MicroRNA in Extracellular Vesicles from Patients with Pulmonary Arterial Hypertension Alters Endothelial Angiogenic Response
Abstract
1. Introduction
2. Results
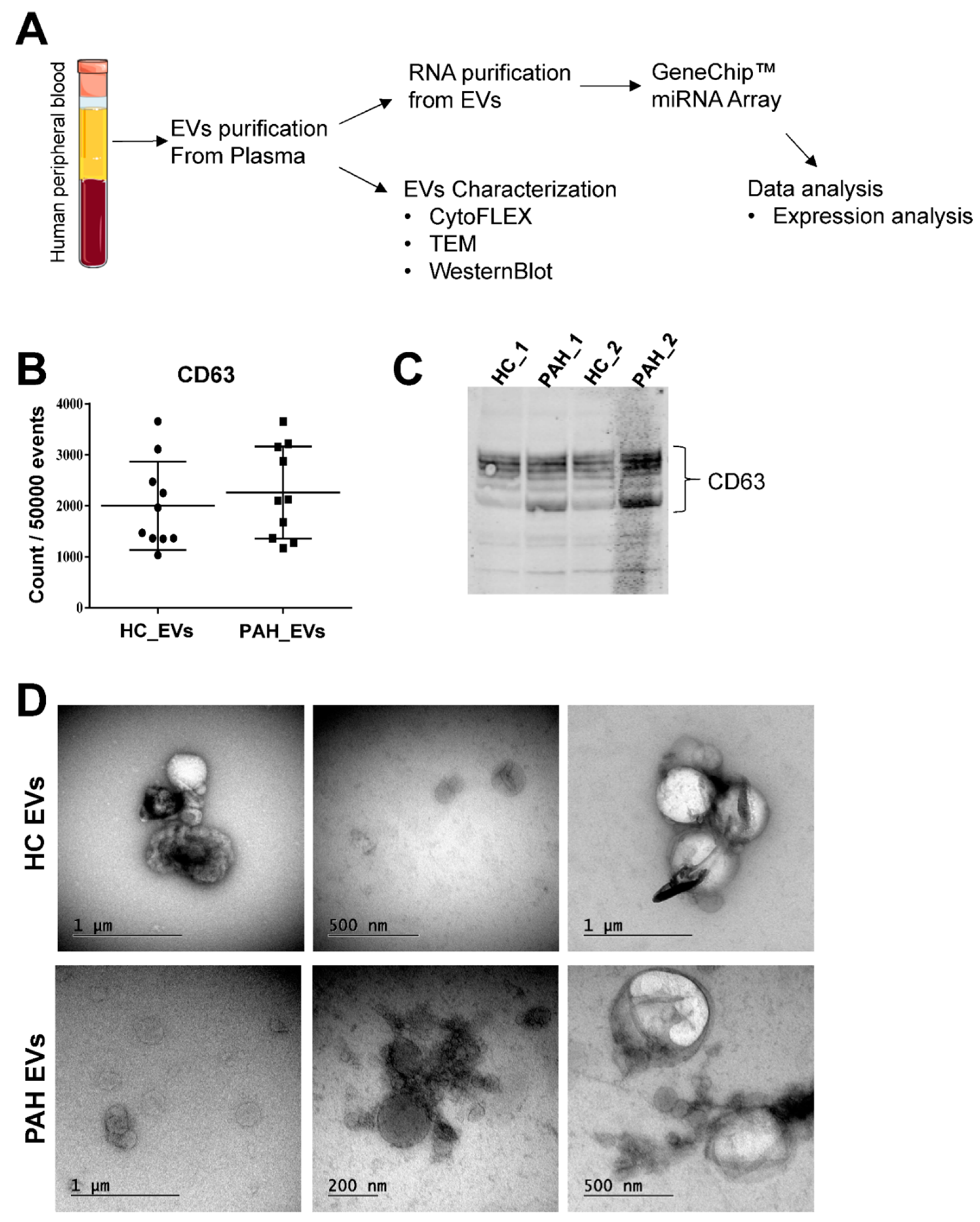
2.1. Characterization of Purified Extracellular Vesicles
2.2. MicroRNAs Identification in EVs from PAH Patients and HC
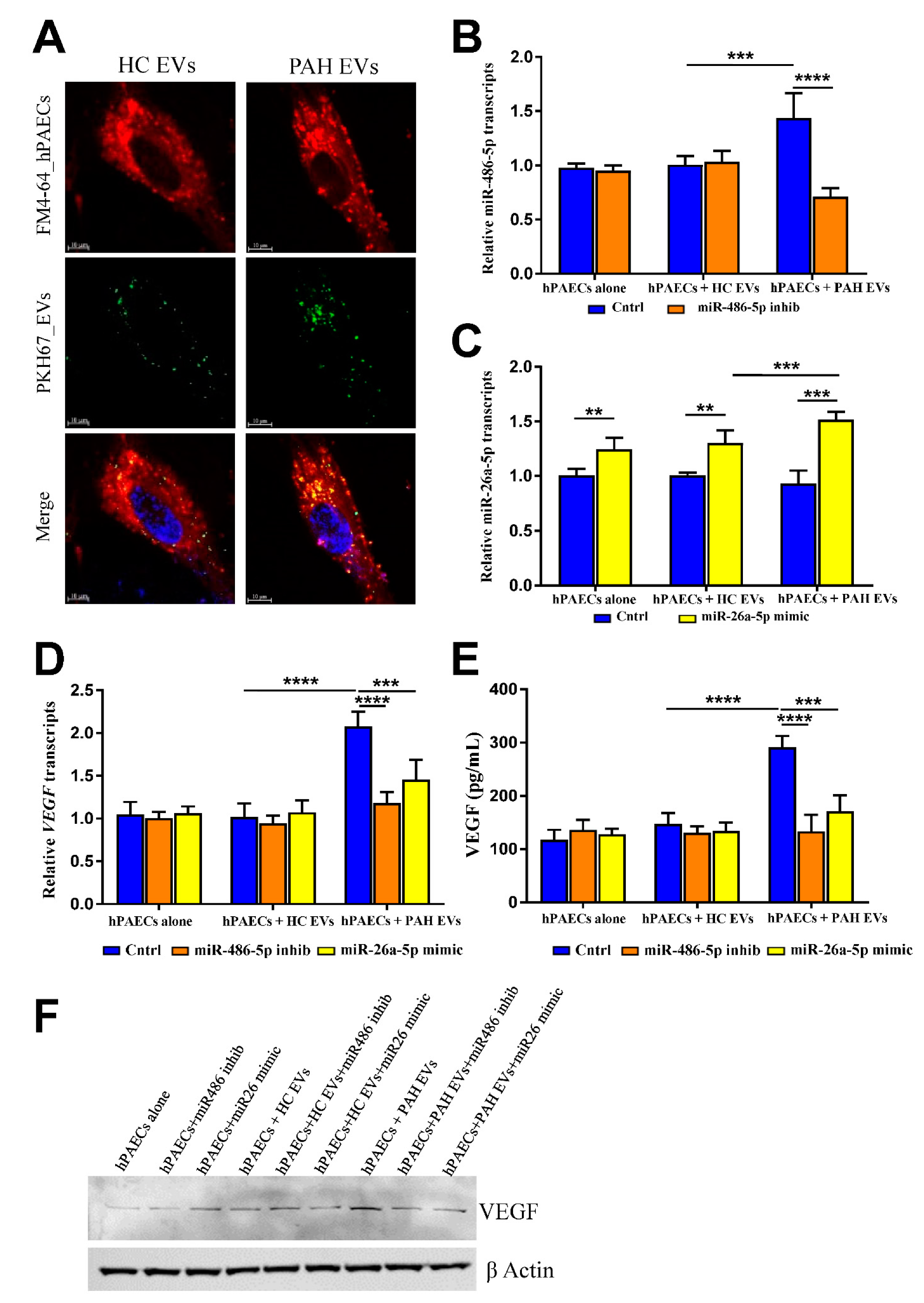
2.3. PAH EVs Derived miRs Effect on hPAECs
2.4. Effect of PAH EVs Derived miRs on hPAEC Tube Formation In Vitro
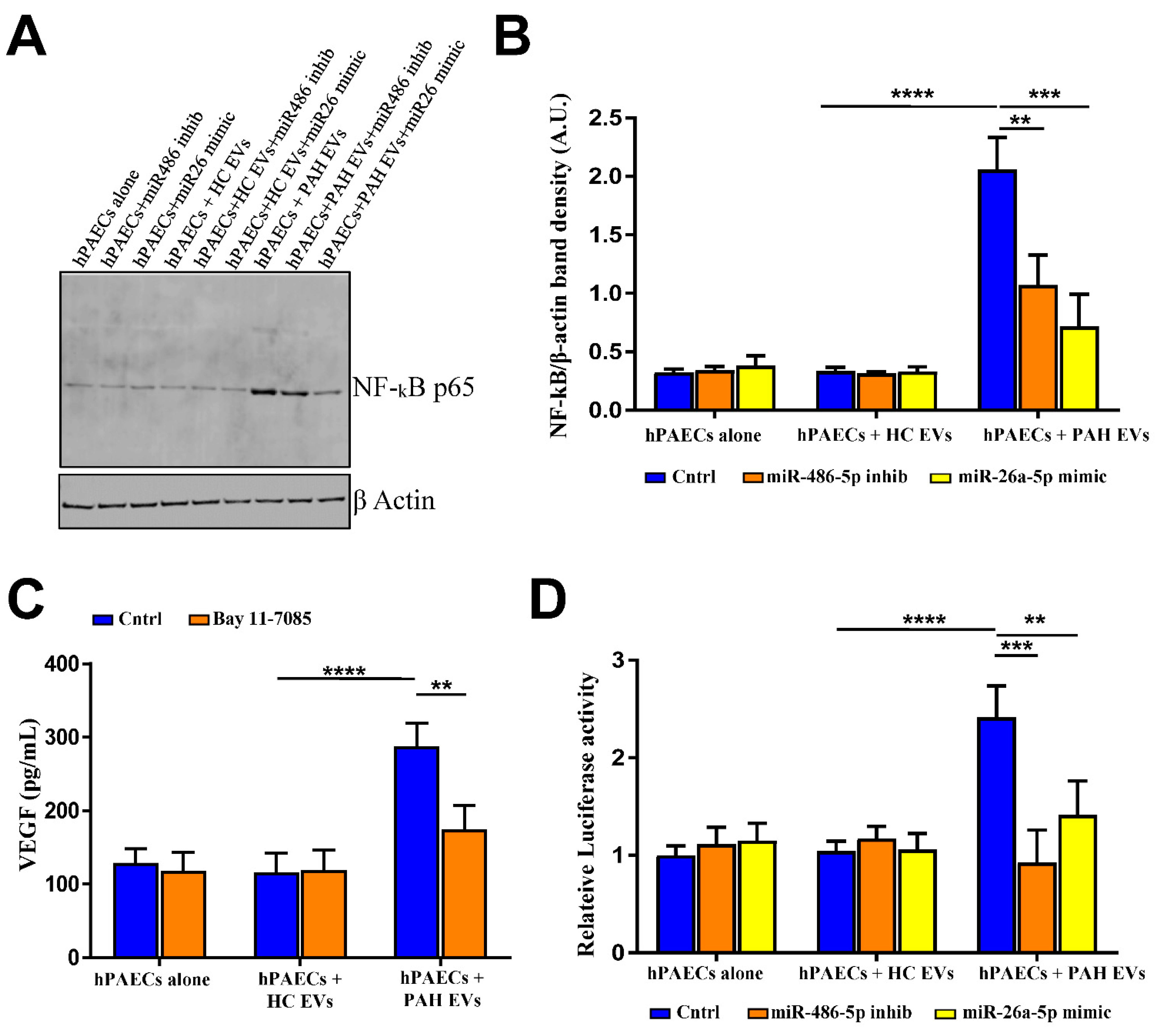
2.5. PAH EVs-Derived miRs Influenced NF-kB-Mediated Cellular Signaling
3. Discussion
4. Methods
4.1. Patient Cohort and Sample Collection
4.2. Blood Collection, Plasma Storage and Preparation of Extracellular Vesicles (EVs)
4.3. Flow Cytometry of Purified Extracellular Vesicles
4.4. Transmission Electron Microscopy (TEM) of Negatively Stained EVs
4.5. RNA Extraction from Extracellular Vesicles
4.6. MicroArray Analysis of microRNA from Purified Extracellular Vesicles
4.7. Microarray Data Analysis
4.8. Functional Enrichment of the miRNA Targets and miRNA
4.9. Cell Culture and miR Silencing or Overexpression
4.10. EV Labeling and Intracellular Visualization Using Immunofluorescence
4.11. RT-PCR Analysis of EV-Derived miRs and Angiogenesis Biomarkers within hPAECs
4.12. VEGF ELISA from Cell Culture Media
4.13. Tube Formation Assay
4.14. Cell Proliferation Assay
4.15. Western Blot of Analysis
4.16. Luciferase Reporter Assay
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schermuly, R.T.; Ghofrani, H.A.; Wilkins, M.R.; Grimminger, F. Mechanisms of Disease: Pulmonary Arterial Hypertension. Nat. Rev. Cardiol. 2011, 8, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, C.J.; Batai, K.; Bleda, M.; Haimel, M.; Southgate, L.; Germain, M.; Pauciulo, M.W.; Hadinnapola, C.; Aman, J.; Girerd, B.; et al. Genetic Determinants of Risk in Pulmonary Arterial Hypertension: International Genome-Wide Association Studies and Meta-Analysis. Lancet Respir. Med. 2019, 7, 227–238. [Google Scholar] [CrossRef]
