NCAPG Promotes Pulmonary Artery Smooth Muscle Cell Proliferation as a Promising Therapeutic Target of Idiopathic Pulmonary Hypertension: Bioinformatics Analysis and Experiment Verification
Abstract
1. Introduction
2. Results
2.1. Identification of DEGs
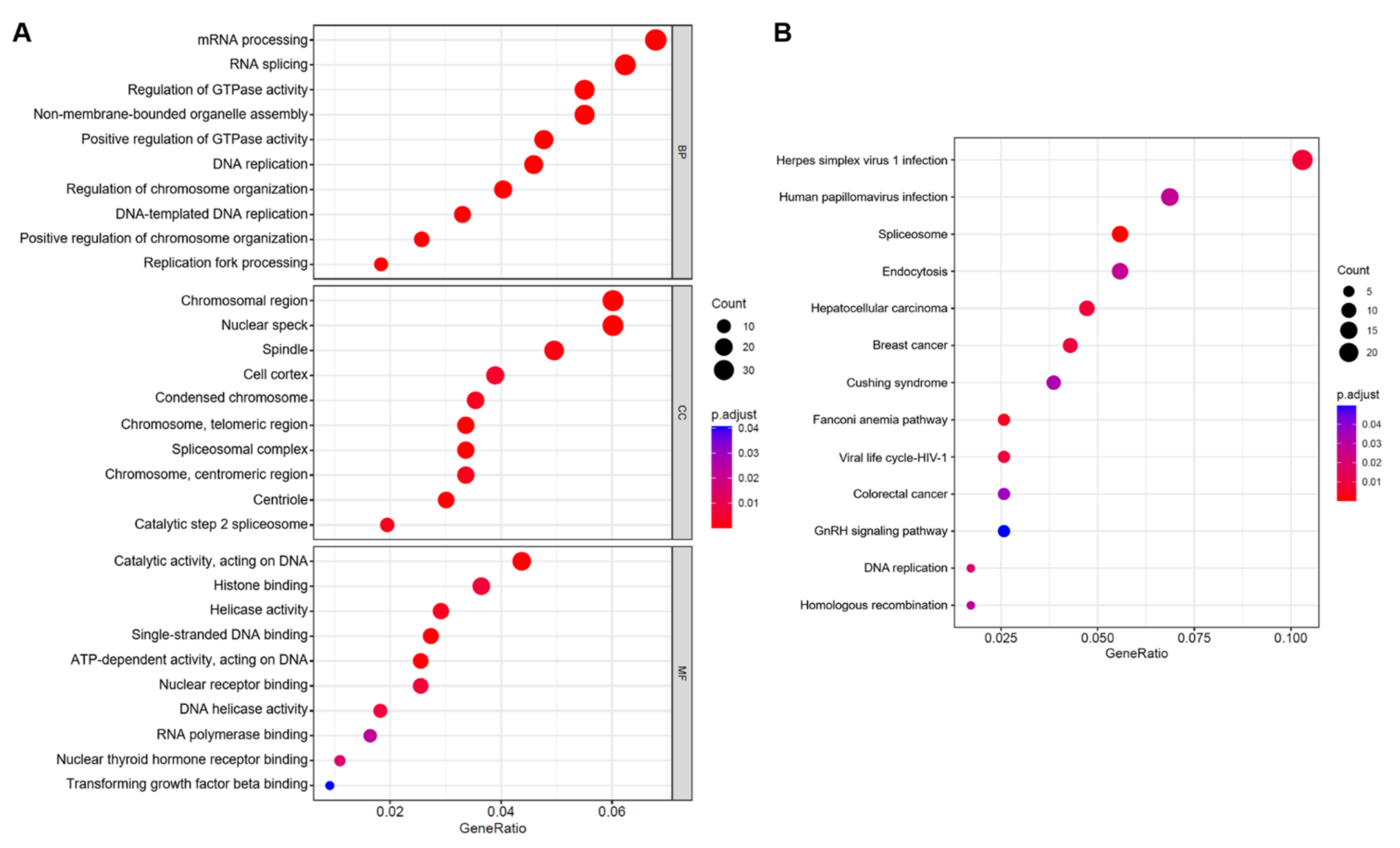
2.2. Enrichment Analysis of DEGs
2.3. PPI Network Establishment and Central Genes Identification
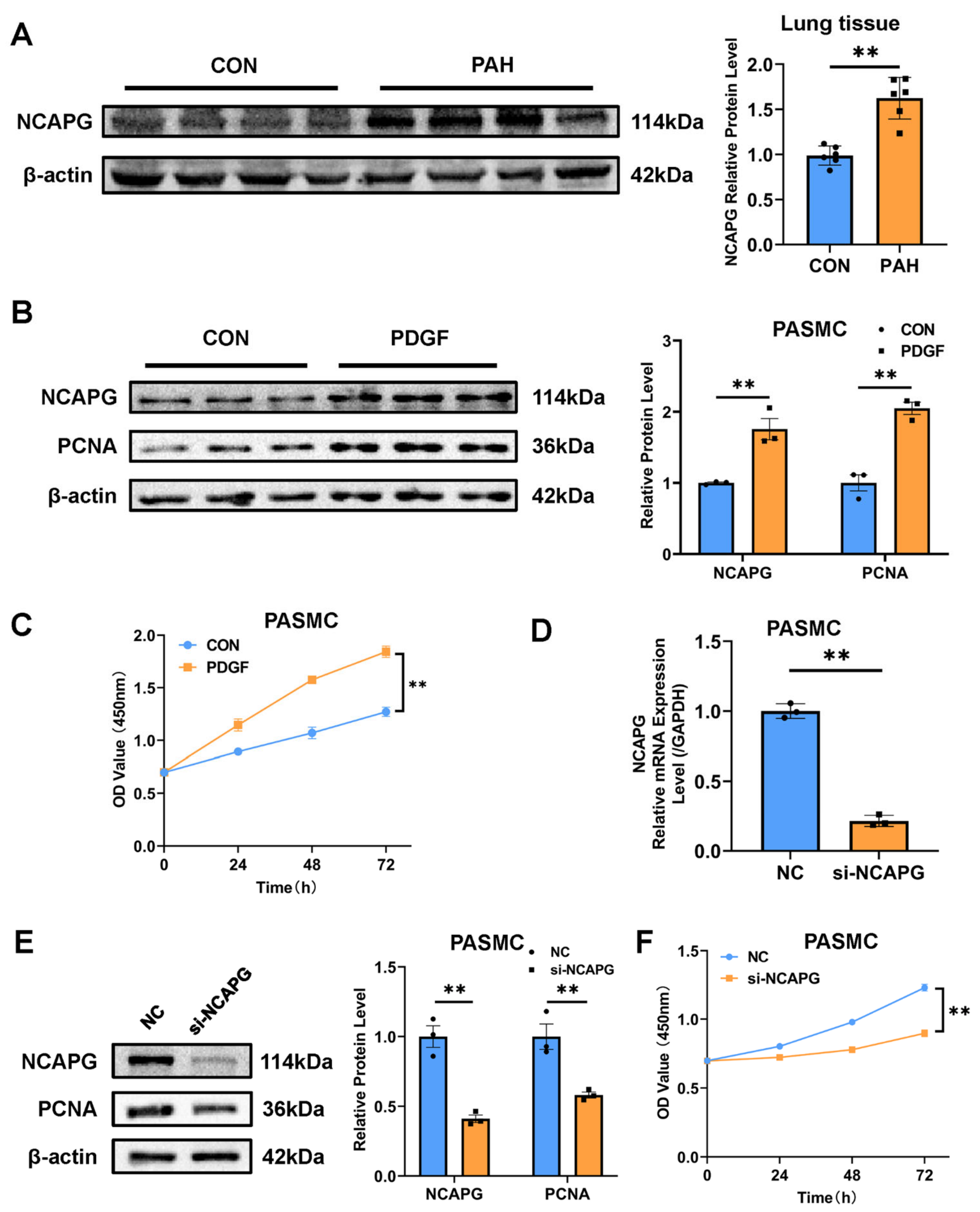
2.4. Validation of Selected Candidate Genes in Animal Models of PAH
2.5. NACPG Is Correlated with Pulmonary Artery Smooth Muscle Cell Proliferation
3. Discussion
4. Materials and Methods
4.1. Data Collection and DEGs Identification
4.2. GO Enrichment Analysis and KEGG Pathway Analysis
4.3. PPI Network Analysis
4.4. PAH Animal Model
4.5. Cell Culture Experiments
4.6. Western Blot Assay
4.7. RNA Extraction and qRT-PCR
4.8. CCK8 Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGoon, M.D.; Benza, R.L.; Escribano-Subias, P.; Jiang, X.; Miller, D.; Peacock, A.J.; Pepke-Zaba, J.; Pulido, T.; Rich, S.; Rosenkranz, S.; et al. Pulmonary Arterial Hypertension: Epidemiology and Registries. J. Am. Coll. Cardiol. 2013, 62, D51–D59. [Google Scholar] [CrossRef] [PubMed]
- Galiè, N.; Humbert, M.; Vachiery, J.L.; Gibbs, S.; Lang, I.; Torbicki, A.; Simonneau, G.; Peacock, A.; Vonk Noordegraaf, A.; Beghetti, M.; et al. 2015 Esc/Ers Guidelines for the Diagnosis and Treatment of Pulmonary Hypertension: The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (Esc) and the European Respiratory Society (Ers): Endorsed By: Association for European Paediatric and Congenital Cardiology (Aepc), International Society for Heart and Lung Transplantation (Ishlt). Eur. Heart J. 2016, 37, 67–119. [Google Scholar] [PubMed]
- Lau, E.; Giannoulatou, E.; Celermajer, D.; Humbert, M. Epidemiology and Treatment of Pulmonary Arterial Hypertension. Nat. Rev. Cardiol. 2017, 14, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Humbert, M.; Guignabert, C.; Bonnet, S.; Dorfmüller, P.; Klinger, J.R.; Nicolls, M.R.; Olschewski, A.J.; Pullamsetti, S.S.; Schermuly, R.T.; Stenmark, K.R.; et al. Pathology and Pathobiology of Pulmonary Hypertension: State of the Art and Research Perspectives. Eur. Respir. J. 2019, 53, 1801887. [Google Scholar] [CrossRef] [PubMed]
- D’Alonzo, G.E.; Barst, R.J.; Ayres, S.M.; Bergofsky, E.H.; Brundage, B.H.; Detre, K.M.; Fishman, A.P.; Goldring, R.M.; Groves, B.M.; Kernis, J.T.; et al. Survival in Patients with Primary Pulmonary Hypertension. Results from a National Prospective Registry. Ann. Intern. Med. 1991, 115, 343–349. [Google Scholar] [CrossRef]
