The NGATHA-like Genes DPA4 and SOD7 Are Not Required for Stem Cell Specification during Embryo Development in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
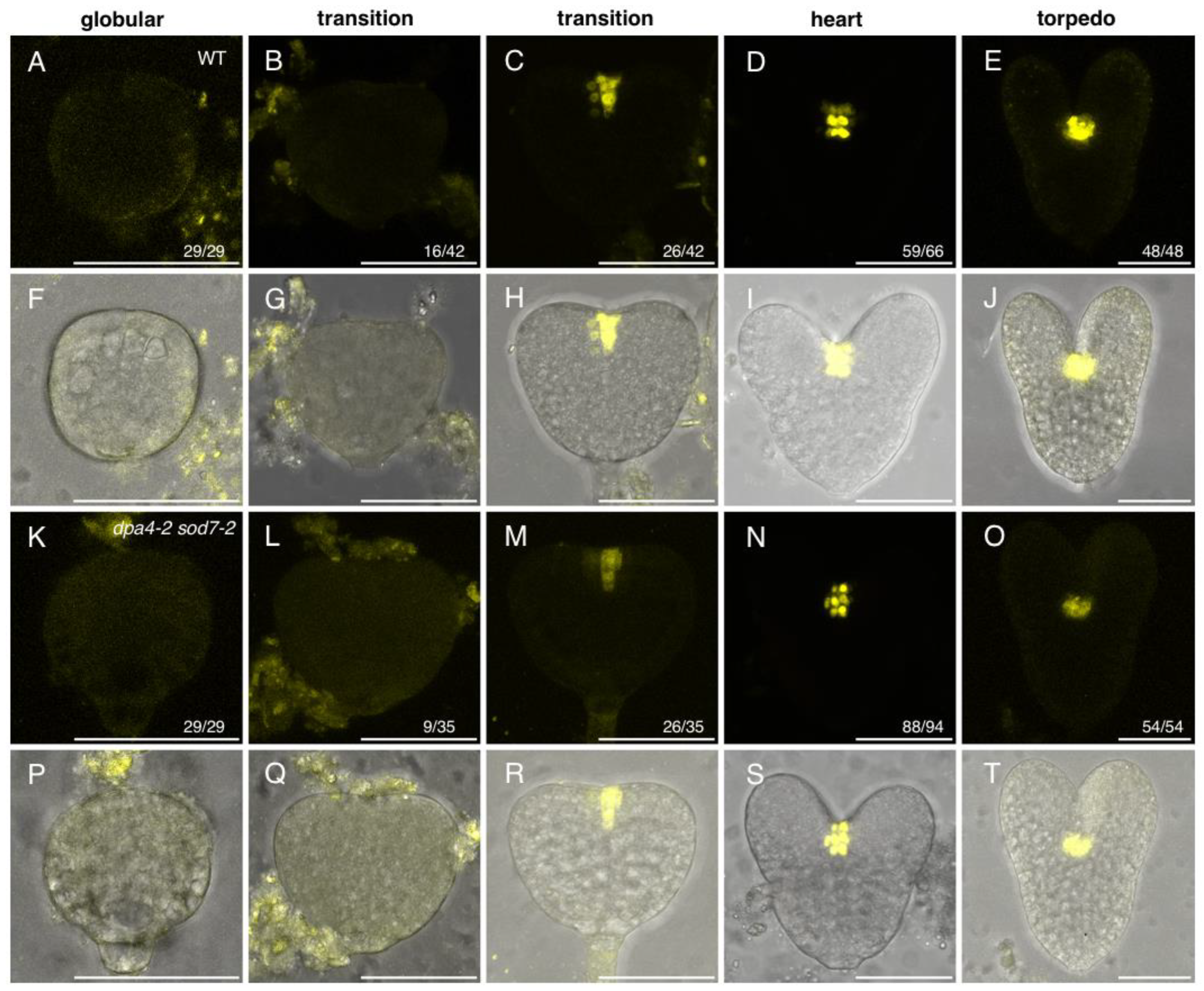
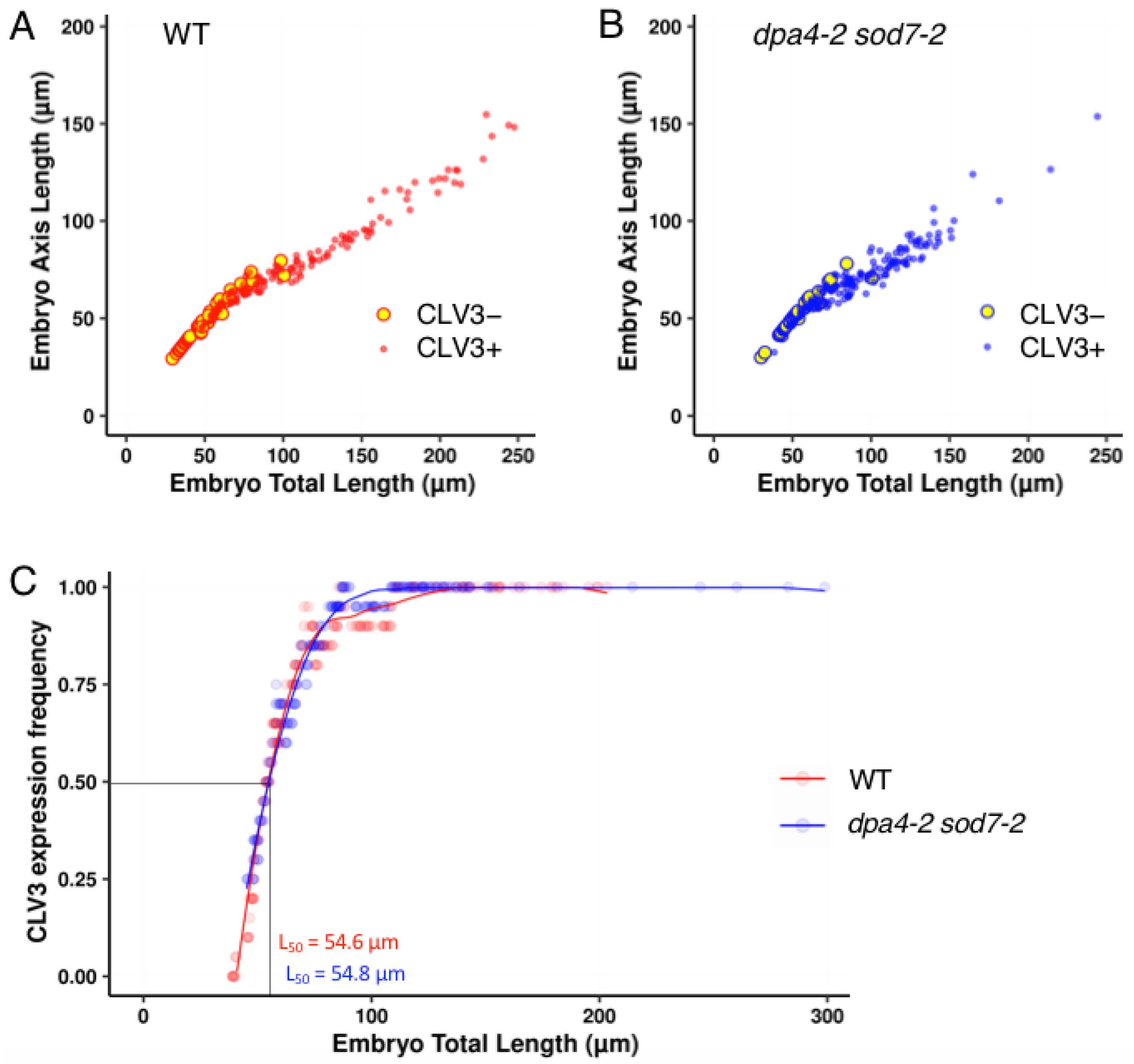
2.1. Expression of a Stem Cell Reporter during Wild-Type and dpa4-2 sod7-2 Embryo Development
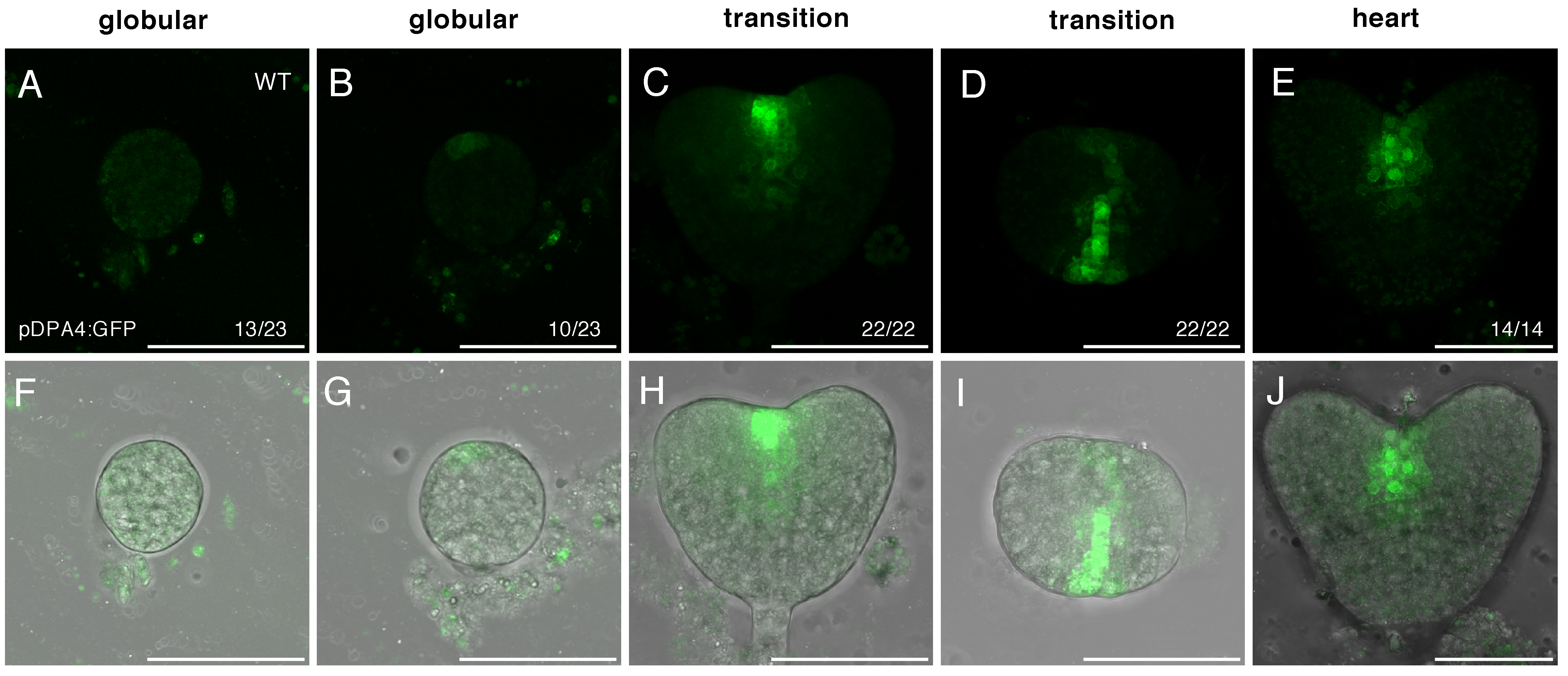
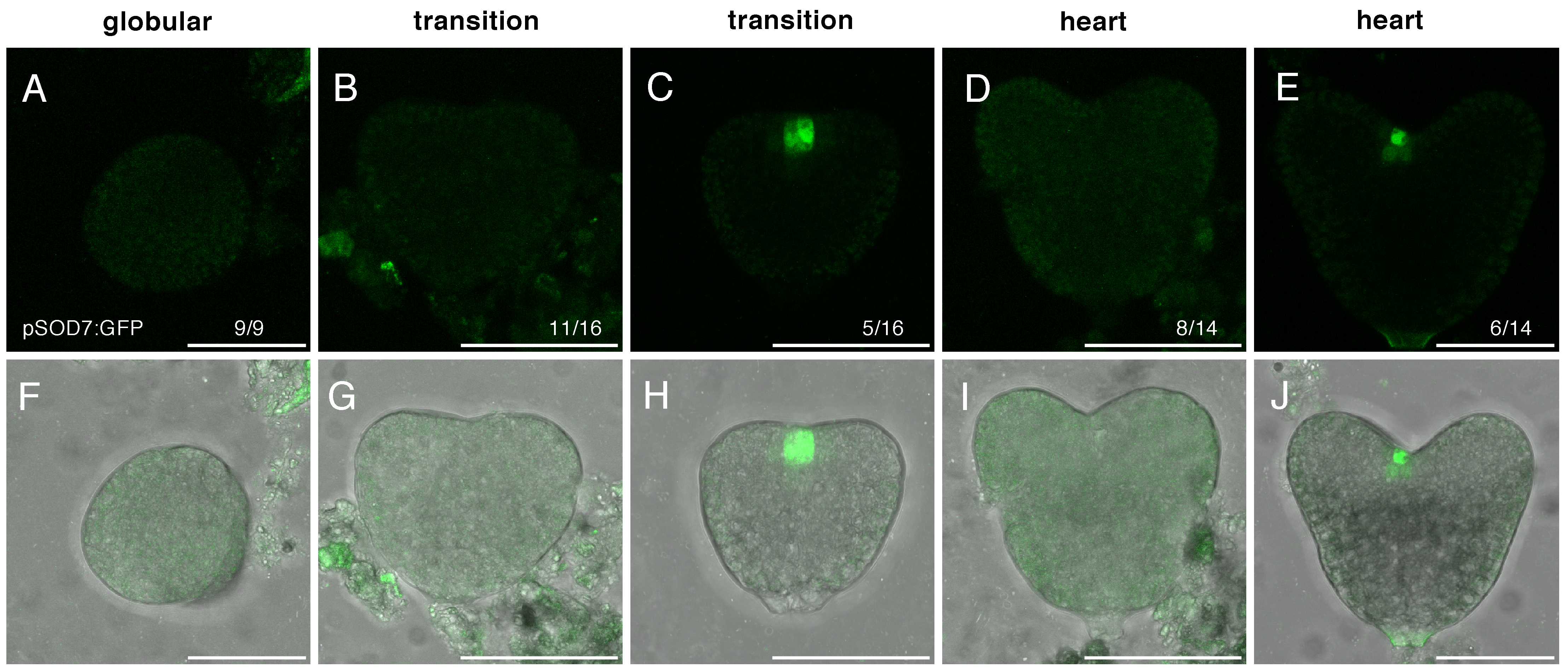
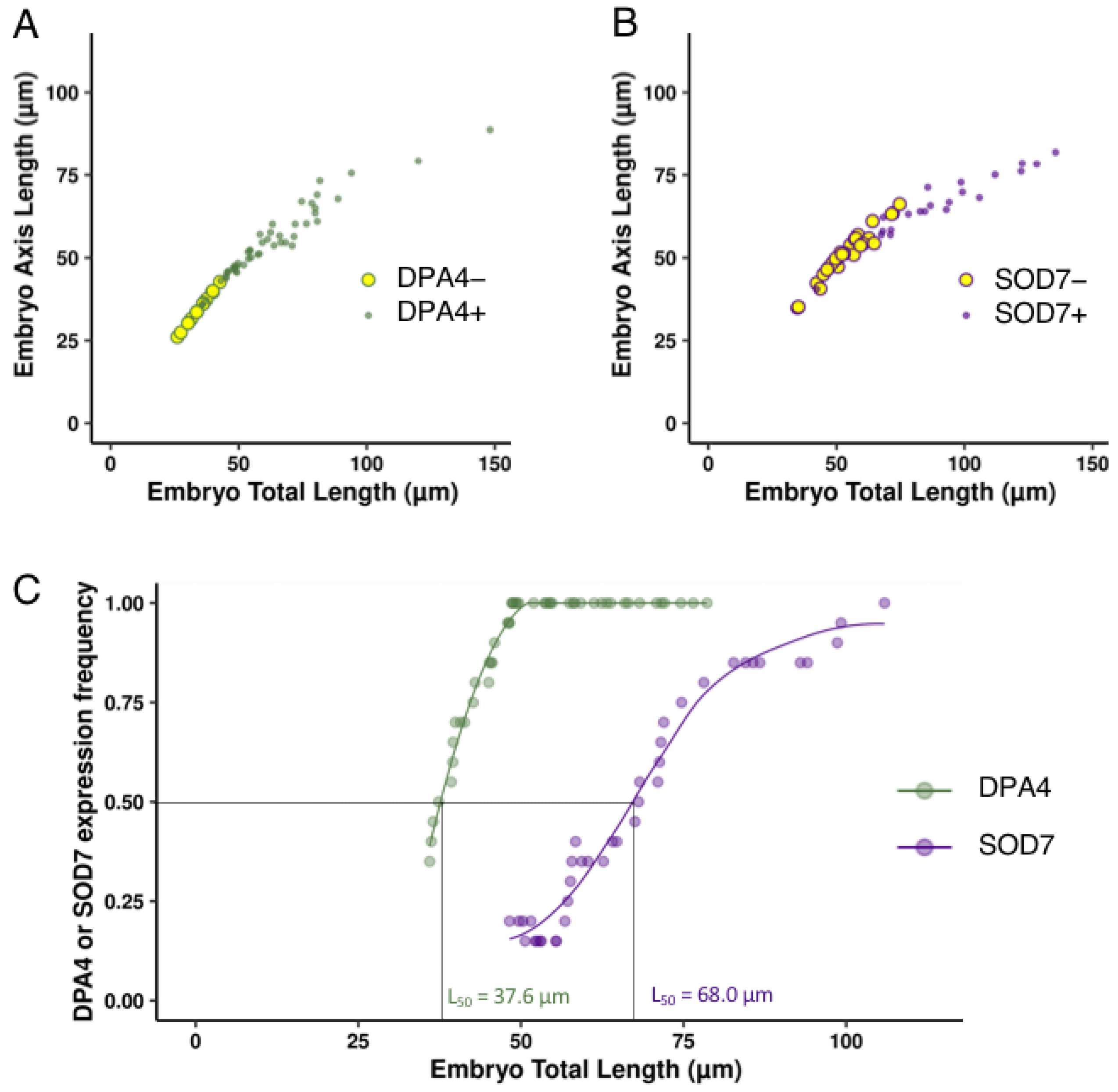
2.2. DPA4 and SOD7 Are Expressed during Embryo Development, but Show Different Dynamics of Expression
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Embryo Dissection and Confocal Microscopy
4.3. Image Analysis
4.4. Data Analysis
4.5. Gene Expression Levels
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dinneny, J.R.; Benfey, P.N. Plant Stem Cell Niches: Standing the Test of Time. Cell 2008, 132, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.R.; Laux, T. Connecting the Paths in Plant Stem Cell Regulation. Trends Cell Biol. 2007, 17, 403–410. [Google Scholar] [CrossRef]
- Bäurle, I.; Laux, T. Apical Meristems: The Plant’s Fountain of Youth. Bioessays 2003, 25, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Laux, T.; Mayer, K.F.; Berger, J.; Jurgens, G. The WUSCHEL Gene Is Required for Shoot and Floral Meristem Integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in Regulating Stem Cell Fate in the Arabidopsis Shoot Meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jonsson, H.; Reddy, G.V.; Jönsson, H.; Venugopala Reddy, G. WUSCHEL Protein Movement Mediates Stem Cell Homeostasis in the Arabidopsis Shoot Apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef] [PubMed]
- Daum, G.; Medzihradszky, A.; Suzaki, T.; Lohmann, J.U. A Mechanistic Framework for Noncell Autonomous Stem Cell Induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14619–14624. [Google Scholar] [CrossRef]