- Thompson, A.A.R.; Lawrie, A. Targeting Vascular Remodeling to Treat Pulmonary Arterial Hypertension. Trends Mol. Med. 2017, 23, 31–45. [Google Scholar] [CrossRef]
- Thenappan, T.; Ormiston, M.L.; Ryan, J.J.; Archer, S.L. Pulmonary Arterial Hypertension: Pathogenesis and Clinical Management. BMJ 2018, 360, j5492. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, L.; Olsson, A.K.; Jogi, P.; Jonmaker, C. How Common Is Severe Pulmonary Hypertension after Pediatric Cardiac Surgery? J. Thorac. Cardiov. Sur. 2002, 123, 1155–1163. [Google Scholar] [CrossRef]
- Khandagale, A.; Aberg, M.; Wikstrom, G.; Lind, S.B.; Shevchenko, G.; Bjorklund, E.; Siegbahn, A.; Christersson, C. Role of Extracellular Vesicles in Pulmonary Arterial Hypertension Modulation of Pulmonary Endothelial Function and Angiogenesis. Arterioscl. Throm. Vas. 2020, 40, 2293–2309. [Google Scholar] [CrossRef]
- Amabile, N.; Heiss, C.; Real, W.M.; Minasi, P.; McGlothlin, D.; Rame, E.J.; Grossman, W.; De Marco, T.; Yeghiazarians, Y. Circulating Endothelial Microparticle Levels Predict Hemodynamic Severity of Pulmonary Hypertension. Am. J. Resp. Crit. Care Med. 2008, 177, 1268–1275. [Google Scholar] [CrossRef]
- Bakouboula, B.; Morel, O.; Faure, A.; Zobairi, F.; Jesel, L.; Trinh, A.; Zupan, M.; Canuet, M.; Grunebaum, L.; Brunette, A.; et al. Procoagulant Membrane Microparticles Correlate with the Severity of Pulmonary Arterial Hypertension. Am. J. Resp. Crit. Care Med. 2008, 177, 536–543. [Google Scholar] [CrossRef]
- Amabile, N.; Heiss, C.; Chang, V.; Angeli, F.S.; Damon, L.; Rame, E.J.; McGlothlin, D.; Grossman, W.; De Marco, T.; Yeghiazarians, Y. Increased CD62e(+) Endothelial Microparticle Levels Predict Poor Outcome in Pulmonary Hypertension Patients. J. Heart Lung Transpl. 2009, 28, 1081–1086. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-Mediated Transfer of Mrnas and Micrornas Is a Novel Mechanism of Genetic Exchange Between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Fu, S.; Zhang, Y.; Li, Y.; Luo, L.; Zhao, Y.; Yao, Y. Extracellular Vesicles in Cardiovascular Diseases. Cell Death Discov. 