- Farber, H.W.; Miller, D.P.; Poms, A.D.; Badesch, D.B.; Frost, A.E.; Rouzic, E.M.-L.; Romero, A.J.; Benton, W.W.; Elliott, C.G.; McGoon, M.D.; et al. Five-Year Outcomes of Patients Enrolled in the Reveal Registry. Chest 2015, 148, 1043–1054. [Google Scholar] [CrossRef]
- Gu, L.; Li, Y.Y.; Gu, L.; Xie, L.; Liu, H.M. Idiopathic Pulmonary Arterial Hypertension and Pulmonary Arterial Hypertension Associated with Congenital Heart Disease in Chinese Children: Similarities, Differences, and Prognostic Factors. Front. Pediatr. 2020, 8, 106. [Google Scholar] [CrossRef]
- Thompson, A.R.; Lawrie, A. Targeting Vascular Remodeling to Treat Pulmonary Arterial Hypertension. Trends Mol. Med. 2017, 23, 31–45. [Google Scholar] [CrossRef]
- Zheng, K.; Yao, S.; Yao, W.; Li, Q.; Wang, Y.; Zhang, L.; Chen, X.; Xiong, H.; Yuan, X.; Wang, Y.; et al. Association between Rsk2 and Clinical Indexes of Primary Breast Cancer: A Meta-Analysis Based on Mrna Microarray Data. Front. Genet. 2021, 12, 770134. [Google Scholar] [CrossRef]
- Ahluwalia, P.; Kolhe, R.; Gahlay, G.K. The Clinical Relevance of Gene Expression Based Prognostic Signatures in Colorectal Cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188513. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, Z.; Li, C.; Wang, R.; Fang, J.; Lu, S.; Guo, J.; Zhu, X.; Wang, X. Integrative Analyses of Gene Expression Profile Reveal Potential Crucial Roles of Mitotic Cell Cycle and Microtubule Cytoskeleton in Pulmonary Artery Hypertension. BMC Med. Genom. 2020, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zheng, X.; Wang, J. Bioinformatic Exploration of the Immune Related Molecular Mechanism Underlying Pulmonary Arterial Hypertension. Bioengineered 2021, 12, 3137–3147. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Xu, L.; Dai, Y.; Yuan, Q.; Zhou, Z. Ecm2 and Glt8d2 in Human Pulmonary Artery Hypertension: Fruits from Weighted Gene Co-Expression Network Analysis. J. Thorac. Dis. 2021, 13, 2242–2254. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Tang, Y.; Zhang, M.; Liang, B.; Wang, M.; Zha, L.; Yu, Z. Integrated Bioinformatic Analysis Reveals Txnrd1 as a Novel Biomarker and Potential Therapeutic Target in Idiopathic Pulmonary Arterial Hypertension. Front. Med. 2022, 9, 894584. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, L.; Ge, H.; Shen, W.; Shan, L. Identification of Autophagy-Related Biomarkers in Patients with Pulmonary Arterial Hypertension Based on Bioinformatics Analysis. Open Med. 2022, 17, 1148–1157. [Google Scholar] [CrossRef]