- Perales, M.; Rodriguez, K.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. Threshold-Dependent Transcriptional Discrimination Underlies Stem Cell Homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, E6298–E6306. [Google Scholar] [CrossRef]
- Rodriguez, K.; Perales, M.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. DNA-Dependent Homodimerization, Sub-Cellular Partitioning, and Protein Destabilization Control WUSCHEL Levels and Spatial Patterning. Proc. Natl. Acad. Sci. USA 2016, 113, E6307–E6315. [Google Scholar] [CrossRef]
- Sloan, J.; Hakenjos, J.P.; Gebert, M.; Ermakova, O.; Gumiero, A.; Stier, G.; Wild, K.; Sinning, I.; Lohmann, J.U. Structural Basis for the Complex DNA Binding Behavior of the Plant Stem Cell Regulator WUSCHEL. Nat. Commun. 2020, 11, 2223. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of Pluripotency Pathways Regulates Stem Cell Maintenance in the Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef]
- Clark, S.E. Cell Signalling at the Shoot Meristem. Nat. Rev. Mol. Cell Biol. 2001, 2, 276–284. [Google Scholar] [CrossRef]
- Brand, U.; Grünewald, M.; Hobe, M.; Simon, R.; Grunewald, M.; Hobe, M.; Simon, R.; Grünewald, M.; Hobe, M.; Simon, R. Regulation of CLV3 Expression by Two Homeobox Genes in Arabidopsis. Plant Physiol. 2002, 129, 565–575. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Bickle, K.L.; Schrage, K.J.; Muskett, P.; Patel, K.; Clark, S.E. The CLAVATA1-Related BAM1, BAM2 and BAM3 Receptor Kinase-like Proteins Are Required for Meristem Function in Arabidopsis. Plant J. 2006, 45, 1–16. [Google Scholar] [CrossRef]
- DeYoung, B.J.; Clark, S.E. BAM Receptors Regulate Stem Cell Specification and Organ Development Through Complex Interactions With CLAVATA Signaling. Genetics 2008, 180, 895–904. [Google Scholar] [CrossRef]