2020, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated Hnrnpa2b1 Controls the Sorting of Mirnas into Exosomes Through Binding to Specific Motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Ruffenach, G.; Mahajan, A.; Eghbali, M.; Umar, S. Novel Biomarkers for Pulmonary Arterial Hypertension. Resp. Res. 2016, 17, 88. [Google Scholar] [CrossRef] [PubMed]
- Rameh, V.; Kossaify, A. Role of Biomarkers in the Diagnosis, Risk Assessment, and Management of Pulmonary Hypertension. Biomark. Insights 2016, 11, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Chang, J.; Zhang, G. Recent Research Progress of MicroRNAs in Hypertension Pathogenesis, with a Focus on the Roles of miRNAs in Pulmonary Arterial Hypertension. Mol. Biol. Rep. 2018, 45, 2883–2896. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An Estimate of the Total Number of True Human MiRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Linneman, J.; Zhong, Y.; Yin, H.; Xia, Q.; Kang, K.; Gou, D. MicroRNAs in Pulmonary Hypertension, from Pathogenesis to Diagnosis and Treatment. Biomolecules 2022, 12, 496. [Google Scholar] [CrossRef]
- Li, Q.J.; Xu, Y.C.; Lv, K.Q.; Wang, Y.C.; Zhong, Z.W.; Xiao, C.C.; Zhu, K.Y.; Ni, C.; Wang, K.; Kong, M.J.; et al. Small Extracellular Vesicles Containing miR-486-5p Promote Angiogenesis after Myocardial Infarction in Mice and Nonhuman Primates. Sci. Transl. Med. 2021, 13, eabb0202. [Google Scholar] [CrossRef]
- Si, Y.; Zhang, H.; Ning, T.; Bai, M.; Wang, Y.; Yang, H.; Wang, X.; Li, J.; Ying, G.; Ba, Y. miR-26a/b Inhibit Tumor Growth and Angiogenesis by Targeting the HGF-VEGF Axis in Gastric Carcinoma. Cell Physiol. Biochem. 2017, 42, 1670–1683. [Google Scholar] [CrossRef]
- Fang, Y.; Dai, X. Emerging Roles of Extracellular Non-Coding RNAs in Vascular Diseases. J. Cardiovasc. Transl. Res. 2022, 15, 492–499. [Google Scholar] [CrossRef]
