A Comprehensive Study of Gradient Conditions for Deep Proteome Discovery in a Complex Protein Matrix
Abstract
1. Introduction
2. Results and Discussion
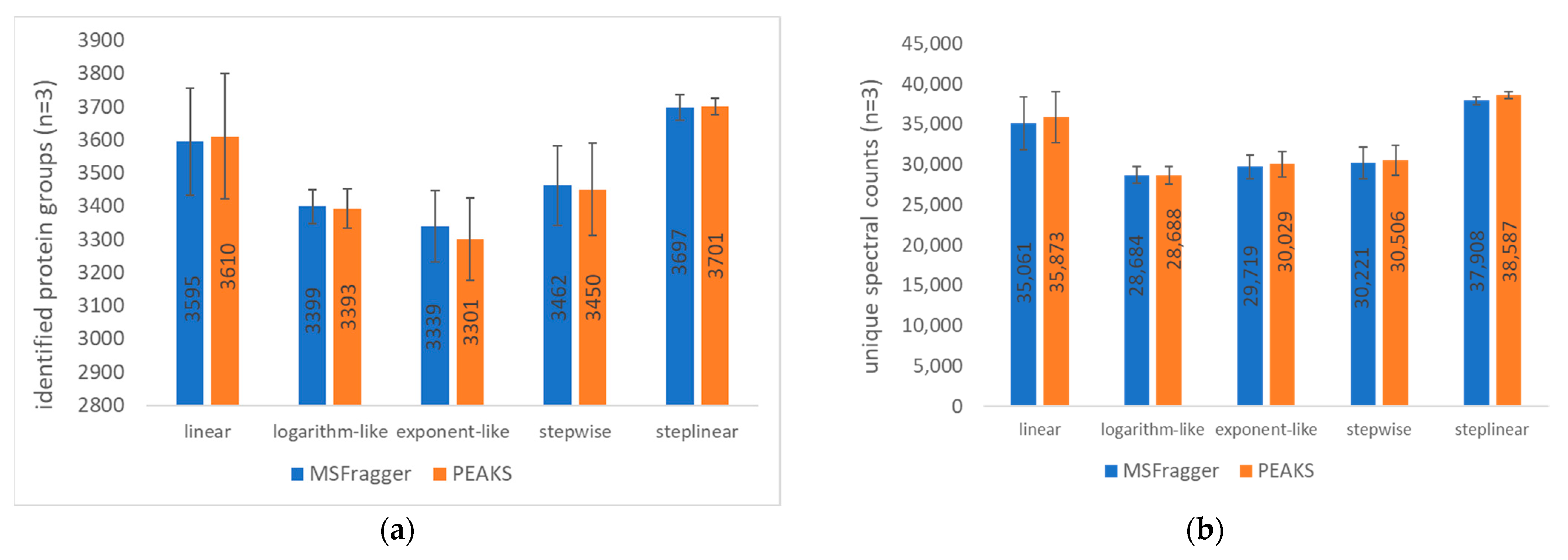
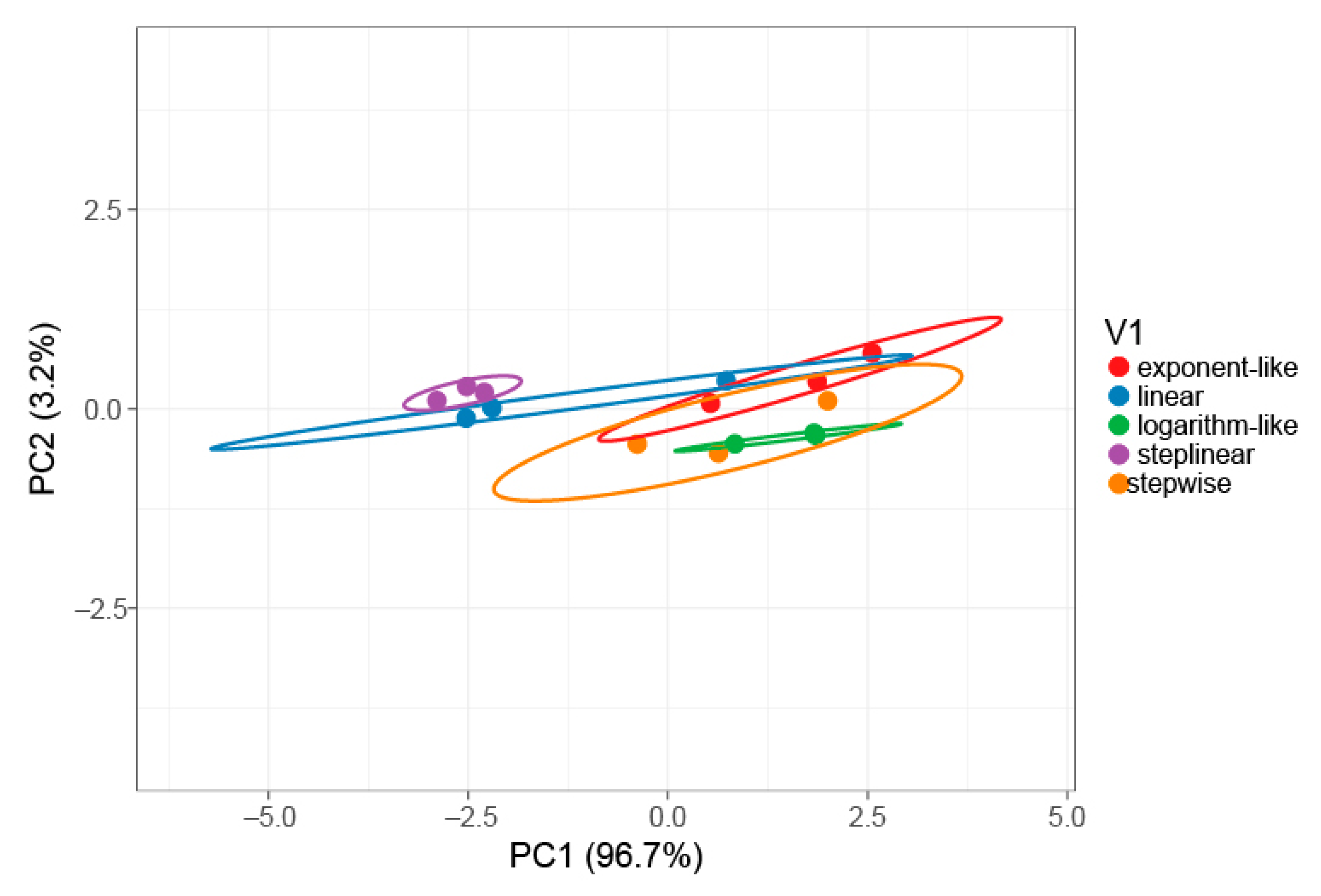
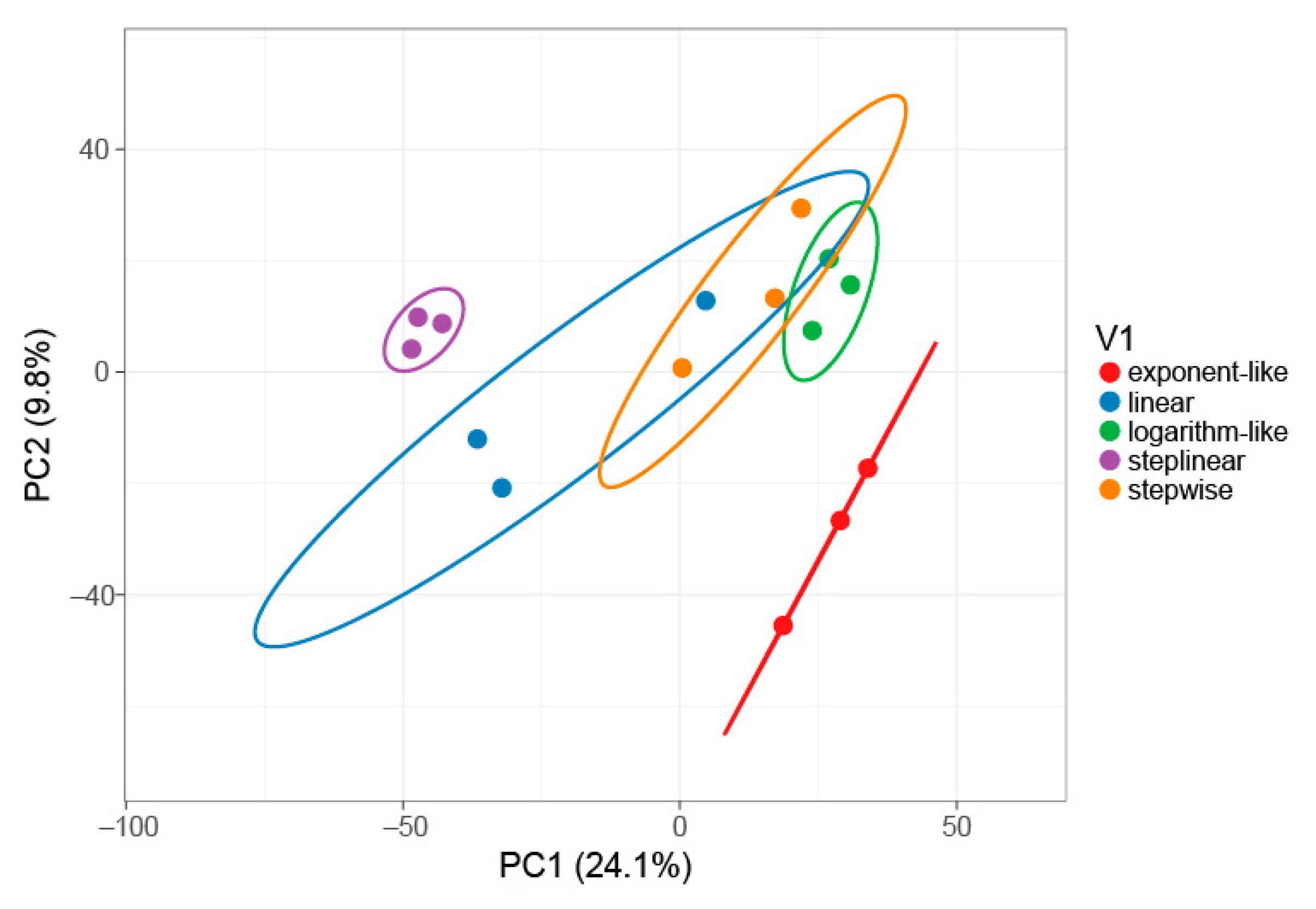
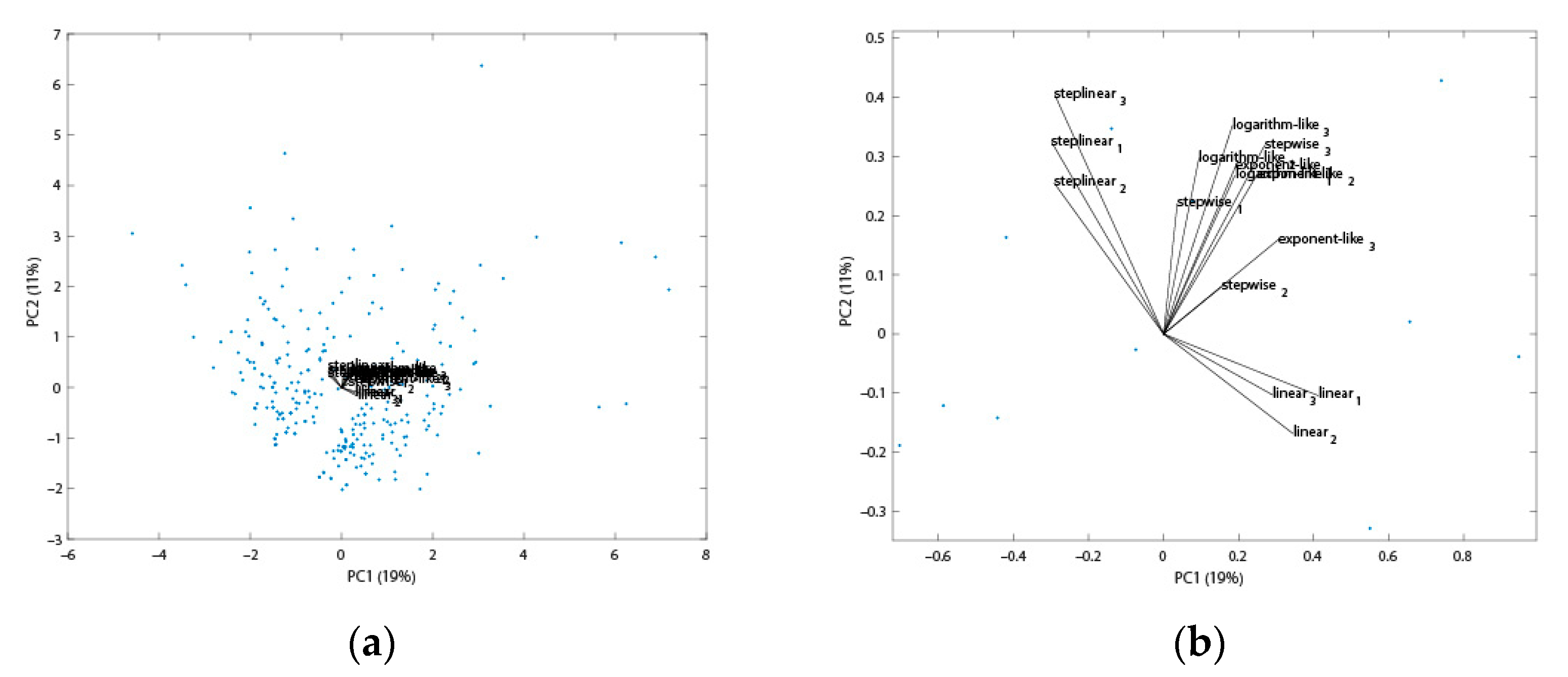
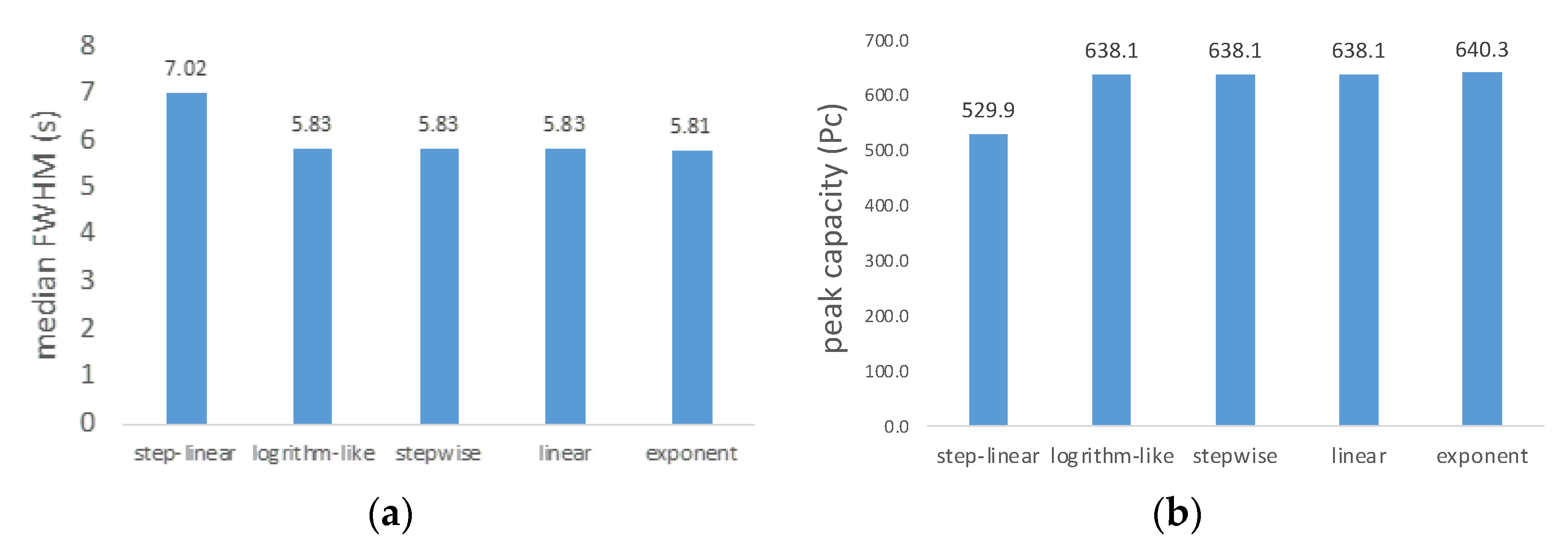
2.1. Gradient Types and Search Engines
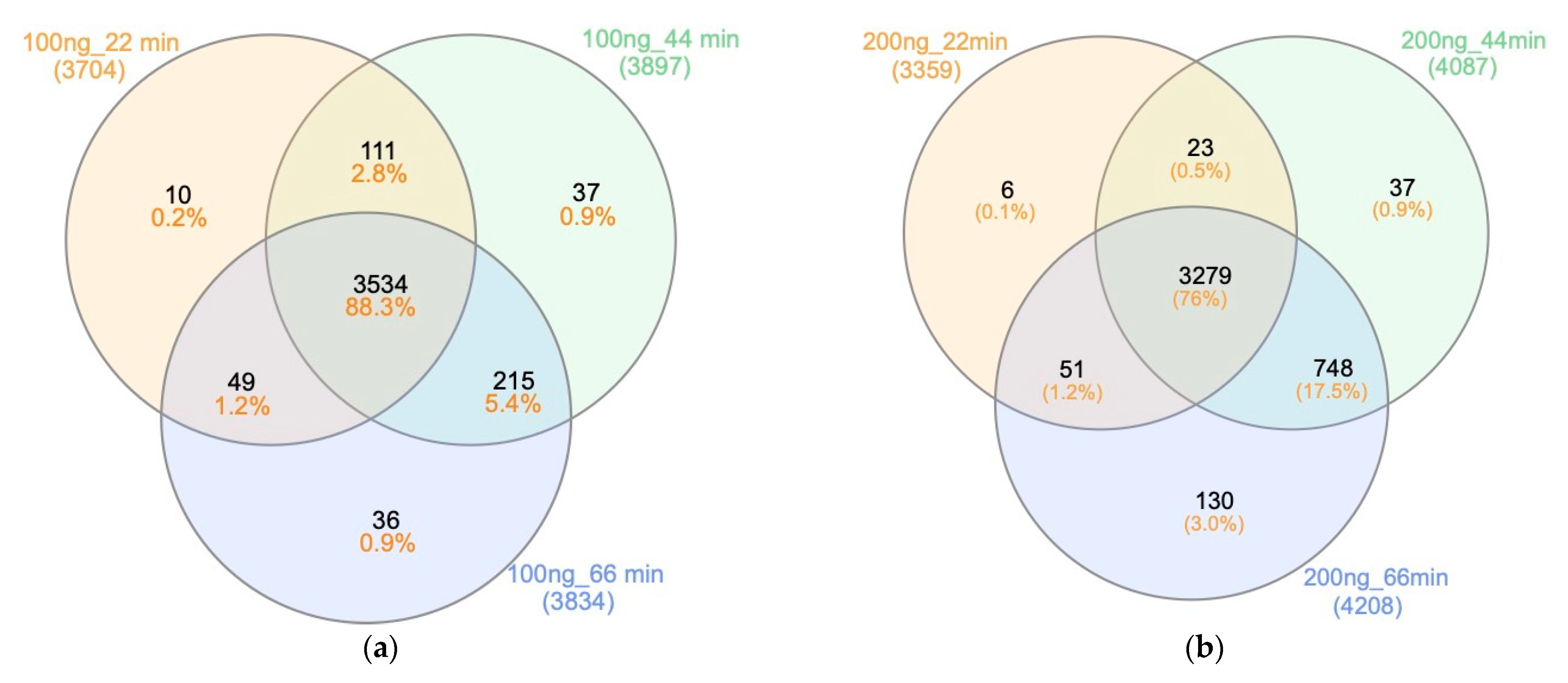
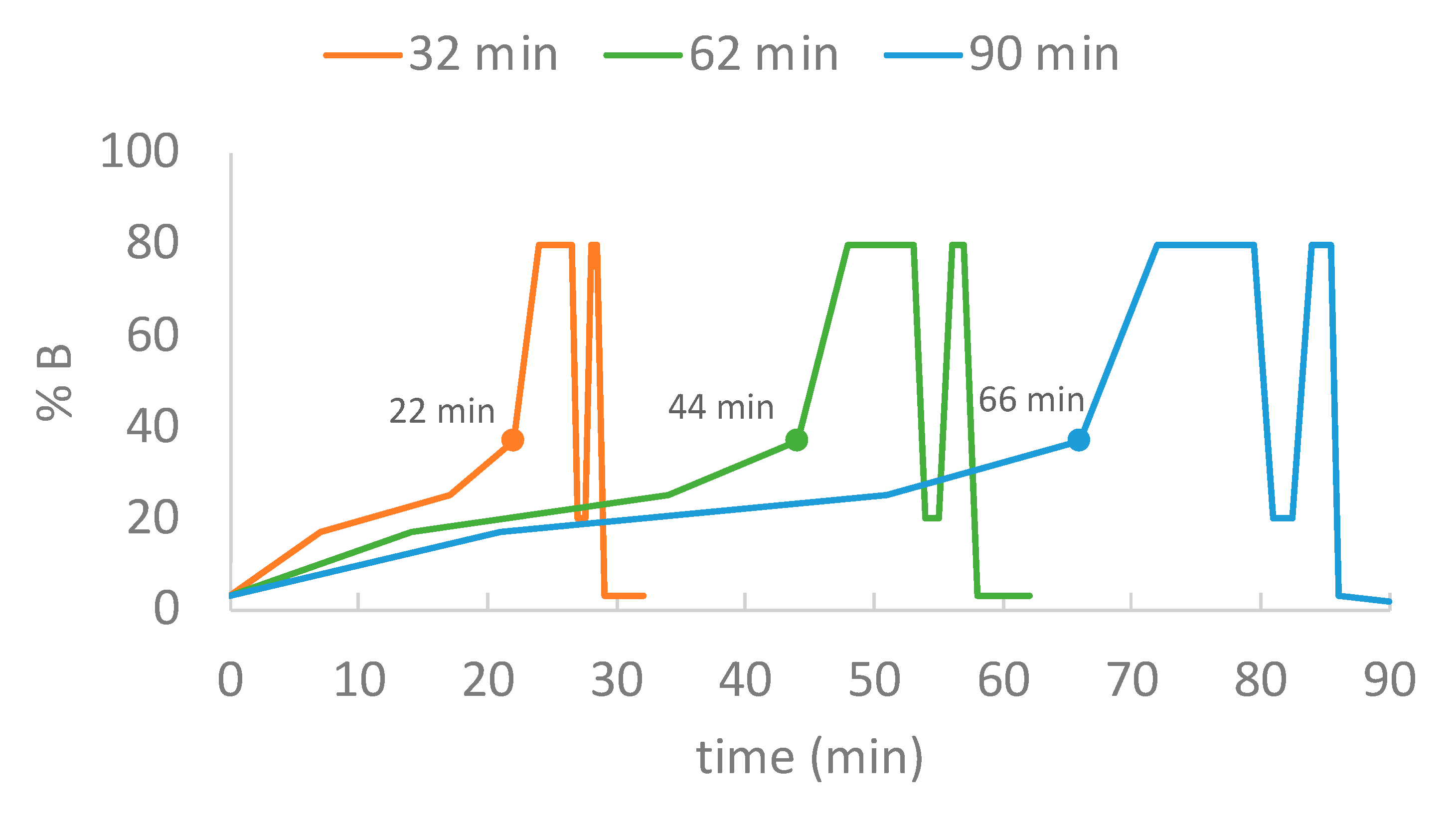
2.2. Gradient Durations, Loading Amount and Search Engines
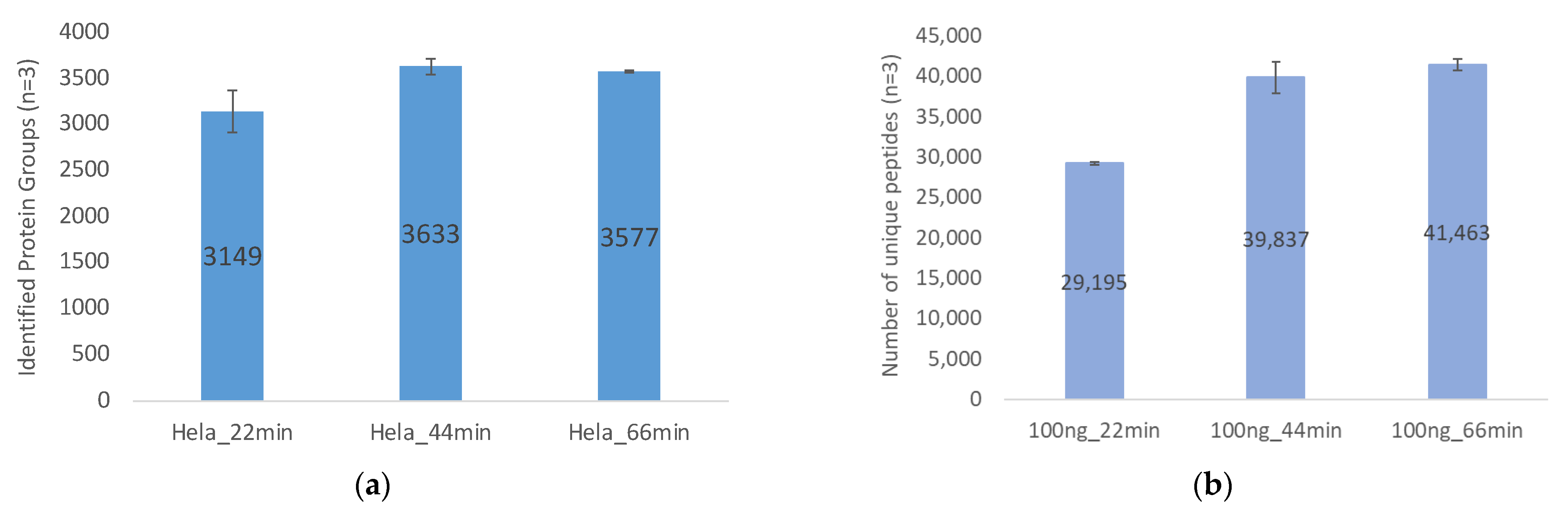
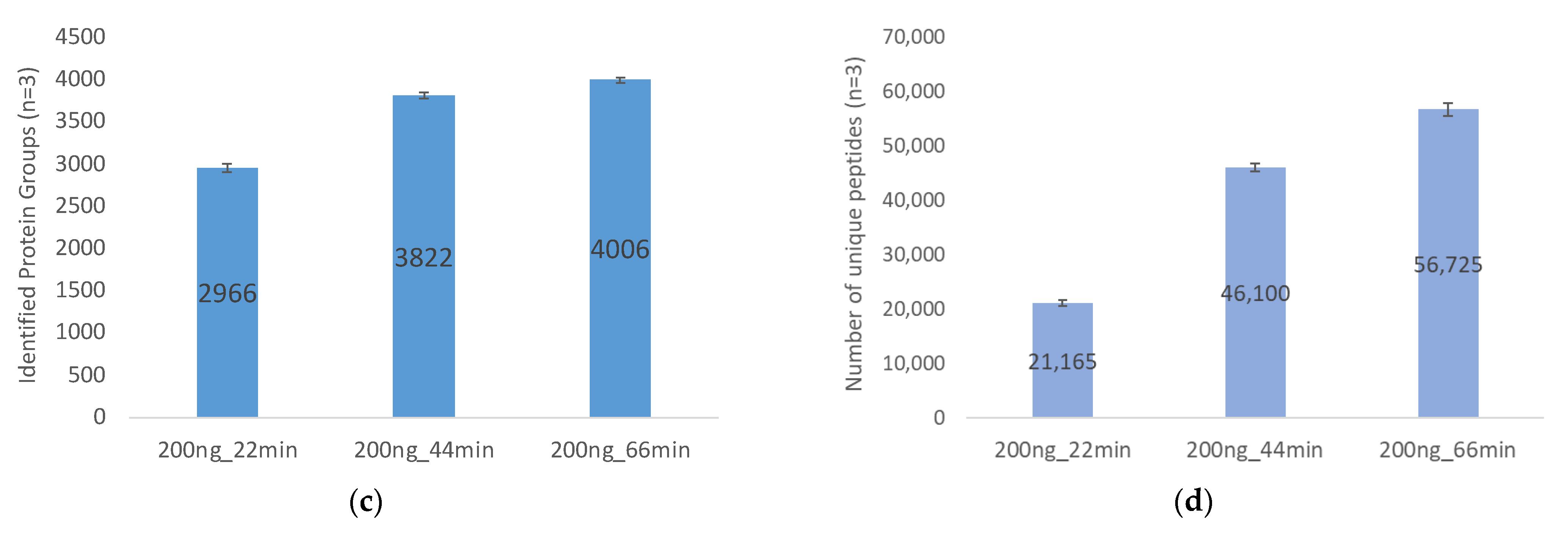
2.2.1. Gradient Durations and Loading Amount