- Müller, R.; Bleckmann, A.; Simon, R. The Receptor Kinase CORYNE of Arabidopsis Transmits the Stem Cell-Limiting Signal CLAVATA3 Independently of CLAVATA1. Plant Cell 2008, 20, 934–946. [Google Scholar] [CrossRef]
- Ogawa, M.; Shinohara, H.; Sakagami, Y.; Matsubayash, Y. Arabidopsis CLV3 Peptide Directly Binds CLV1 Ectodomain. Science 2008, 319, 294. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL Controls Meristem Function by Direct Regulation of Cytokinin-Inducible Response Regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef]
- Wang, L.; Mai, Y.X.; Zhang, Y.C.; Luo, Q.; Yang, H.Q. MicroRNA171c-Targeted SCL6-II, SCL6-III, and SCL6-IV Genes Regulate Shoot Branching in Arabidopsis. Mol. Plant 2010, 3, 794–806. [Google Scholar] [CrossRef]
- Han, H.; Yan, A.; Li, L.; Zhu, Y.; Feng, B.; Liu, X.; Zhou, Y. A Signal Cascade Originated from Epidermis Defines Apical-Basal Patterning of Arabidopsis Shoot Apical Meristems. Nat. Commun. 2020, 11, 1214. [Google Scholar] [CrossRef]
- Han, H.; Geng, Y.; Guo, L.; Yan, A.; Meyerowitz, E.M.; Liu, X.; Zhou, Y. The Overlapping and Distinct Roles of HAM Family Genes in Arabidopsis Shoot Meristems. Front. Plant Sci. 2020, 11, 541968. [Google Scholar] [CrossRef]
- Chickarmane, V.S.; Gordon, S.P.; Tarr, P.T.; Heisler, M.G.; Meyerowitz, E.M. Cytokinin Signaling as a Positional Cue for Patterning the Apical-Basal Axis of the Growing Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2012, 109, 4002–4007. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of Plant Stem Cell Function by Conserved Interacting Transcriptional Regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef]