- Wei, R.-Q.; Zhang, W.-M.; Liang, Z.; Piao, C.; Zhu, G. Identification of Signal Pathways and Hub Genes of Pulmonary Arterial Hypertension by Bioinformatic Analysis. Can. Respir. J. 2022, 2022, 1394088. [Google Scholar] [CrossRef]
- Gräf, S.; Haimel, M.; Bleda, M.; Hadinnapola, C.; Southgate, L.; Li, W.; Hodgson, J.; Liu, B.; Salmon, R.M.; Southwood, M.; et al. Identification of Rare Sequence Variation Underlying Heritable Pulmonary Arterial Hypertension. Nat. Commun. 2018, 9, 1416. [Google Scholar] [CrossRef]
- Kariotis, S.; Jammeh, E.; Swietlik, E.M.; Pickworth, J.A.; Rhodes, C.J.; Otero, P.; Wharton, J.; Iremonger, J.; Dunning, M.J.; Pandya, D.; et al. Biological Heterogeneity in Idiopathic Pulmonary Arterial Hypertension Identified through Unsupervised Transcriptomic Profiling of Whole Blood. Nat. Commun. 2021, 12, 7104. [Google Scholar] [CrossRef]
- Wu, X.; Miao, J.; Jiang, J.; Liu, F. Analysis of Methylation Profiling Data of Hyperplasia and Primary and Metastatic Endometrial Cancers. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 217, 161–166. [Google Scholar] [CrossRef]
- Mallik, M.K. An Attempt to Understand Glioma Stem Cell Biology through Centrality Analysis of a Protein Interaction Network. J. Theor. Biol. 2018, 438, 78–91. [Google Scholar] [CrossRef]
- Owa, M.; Dynlacht, B. A Non-Canonical Function for Centromere-Associated Protein-E Controls Centrosome Integrity and Orientation of Cell Division. Commun. Biol. 2021, 4, 358. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; Maiato, H. The Tubulin Code: A Navigation System for Chromosomes During Mitosis. Trends Cell Biol. 2016, 26, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Xie, M.; Liu, X.; He, Y. Cenpe Contributes to Pulmonary Vascular Remodeling in Pulmonary Hypertension. Biochem. Biophys. Res. Commun. 2021, 557, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Grue, P.; Grässer, A.; Sehested, M.; Jensen, P.B.; Uhse, A.; Straub, T.; Ness, W.; Boege, F. Essential Mitotic Functions of DNA Topoisomerase Iialpha Are Not Adopted by Topoisomerase Iibeta in Human H69 Cells. J. Biol. Chem. 1998, 273, 33660–33666. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Berger, J.M. Cell Cycle-Dependent Control and Roles of DNA Topoisomerase Ii. Genes 2019, 10, 859. [Google Scholar] [CrossRef]