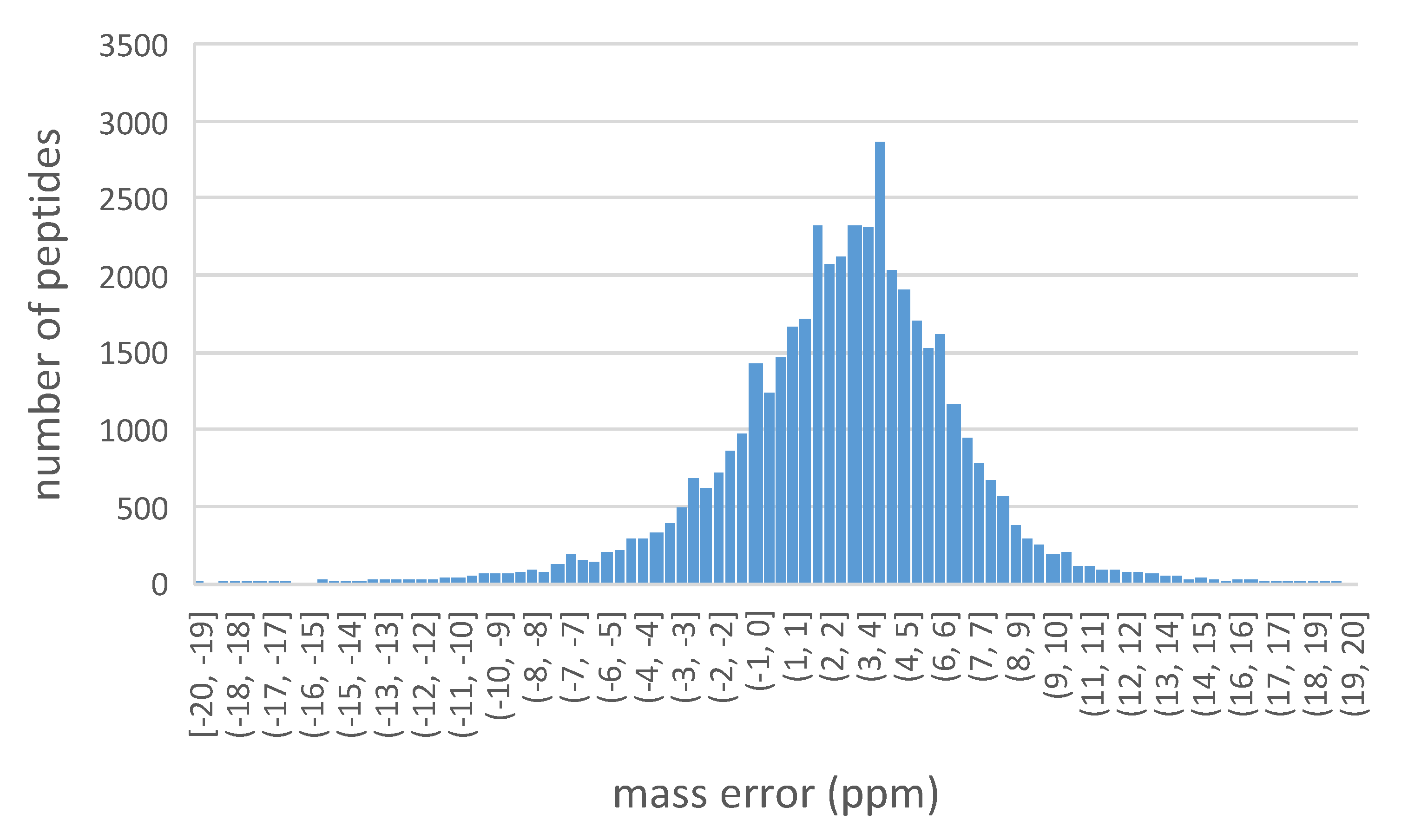
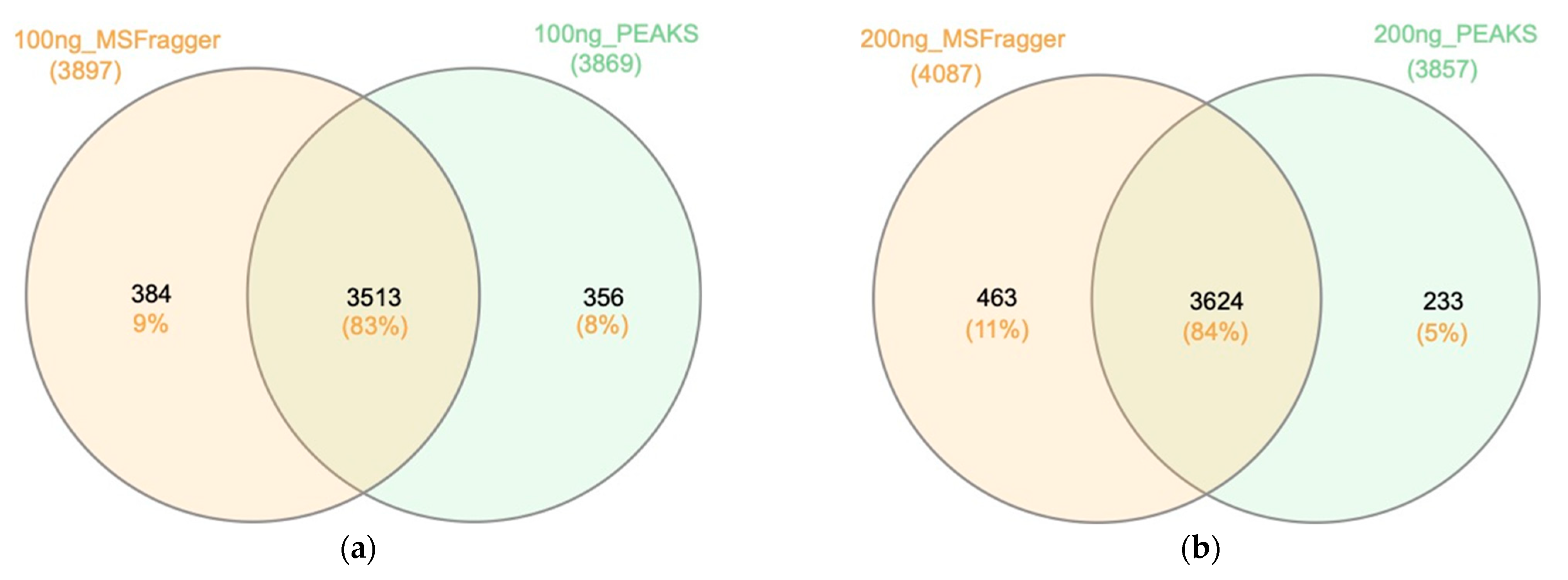
2.2.2. Search Engines
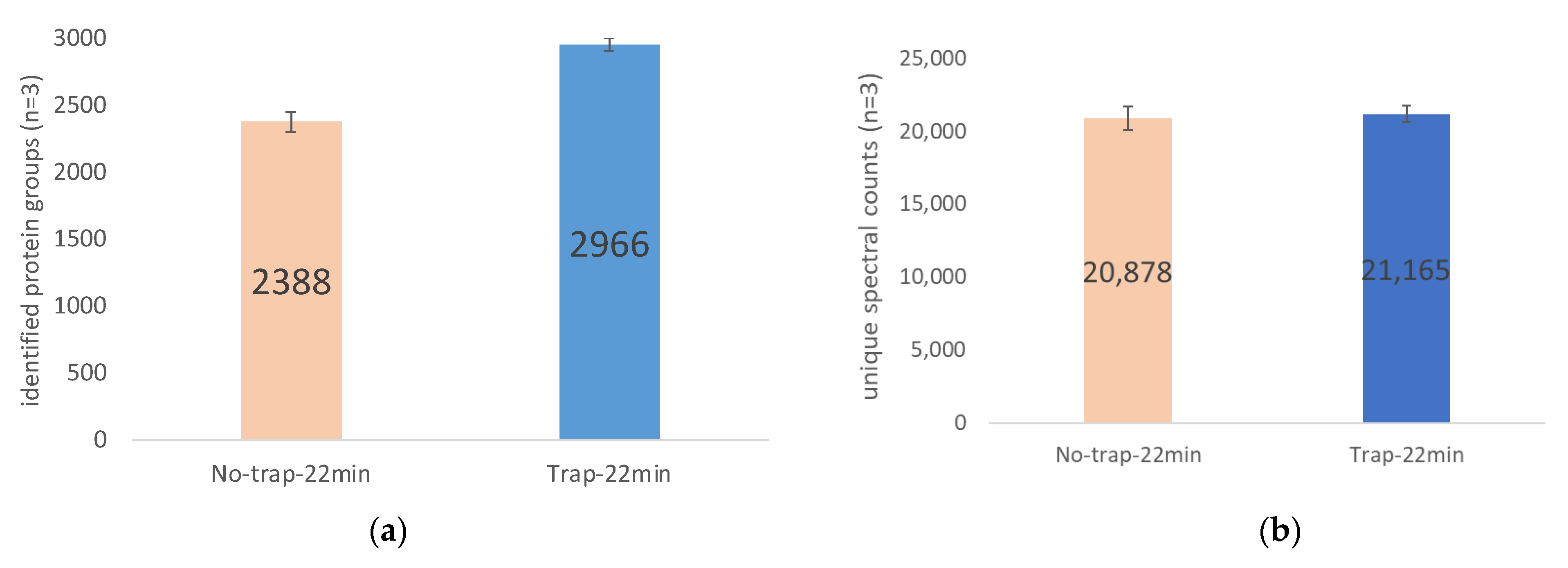
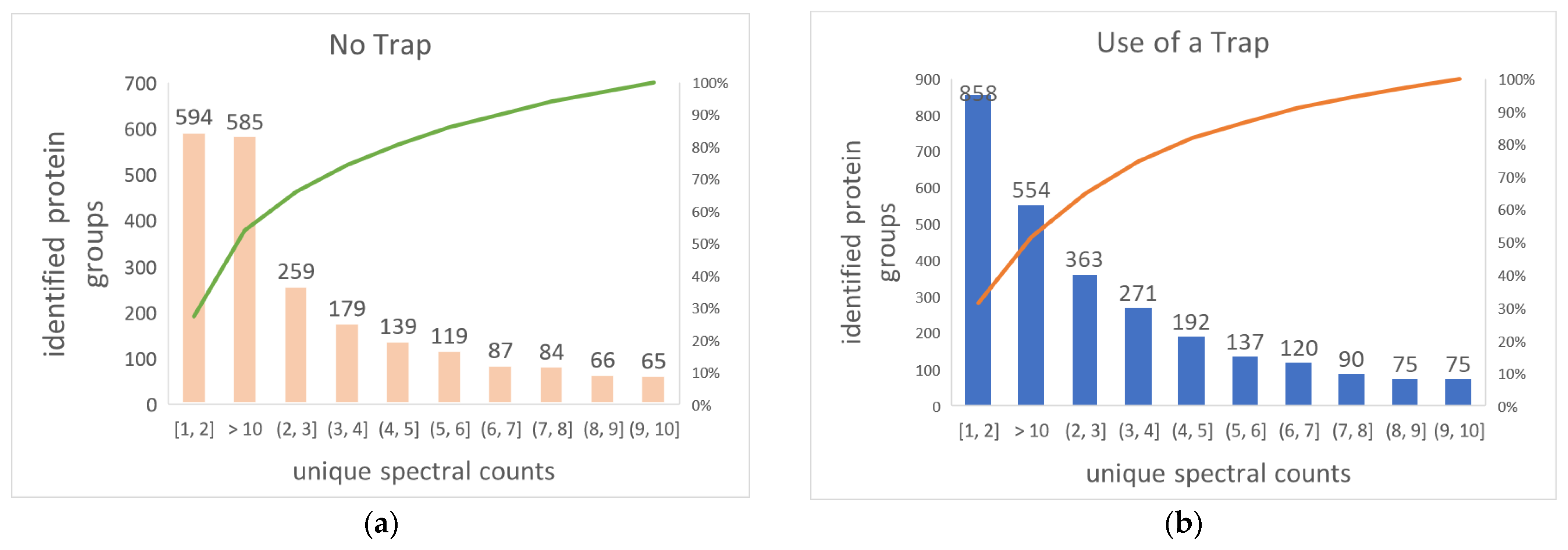
2.3. The Use of Trap Column Versus No-Trap Column
3. Materials and Methods
3.1. HeLa Digest
3.2. Nano-LC
3.3. Nano Electrospray Ionization (NSI)
3.4. Tims-Q-ToF
3.5. Liquid Chromatography Gradients
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shah, T.R.; Misra, A. Proteomics Challenges in Delivery of Therapeutic Genomics and Proteomics; Elsevier: London, UK, 2011. [Google Scholar]
- Chadwick, R.; Callahan, D.; Singer, P. Encyclopedia of Applied Ethics; Academic Press: London, UK, 2012. [Google Scholar]