- Gruel, J.; Landrein, B.; Tarr, P.; Schuster, C.; Refahi, Y.; Sampathkumar, A.; Hamant, O.; Meyerowitz, E.M.; Jönsson, H. An Epidermis-Driven Mechanism Positions and Scales Stem Cell Niches in Plants. Sci. Adv. 2016, 2, e1500989. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. Hairy Meristem with Wuschel Confines Clavata3 Expression to the Outer Apical Meristem Layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef]
- Shi, B.; Guo, X.; Wang, Y.; Xiong, Y.; Wang, J.; Hayashi, K.; Lei, J.; Zhang, L.; Jiao, Y. Feedback from Lateral Organs Controls Shoot Apical Meristem Growth by Modulating Auxin Transport. Dev. Cell 2018, 44, 204–216.e6. [Google Scholar] [CrossRef]
- Takanashi, H.; Sumiyoshi, H.; Mogi, M.; Hayashi, Y.; Ohnishi, T.; Tsutsumi, N. MiRNAs Control HAM1 Functions at the Single-Cell-Layer Level and Are Essential for Normal Embryogenesis in Arabidopsis. Plant Mol. Biol. 2018, 96, 627–640. [Google Scholar] [CrossRef]
- Ma, Y.; Miotk, A.; Šutiković, Z.; Ermakova, O.; Wenzl, C.; Medzihradszky, A.; Gaillochet, C.; Forner, J.; Utan, G.; Brackmann, K.; et al. WUSCHEL Acts as an Auxin Response Rheostat to Maintain Apical Stem Cells in Arabidopsis. Nat. Commun. 2019, 10, 5093. [Google Scholar] [CrossRef]
- Galvan-Ampudia, C.S.; Cerutti, G.; Legrand, J.; Brunoud, G.; Martin-Arevalillo, R.; Azais, R.; Bayle, V.; Moussu, S.; Wenzl, C.; Jaillais, Y.; et al. Temporal Integration of Auxin Information for the Regulation of Patterning. eLife 2020, 9, e55832. [Google Scholar] [CrossRef]
- Yoshida, S.; Mandel, T.; Kuhlemeier, C. Stem Cell Activation by Light Guides Plant Organogenesis. Genes Dev. 2011, 25, 1439–1450. [Google Scholar] [CrossRef]
- Pfeiffer, A.; Janocha, D.; Dong, Y.; Medzihradszky, A.; Schöne, S.; Daum, G.; Suzaki, T.; Forner, J.; Langenecker, T.; Rempel, E.; et al. Integration of Light and Metabolic Signals for Stem Cell Activation at the Shoot Apical Meristem. eLife 2016, 5, e17023. [Google Scholar] [CrossRef] [PubMed]
