disruption of intraflagellar protein transport in photoreceptor cilia causes Leber congenital amaurosis in humans and mice. J. Clin. Investig. 2011, 121, 2169–2180. [Google Scholar] [CrossRef]
- Qu, Z.; Yimer, T.A.; Xie, S.; Wong, F.; Yu, S.; Liu, X.; Han, S.; Ma, J.; Lu, Z.; Hu, X.; et al. Knocking out lca5 in zebrafish causes cone-rod dystrophy due to impaired outer segment protein trafficking. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2694–2705. [Google Scholar] [CrossRef]
- Vojtek, A.B.; Hollenberg, S.M. Ras-Raf interaction: Two-hybrid analysis. Methods Enzymol. 1995, 255, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Bartel, P.L.; Chein, C.T.; Sternglanz, R.; Fields, S. Cellular Interactions in Development: A Practical Approach; Hartley, D.A., Ed.; Oxford University Press: Oxford, UK, 1993. [Google Scholar]
- Fromont-Racine, M.; Rain, J.C.; Legrain, P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997, 16, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Colland, F.; Jacq, X.; Trouplin, V.; Mougin, C.; Groizeleau, C.; Hamburger, A.; Meil, A.; Wojcik, J.; Legrain, P.; Gauthier, J.M. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004, 14, 1324–1332. [Google Scholar] [CrossRef] [PubMed]




| Protein Name (Gene Name) | Confidence of Interaction * |
|---|---|
| C22orf19 (THOC5) | Moderate |
| CASP8-associated protein 2 (CASP8AP2) | Moderate |
| Chromodomain helicase DNA-binding protein 3 (CHD3) | Good |
| Cone–rod homeobox (CRX) | Moderate |
| Death domain-associated protein (DAXX) | Moderate |
| Family with sequence similarity 161, member A (FAM161A) | Moderate |
| Homeodomain-interacting protein kinase 2, variant 2 (HIPK2) | Very High |
| Heat shock transcription factor 2 (HSF2) | Moderate |
| Mitochondrial ribosomal protein S6 (MRPS6) | Moderate |
| Plectin (PLEC) | Moderate |
| Protein tyrosine kinase 2 Beta (PTK2B) | Good |
| RING finger protein 111 (RNF111) | Moderate |
| Synaptotagmin-binding cytoplasmic RNA- interacting protein (SYNCRIP) | Moderate |
| Zinc finger protein 106 (ZNF106) | Moderate |
| Non-receptor tyrosine kinase, Src (nRTK) | Moderate |
| Ankyrin repeat domain 11 (ANKRD11) | Moderate |
| RNA-binding fox-1 homolog 1 (RBFOX1) | Moderate |
| N-acetylneuraminate pyruvate lyase (NPL) | Moderate |
| Thioredoxin reductase 2 (TXNRD2) | Moderate |
| DEAD box helicase 17 (DDX17) | Moderate |
| SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (SMARCAD1) | Moderate |
| Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1/NEAT2) | Moderate |
| Calcium/calmodulin-dependent protein kinase type 1D (CAMK1D) | Moderate |
| Phosphodiesterase 4D (PDE4D) | Moderate |
| Tripartite motif-containing 37 (TRIM37) | Moderate |
| FSHD region gene 1 (FRG1) | Moderate |
| Synapsin III (SYN3) | Moderate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, J.; Sager, R.; Sasaki, E.; Hu, H. Interactions between C8orf37 and FAM161A, Two Ciliary Proteins Essential for Photoreceptor Survival. Int. J. Mol. Sci. 2022, 23, 12033. https://doi.org/10.3390/ijms231912033
Liu Y, Chen J, Sager R, Sasaki E, Hu H. Interactions between C8orf37 and FAM161A, Two Ciliary Proteins Essential for Photoreceptor Survival. International Journal of Molecular Sciences. 2022; 23(19):12033. https://doi.org/10.3390/ijms231912033
Chicago/Turabian StyleLiu, Yu, Jinjun Chen, Rachel Sager, Erika Sasaki, and Huaiyu Hu. 2022. "Interactions between C8orf37 and FAM161A, Two Ciliary Proteins Essential for Photoreceptor Survival" International Journal of Molecular Sciences 23, no. 19: 12033. https://doi.org/10.3390/ijms231912033
APA StyleLiu, Y., Chen, J., Sager, R., Sasaki, E., & Hu, H. (2022). Interactions between C8orf37 and FAM161A, Two Ciliary Proteins Essential for Photoreceptor Survival. International Journal of Molecular Sciences, 23(19), 12033. https://doi.org/10.3390/ijms231912033