- Padmanabhan, S. Handbook of Pharmacogenomics and Stratified Medicine; Academic Press: London, UK, 2014. [Google Scholar]
- Yu, L.-R.; Stewart, N.A.; Veenstra, T.D. Proteomics: The deciphering of the functional genome. In Essentials of Genomic and Personalized Medicine; Academic Press: London, UK, 2010; pp. 89–96. [Google Scholar]
- Coorssen, J.R. Top-down proteomics: 2D gels are an integral part of the process. In Farm Animal Proteomics 2013; Springer: Berlin/Heidelberg, Germany; Wageningen Academic Publishers: Wageningen, The Netherlands, 2013; pp. 31–33. [Google Scholar]
- Manadas, B.; Mendes, V.M.; English, J.; Dunn, M.J. Peptide fractionation in proteomics approaches. Expert Rev. Proteom. 2010, 7, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Aslanian, A.; Yates, J., III. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [Google Scholar] [CrossRef]
- Aballo, T.J.; Roberts, D.S.; Melby, J.A.; Buck, K.M.; Brown, K.A.; Ge, Y. Ultrafast and Reproducible Proteomics from Small Amounts of Heart Tissue Enabled by Azo and timsTOF Pro. J. Proteome Res. 2021, 20, 4203–4211. [Google Scholar] [CrossRef] [PubMed]
- Tose, L.V.; Benigni, P.; Leyva, D.; Sundberg, A.; Ramírez, C.E.; Ridgeway, M.E.; Park, M.A.; Romão, W.; Jaffé, R.; Fernandez-Lima, F. Coupling trapped