- Landrein, B.; Formosa-Jordan, P.; Malivert, A.; Schuster, C.; Melnyk, C.W.; Yang, W.; Turnbull, C.; Meyerowitz, E.M.; Locke, J.C.W.; Jönsson, H. Nitrate Modulates Stem Cell Dynamics in Arabidopsis Shoot Meristems through Cytokinins. Proc. Natl. Acad. Sci. USA 2018, 115, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Barton, M.K. Initiation of Axillary and Floral Meristems in Arabidopsis. Dev. Biol. 2000, 218, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Kohlen, W.; Rossmann, S.; Vernoux, T.; Theres, K. Auxin Depletion from the Leaf Axil Conditions Competence for Axillary Meristem Formation in Arabidopsis and Tomato. Plant Cell 2014, 26, 2068–2079. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An Organ Boundary-enriched Gene Regulatory Network Uncovers Regulatory Hierarchies Underlying Axillary Meristem Initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef]
- Xin, W.; Wang, Z.; Liang, Y.; Wang, Y.; Hu, Y. Dynamic Expression Reveals a Two-Step Patterning of WUS and CLV3 during Axillary Shoot Meristem Formation in Arabidopsis. J. Plant Physiol. 2017, 214, 1–6. [Google Scholar] [CrossRef]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of MiR164, CUP-SHAPED COTYLEDON Genes and LATERAL SUPPRESSOR Controls Axillary Meristem Formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 Regulates Postembryonic Shoot Meristem and Organ Boundary Formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Shi, B.; Yu, T.; Qi, J.; Meyerowitz, E.M.; Jiao, Y. The Stem Cell Niche in Leaf Axils Is Established by Auxin and Cytokinin in Arabidopsis. Plant Cell 2014, 26, 2055–2067. [Google Scholar] [CrossRef]
- Zhang, C.; Fan, L.; Le, B.H.; Ye, P.; Mo, B.; Chen, X. Regulation of ARGONAUTE10 Expression Enables Temporal and Spatial Precision in Axillary Meristem Initiation in Arabidopsis. Dev. Cell 2020, 55, 603–616.e5. [Google Scholar] [CrossRef]