- Kolberg, M.; Høland, M.; Lind, G.E.; Ågesen, T.H.; Skotheim, R.I.; Hall, K.S.; Mandahl, N.; Smeland, S.; Mertens, F.; Davidson, B.; et al. Protein Expression of Birc5, Tk1, and Top2a in Malignant Peripheral Nerve Sheath Tumours—A Prognostic Test after Surgical Resection. Mol. Oncol. 2015, 9, 1129–1139. [Google Scholar] [CrossRef]
- Pei, Y.-F.; Yin, X.-M.; Liu, X.-Q. Top2a Induces Malignant Character of Pancreatic Cancer through Activating Β-Catenin Signaling Pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 197–207. [Google Scholar] [CrossRef]
- Brase, J.C.; Schmidt, M.; Fischbach, T.; Sültmann, H.; Bojar, H.; Koelbl, H.; Hellwig, B.; Rahnenführer, J.; Hengstler, J.G.; Gehrmann, M.C. Erbb2 and Top2a in Breast Cancer: A Comprehensive Analysis of Gene Amplification, Rna Levels, and Protein Expression and Their Influence on Prognosis and Prediction. Clin. Cancer Res. 2010, 16, 2391–2401. [Google Scholar] [CrossRef]
- Kou, F.; Sun, H.; Wu, L.; Li, B.; Zhang, B.; Wang, X.; Yang, L. Top2a Promotes Lung Adenocarcinoma Cells’ Malignant Progression and Predicts Poor Prognosis in Lung Adenocarcinoma. J. Cancer 2020, 11, 2496–2508. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, S.S.; Feng, Y.Y.; Wang, H.L. Identification of Novel Biomarkers Involved in Pulmonary Arterial Hypertension Based on Multiple-Microarray Analysis. Biosci. Rep. 2020, 40, BSR20202346. [Google Scholar] [CrossRef]
- Bikeye, S.-N.N.; Colin, C.; Marie, Y.; Vampouille, R.; Ravassard, P.; Rousseau, A.; Boisselier, B.; Idbaih, A.; Calvo, C.F.; Leuraud, P.; et al. ASPM-Associated Stem Cell Proliferation Is Involved in Malignant Progression of Gliomas and Constitutes an Attractive Therapeutic Target. Cancer Cell Int. 2010, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Pan, H.W.; Liu, S.H.; Jeng, Y.M.; Hu, F.C.; Peng, S.Y.; Lai, P.L.; Hsu, H.C. Aspm Is a Novel Marker for Vascular Invasion, Early Recurrence, and Poor Prognosis of Hepatocellular Carcinoma. Clin. Cancer Res. 2008, 14, 4814–4820. [Google Scholar] [CrossRef] [PubMed]
- Brüning-Richardson, A.; Bond, J.; Alsiary, R.; Richardson, J.; Cairns, D.A.; McCormack, L.; Hutson, R.; Burns, P.; Wilkinson, N.; Hall, G.D.; et al. Aspm and Microcephalin Expression in Epithelial Ovarian Cancer Correlates with Tumour Grade and Survival. Br. J. Cancer 2011, 104, 1602–1610. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hsu, C.; Wang, T.; Li, C.