ion mobility spectrometry to mass spectrometry: Trapped ion mobility spectrometry–time-of-flight mass spectrometry versus trapped ion mobility spectrometry—Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2018, 32, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Brunner, A.-D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M.A. Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 2018, 17, 2534–2545. [Google Scholar] [CrossRef]
- Meier, F.; Beck, S.; Grassl, N.; Lubeck, M.; Park, M.A.; Raether, O.; Mann, M. Parallel accumulation–serial fragmentation (PASEF): Multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. J. Proteome Res. 2015, 14, 5378–5387. [Google Scholar] [CrossRef]
- Mitulović, G.; Mechtler, K. HPLC techniques for proteomics analysis—A short overview of latest developments. Brief. Funct. Genom. 2006, 5, 249–260. [Google Scholar] [CrossRef]
- Van Puyvelde, B.; Daled, S.; Willems, S.; Gabriels, R.; Gonzalez de Peredo, A.; Chaoui, K.; Mouton-Barbosa, E.; Bouyssié, D.; Boonen, K.; Hughes, C.J. A comprehensive LFQ benchmark dataset on modern day acquisition strategies in proteomics. Sci. Data 2022, 9, 126. [Google Scholar] [CrossRef]
- Babushok, V.I.; Zenkevich, I.G. Retention characteristics of peptides in RP-LC: Peptide retention prediction. Chromatographia 2010, 72, 781–797. [Google Scholar] [CrossRef]