- Corssac, G.B.; Bonetto, J.P.; Campos-Carraro, C.; Cechinel, L.R.; Zimmer, A.; Parmeggiani, B.; Grings, M.; Carregal, V.M.; Massensini, A.R.; Siqueira, I.; et al. Pulmonary Arterial Hypertension Induces the Release of Circulating Extracellular Vesicles with Oxidative Content and Alters Redox and Mitochondrial Homeostasis in the Brains of Rats. Hypertens. Res. 2021, 44, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.F.; Chen, T.J.; Raj, J.U. MicroRNAs in Pulmonary Arterial Hypertension. Am. J. Resp. Cell Mol. 2015, 52, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Davis-Dusenbery, B.N.; Wu, C.; Hata, A. Micromanaging Vascular Smooth Muscle Cell Differentiation and Phenotypic Modulation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2370–2377. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, M. Molecular Pathogenesis of Pulmonary Arterial Hypertension. J. Clin. Investig. 2008, 118, 2372–2379. [Google Scholar] [CrossRef]
- Rhodes, C.J.; Wharton, J.; Boon, R.A.; Roexe, T.; Tsang, H.; Wojciak-Stothard, B.; Chakrabarti, A.; Howard, L.S.; Gibbs, J.S.R.; Lawrie, A.; et al. Reduced MicroRNA-150 Is Associated with Poor Survival in Pulmonary Arterial Hypertension. Am. J. Resp. Crit. Care. 2013, 187, 294–302. [Google Scholar] [CrossRef]
- Courboulin, A.; Paulin, R.; Giguere, N.J.; Saksouk, N.; Perreault, T.; Meloche, J.; Paquet, E.R.; Biardel, S.; Provencher, S.; Cote, J.; et al. Role for miR-204 in Human Pulmonary Arterial Hypertension. J. Exp. Med. 2011, 208, 535–548. [Google Scholar] [CrossRef]
- Pullamsetti, S.S.; Doebele, C.; Fischer, A.; Savai, R.; Kojonazarov, B.; Dahal, B.K.; Ghofrani, H.A.; Weissmann, N.; Grimminger, F.; Bonauer, A.; et al. Inhibition of MicroRNA-17 Improves Lung and Heart Function in Experimental Pulmonary Hypertension. Am. J. Resp. Crit. Care Med. 2012, 185, 409–419. [Google Scholar] [CrossRef]