- Wang, J.; Tian, C.; Zhang, C.; Shi, B.; Cao, X.; Zhang, T.Q.; Zhao, Z.; Wang, J.W.; Jiao, Y. Cytokinin Signaling Activates WUSCHEL Expression during Axillary Meristem Initiation. Plant Cell 2017, 29, 1373–1387. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Maugarny-Calès, A.; Adroher, B.; Chelysheva, L.; Li, Y.; Burguet, J.; Bågman, A.-M.; Smit, M.E.; Brady, S.M.; Li, Y.; et al. De Novo Stem Cell Establishment in Meristems Requires Repression of Organ Boundary Cell Fate. Plant Cell 2022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tucker, E.; Hermann, M.; Laux, T. A Molecular Framework for the Embryonic Initiation of Shoot Meristem Stem Cells. Dev. Cell 2017, 40, 264–277.e4. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Tasaka, M. Shoot Apical Meristem and Cotyledon Formation during Arabidopsis Embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS Genes. Development 1999, 126, 1563–1570. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription Factors SOD7/NGAL2 and DPA4/NGAL3 Act Redundantly to Regulate Seed Size by Directly Repressing KLU Expression in Arabidopsis thaliana. Plant Cell 2015, 1, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, N.; Zhang, Q.; Zhou, G.; Tian, H.; Hussain, S.; Ahmed, S.; Wang, T.; Wang, S. Genome Editing to Integrate Seed Size and Abiotic Stress Tolerance Traits in Arabidopsis Reveals a Role for DPA4 and SOD7 in the Regulation of Inflorescence Architecture. Int. J. Mol. Sci. 2019, 20, 2695. [Google Scholar] [CrossRef] [PubMed]