-R.; Hou, Y.; Chu, J.; Lee, C.; Liu, M.; Su, J.J.; Jian, K.; et al. Tsai. A Gene Expression Signature of Epithelial Tubulogenesis and a Role for Aspm in Pancreatic Tumor Progression. Gastroenterology 2013, 145, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Liu, L.; Zhao, A.; Pfeifer, G.P.; Xu, X. The Abnormal Spindle-like, Microcephaly-Associated (Aspm) Gene Encodes a Centrosomal Protein. Cell Cycle 2005, 4, 1227–1229. [Google Scholar] [CrossRef]
- Eberlein, A.; Takasuga, A.; Setoguchi, K.; Pfuhl, R.; Flisikowski, K.; Fries, R.; Klopp, N.; Fürbass, R.; Weikard, R.; Kühn, C. Dissection of Genetic Factors Modulating Fetal Growth in Cattle Indicates a Substantial Role of the Non-Smc Condensin I Complex, Subunit G (Ncapg) Gene. Genetics 2009, 183, 951–964. [Google Scholar] [CrossRef]
- Song, B.; Du, J.; Song, D.-F.; Ren, J.-C.; Feng, Y. Dysregulation of Ncapg, Knl1, Mir-148a-3p, Mir-193b-3p, and Mir-1179 May Contribute to the Progression of Gastric Cancer. Biol. Res. 2018, 51, 44. [Google Scholar] [CrossRef]
- Wu, Y.; Lin, Y.; Pan, J.; Tu, X.; Xu, Y.; Li, H.; Chen, Y. Ncapg Promotes the Progression of Lung Adenocarcinoma via the Tgf-Β Signaling Pathway. Cancer Cell Int. 2021, 21, 443. [Google Scholar] [CrossRef]
- Yamamoto, S.; Takayama, K.I.; Obinata, D.; Fujiwara, K.; Ashikari, D.; Takahashi, S.; Inoue, S. Identification of New Octamer Transcription Factor 1-Target Genes Upregulated in Castration-Resistant Prostate Cancer. Cancer Sci. 2019, 110, 3476–3485. [Google Scholar] [CrossRef]
- Chen, J.; Qian, X.; He, Y.; Han, X.; Pan, Y. Novel Key Genes in Triple-Negative Breast Cancer Identified by Weighted Gene Co-Expression Network Analysis. J. Cell. Biochem. 2019, 120, 16900–16912. [Google Scholar] [CrossRef]
- Gong, C.; Ai, J.; Fan, Y.; Gao, J.; Liu, W.; Feng, Q.; Liao, W.; Wu, L. Ncapg Promotes the Proliferation of Hepatocellular Carcinoma through Pi3k/Akt Signaling. Onco Targets Ther. 2019, 12, 8537–8552. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.; Cecchini, M.J.; Joseph, M.; Granton, J.T. Osteopontin Lung Gene Expression Is a Marker of Disease Severity in Pulmonary Arterial Hypertension. Respirology 2019, 24, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Halliday, S.J.; Matthews, D.T.; Talati, M.H.; Austin, E.D.; Su, Y.R.; Absi, T.S.; Fortune, N.L.; Gailani, D.; Matafonov, A.; West, J.D.; et al. A Multifaceted Investigation into Molecular Associations of Chronic Thromboembolic Pulmonary Hypertension Pathogenesis. JRSM Cardiovasc. Dis. 2020, 9, 2048004020906994. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. Networkanalyst 3.0: A Visual Analytics Platform for Comprehensive Gene Expression Profiling and Meta-Analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. Clusterprofiler: An R Package for Comparing Biological Themes among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. String V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X. Cytonca: A Cytoscape Plugin for Centrality Analysis and Evaluation of Protein Interaction Networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, L.; Zhang, Y.; Wang, G.; Han, D.; Ke, R.; Li, S.; Feng, W.; Li, M. Activation of Ampk Inhibits Pulmonary Arterial Smooth Muscle Cells Proliferation. Exp. Lung Res. 2014, 40, 251–258. [Google Scholar] [CrossRef]



| References | Sample | GEO | Platform | IPAH | Control | Other (Not Using) |
|---|---|---|---|---|---|---|
| Mura, M. et al. | Lung tissue | GSE113439 | GPL6244 | 6 | 11 | 9 |
| Halliday, S. et al. | Lung tissue | GSE130391 | GPL570 | 4 | 4 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, B.; Li, Y.; Shi, X.; Liu, P.; Zhang, Y.; Tian, H. NCAPG Promotes Pulmonary Artery Smooth Muscle Cell Proliferation as a Promising Therapeutic Target of Idiopathic Pulmonary Hypertension: Bioinformatics Analysis and Experiment Verification. Int. J. Mol. Sci. 2022, 23, 11762. https://doi.org/10.3390/ijms231911762
Fu B, Li Y, Shi X, Liu P, Zhang Y, Tian H. NCAPG Promotes Pulmonary Artery Smooth Muscle Cell Proliferation as a Promising Therapeutic Target of Idiopathic Pulmonary Hypertension: Bioinformatics Analysis and Experiment Verification. International Journal of Molecular Sciences. 2022; 23(19):11762. https://doi.org/10.3390/ijms231911762
Chicago/Turabian StyleFu, Bowen, You Li, Xiaobo Shi, Peng Liu, Yiman Zhang, and Hongyan Tian. 2022. "NCAPG Promotes Pulmonary Artery Smooth Muscle Cell Proliferation as a Promising Therapeutic Target of Idiopathic Pulmonary Hypertension: Bioinformatics Analysis and Experiment Verification" International Journal of Molecular Sciences 23, no. 19: 11762. https://doi.org/10.3390/ijms231911762
APA StyleFu, B., Li, Y., Shi, X., Liu, P., Zhang, Y., & Tian, H. (2022). NCAPG Promotes Pulmonary Artery Smooth Muscle Cell Proliferation as a Promising Therapeutic Target of Idiopathic Pulmonary Hypertension: Bioinformatics Analysis and Experiment Verification. International Journal of Molecular Sciences, 23(19), 11762. https://doi.org/10.3390/ijms231911762