- Caruso, P.; Dempsie, Y.; Stevens, H.C.; McDonald, R.A.; Long, L.; Lu, R.F.; White, K.; Mair, K.M.; McClure, J.D.; Southwood, M.; et al. A Role for miR-145 in Pulmonary Arterial Hypertension Evidence from Mouse Models and Patient Samples. Circ. Res. 2012, 111, 290–300. [Google Scholar] [CrossRef]
- Caruso, P.; MacLean, M.R.; Khanin, R.; McClure, J.; Soon, E.; Southgate, M.; MacDonald, R.A.; Greig, J.A.; Robertson, K.E.; Masson, R.; et al. Dynamic Changes in Lung MicroRNA Profiles During the Development of Pulmonary Hypertension due to Chronic Hypoxia and Monocrotaline. Arterioscl. Throm. Vas. 2010, 30, 716–723. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, Y.J.; Wu, B. MicroRNA-146-5p Promotes Pulmonary Artery Endothelial Cell Proliferation under Hypoxic Conditions through Regulating USP3. Dis. Markers 2021, 2021, 3668422. [Google Scholar] [CrossRef]
- Deng, L.; Blanco, F.J.; Stevens, H.; Lu, R.F.; Caudrillier, A.; McBride, M.; McClure, J.D.; Grant, J.; Thomas, M.; Frid, M.; et al. MicroRNA-143 Activation Regulates Smooth Muscle and Endothelial Cell Crosstalk in Pulmonary Arterial Hypertension. Circ. Res. 2015, 117, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhang, H.; Xiang, B.; Zhang, W.; Gong, F.; Li, S.; Chen, H.; Luo, X.; Deng, J.; You, Y.; et al. Hyperoxia-Induced miR-342-5p Down-Regulation Exacerbates Neonatal Bronchopulmonary Dysplasia Via the Raf1 Regulator Spred3. Br. J. Pharmacol. 2021, 178, 2266–2283. [Google Scholar] [CrossRef] [PubMed]
- Chai, Z.T.; Kong, J.; Zhu, X.D.; Zhang, Y.Y.; Lu, L.; Zhou, J.M.; Wang, L.R.; Zhang, K.Z.; Zhang, Q.B.; Ao, J.Y.; et al. MicroRNA-26a Inhibits Angiogenesis by Down-Regulating VEGFA through the PIK3C2 alpha/Akt/HIF-1 Alpha Pathway in Hepatocellular Carcinoma. PloS ONE 2013, 8, e77957. [Google Scholar] [CrossRef] [PubMed]
- Voelkel, N.F.; Gomez-Arroyo, J. The Role of Vascular Endothelial Growth Factor in Pulmonary Arterial Hypertension the Angiogenesis Paradox. Am. J. Resp. Cell Mol. 2014, 51, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wen, H.; Huang, J.; Liao, P.; Liao, H.; Tu, J.; Zeng, Y. Extracellular Vesicle-Enclosed miR-486-5p Mediates Wound Healing with Adipose-Derived Stem Cells by Promoting Angiogenesis. J. Cell Mol. Med. 2020, 24, 9590–9604. [Google Scholar] [CrossRef]