- Romanel, E.A.C.; Schrago, C.G.; Counago, R.M.; Russo, C.A.M.; Alves-Ferreira, M.; Couñago, R.M.; Russo, C.A.M.; Alves-Ferreira, M. Evolution of the B3 DNA Binding Superfamily: New Insights into REM Family Gene Diversification. PLoS ONE 2009, 4, e5791. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, K.; Peterson, K.; Jack, T. The Plant B3 Superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Meng, J.; Wang, F.; Shou, B.; Chen, Y.; Xue, H.; Zhao, J.; Qi, Y.; An, L.; Yu, F.; et al. NGATHA-likes Control Leaf Margin Development by Repressing Cup-Shaped Cotyledon2 Transcription1. Plant Physiol. 2020, 184, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Belmonte, M.F.; Kirkbride, R.C.; Stone, S.L.; Pelletier, J.M.; Bui, A.Q.; Yeung, E.C.; Hashimoto, M.; Fei, J.; Harada, C.M.; Munoz, M.D.; et al. Comprehensive Developmental Profiles of Gene Activity in Regions and Subregions of the Arabidopsis Seed. Proc. Natl. Acad. Sci. USA 2013, 110, E435–E444. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Laufs, P. Meristem Initiation and de Novo Stem Cell Formation. Front. Plant Sci. 2022, 13, 891228. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Team, R.C. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.r-project.org/ (accessed on 15 August 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Zeileis, A.; Grothendieck, G. Zoo: S3 Infrastructure for Regular and Irregular Time Series. J. Stat. Softw. 2005, 14, 1–27. [Google Scholar] [CrossRef]

| Genotype | Stage | |||||||
|---|---|---|---|---|---|---|---|---|
| Globular | Transition | Heart | Torpedo | |||||
| WT | 0% | (0/29) | 62% | (26/42) | 89% | (59/66) | 100% | (48/48) |
| dpa4-2 sod7-2 | 0% | (0/29) | 74% | (26/35) | 94% | (88/94) | 100% | (54/54) |
| p-value | - | 0.3297 | 0.386 | - | ||||
| Genotype | Embryo Total Length (µm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ]0–50] | ]50–75] | ]75–100] | ]100–150] | ]150–200] | ]200–250] | |||||||
| WT | 9% | (3/32) | 68% | (38/56) | 88% | (29/33) | 97% | (34/35) | 100% | (18/18) | 100% | (11/11) |