- Jo, H.N.; Kang, H.; Lee, A.; Choi, J.; Chang, W.; Lee, M.S.; Kim, J. Endothelial miR-26a Regulates VEGF-Nogo-B Receptor-Mediated Angiogenesis. BMB Rep. 2017, 50, 384–389. [Google Scholar] [CrossRef]
- Yan, X.C.; Cao, J.; Liang, L.; Wang, L.; Gao, F.; Yang, Z.Y.; Duan, J.L.; Chang, T.F.; Deng, S.M.; Liu, Y.; et al. miR-342-5p Is a Notch Downstream Molecule and Regulates Multiple Angiogenic Pathways Including Notch, Vascular Endothelial Growth Factor and Transforming Growth Factor Beta Signaling. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, Y.; Zhang, Q.; Liu, J.J.; Li, T.J.; Yang, J.R.; Zeng, C.X.; Zhuang, S.M. MicroRNA-26a/b and their Host Genes Cooperate to Inhibit the G1/S Transition by Activating the pRb Protein. Nucleic Acids Res. 2012, 40, 4615–4625. [Google Scholar] [CrossRef]
- Sun, X.H.; Wang, X.; Zhang, Y.; Hui, J. Exosomes of Bone-Marrow Stromal Cells Inhibit Cardiomyocyte Apoptosis under Ischemic and Hypoxic Conditions Via miR-486-5p Targeting the PTEN/PI3K/AKT Signaling Pathway. Thromb. Res. 2019, 177, 23–32. [Google Scholar] [CrossRef]
- Tian, F.; Wang, J.; Ouyang, T.L.; Lu, N.; Lu, J.F.; Shen, Y.T.; Bai, Y.F.; Xie, X.Y.; Ge, Q.Y. MiR-486-5p Serves as a Good Biomarker in Nonsmall Cell Lung Cancer and Suppresses Cell Growth with the Involvement of a Target PIK3R1. Front Genet. 2019, 10, 688. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wong, W.H. Model-Based Analysis of Oligonucleotide Arrays: Expression Index Computation and Outlier Detection. Proc. Natl. Acad. Sci. USA 2001, 98, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Su, N.; Liu, L.; Zhang, J.; Wang, H.; Zhang, W.; Gui, J.; Yu, K.; Li, J.; Le, T.D. miRBaseConverter: An R/Bioconductor Package for Converting and Retrieving miRNA Name, Accession, Sequence and Family Information in Different Versions of miRBase. BMC Bioinform. 2018, 19, 514. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust Hyperparameter Estimation Protects against Hypervariable Genes and Improves Power to Detect Differential Expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Zagganas, K.; Vergoulis, T.; Paraskevopoulou, M.D.; Vlachos, I.S.; Skiadopoulos, S.; Dalamagas, T. BUFET: Boosting the Unbiased miRNA Functional Enrichment Analysis Using Bitsets. BMC Bioinform. 2017, 18, 399. [Google Scholar] [CrossRef]
- Bleazard, T.; Lamb, J.A.; Griffiths-Jones, S. Bias in microRNA Functional Enrichment Analysis. Bioinformatics 2015, 31, 1592–1598. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2—An R Package for Gene List Functional Enrichment Analysis and Namespace Conversion Toolset g: Profiler. F1000Research 2020, 9. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Kogure, T.; Lin, W.L.; Yan, I.K.; Braconi, C.; Patel, T. Intercellular Nanovesicle-Mediated microRNA Transfer: A Mechanism of Environmental Modulation of Hepatocellular Cancer Cell Growth. Hepatology 2011, 54, 1237–1248. [Google Scholar] [CrossRef] [PubMed]


| Transcript | Type | Log Fold Change | Adj. P-Value |
|---|---|---|---|
| Down-regulated | |||
| hsa-miR-6089 | miRNA | −2.78175 | 0.003676 |
| hsa-miR-3665 | miRNA | −2.5509 | 0.006932 |
| hsa-miR-6090 | miRNA | −2.54716 | 0.003718 |
| hsa-miR-6087 | miRNA | −2.13755 | 0.01727 |
| hsa-miR-6125 | miRNA | −2.1033 | 0.005246 |
| hsa-miR-26a-5p | miRNA | −2.07892 | 0.009452 |
| hsa-miR-7704 | miRNA | −1.9402 | 0.029668 |
| hsa-miR-4466 | miRNA | −1.86555 | 0.009452 |
| hsa-miR-6088 | miRNA | −1.85115 | 0.01727 |
| hsa-miR-4516 | miRNA | −1.73597 | 0.031291 |
| hsa-miR-103a-3p | miRNA | −1.73382 | 0.006913 |
| hsa-miR-6869-5p | miRNA | −1.68796 | 0.031291 |
| hsa-miR-1915-3p | miRNA | −1.62478 | 0.011043 |
| hsa-miR-3960 | miRNA | −1.55252 | 0.00461 |
| hsa-miR-342-3p | miRNA | −1.1534 | 0.009452 |
| hsa-miR-17-5p | miRNA | −1.12446 | 0.017975 |
| hsa-miR-2861 | miRNA | −1.07266 | 0.049252 |