| dpa4-2 sod7-2 | 23% | (5/22) | 63% | (43/68) | 98% | (41/42) | 98% | (57/58) | 100% | (5/5) | 100% | (2/2) |
| p-value | 0.2479 | 0.705 | 0.1626 | 1 | - | - | ||||||
| Gene | Stage | |||||||
|---|---|---|---|---|---|---|---|---|
| Globular | Transition | Heart | Torpedo | |||||
| DPA4 | 43% | (10/23) | 100% | (22/22) | 100% | (14/14) | 100% | (9/9) |
| SOD7 | 0% | (0/9) | 31% | (5/16) | 43% | (6/14) | 100% | (19/19) |
| Gene | Embryo Total Length (µm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ]0–50] | ]50–75] | ]75–100] | ]100–150] | ]150–200] | ||||||
| DPA4 | 63% | (22/35) | 100% | (21/21) | 100% | (9/9) | 100% | (2/2) | 100% | (1/1) |
| SOD7 | 18% | (2/11) | 30% | (8/27) | 100% | (9/9) | 100% | (4/4) | 100% | (1/1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicolas, A.; Laufs, P. The NGATHA-like Genes DPA4 and SOD7 Are Not Required for Stem Cell Specification during Embryo Development in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 12007. https://doi.org/10.3390/ijms231912007
Nicolas A, Laufs P. The NGATHA-like Genes DPA4 and SOD7 Are Not Required for Stem Cell Specification during Embryo Development in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(19):12007. https://doi.org/10.3390/ijms231912007
Chicago/Turabian StyleNicolas, Antoine, and Patrick Laufs. 2022. "The NGATHA-like Genes DPA4 and SOD7 Are Not Required for Stem Cell Specification during Embryo Development in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 19: 12007. https://doi.org/10.3390/ijms231912007
APA StyleNicolas, A., & Laufs, P. (2022). The NGATHA-like Genes DPA4 and SOD7 Are Not Required for Stem Cell Specification during Embryo Development in Arabidopsis thaliana. International Journal of Molecular Sciences, 23(19), 12007. https://doi.org/10.3390/ijms231912007