| hsa-miR-3178 | miRNA | −0.85985 | 0.029668 |
| hsa-miR-4687-3p | miRNA | −0.82483 | 0.049252 |
| hsa-miR-6724-5p | miRNA | −0.7889 | 0.033406 |
| hsa-miR-4787-5p | miRNA | −0.74063 | 0.031291 |
| hsa-miR-6793-3p | miRNA | −0.40702 | 0.048881 |
| Up-regulated | |||
| hsa-mir-320e | Stem-loop | 0.379859 | 0.009452 |
| hsa-mir-6722 | Stem-loop | 0.380027 | 0.018802 |
| hsa-miR-8075 | miRNA | 0.402266 | 0.01727 |
| hsa-miR-6732-5p | miRNA | 0.561575 | 0.000734 |
| hsa-miR-4793-3p | miRNA | 0.730484 | 0.029668 |
| hsa-miR-486-5p | miRNA | 1.82015 | 0.03513 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khandagale, A.; Corcoran, P.; Nikpour, M.; Isaksson, A.; Wikström, G.; Siegbahn, A.; Christersson, C. MicroRNA in Extracellular Vesicles from Patients with Pulmonary Arterial Hypertension Alters Endothelial Angiogenic Response. Int. J. Mol. Sci. 2022, 23, 11964. https://doi.org/10.3390/ijms231911964
Khandagale A, Corcoran P, Nikpour M, Isaksson A, Wikström G, Siegbahn A, Christersson C. MicroRNA in Extracellular Vesicles from Patients with Pulmonary Arterial Hypertension Alters Endothelial Angiogenic Response. International Journal of Molecular Sciences. 2022; 23(19):11964. https://doi.org/10.3390/ijms231911964
Chicago/Turabian StyleKhandagale, Avinash, Padraic Corcoran, Maryam Nikpour, Anders Isaksson, Gerhard Wikström, Agneta Siegbahn, and Christina Christersson. 2022. "MicroRNA in Extracellular Vesicles from Patients with Pulmonary Arterial Hypertension Alters Endothelial Angiogenic Response" International Journal of Molecular Sciences 23, no. 19: 11964. https://doi.org/10.3390/ijms231911964
APA StyleKhandagale, A., Corcoran, P., Nikpour, M., Isaksson, A., Wikström, G., Siegbahn, A., & Christersson, C. (2022). MicroRNA in Extracellular Vesicles from Patients with Pulmonary Arterial Hypertension Alters Endothelial Angiogenic Response. International Journal of Molecular Sciences, 23(19), 11964. https://doi.org/10.3390/ijms231911964