- Gilar, M.; Xie, H.; Jaworski, A. Utility of retention prediction model for investigation of peptide separation selectivity in reversed-phase liquid chromatography: Impact of concentration of trifluoroacetic acid, column temperature, gradient slope and type of stationary phase. Anal. Chem. 2010, 82, 265–275. [Google Scholar] [CrossRef]
- Hsieh, E.J.; Bereman, M.S.; Durand, S.; Valaskovic, G.A.; MacCoss, M.J. Effects of column and gradient lengths on peak capacity and peptide identification in nanoflow LC-MS/MS of complex proteomic samples. J. Am. Soc. Mass Spectrom. 2012, 24, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Bączek, T.; Kaliszan, R. Predictions of peptides’ retention times in reversed-phase liquid chromatography as a new supportive tool to improve protein identification in proteomics. Proteomics 2009, 9, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Young, R.; Straubinger, R.M.; Page, B.; Cao, J.; Wang, H.; Yu, H.; Canty, J.M., Jr.; Qu, J. A straightforward and highly efficient precipitation/on-pellet digestion procedure coupled with a long gradient nano-LC separation and Orbitrap mass spectrometry for label-free expression profiling of the swine heart mitochondrial proteome. J. Proteome Res. 2009, 8, 2838–2850. [Google Scholar] [CrossRef]
- Kouba, V.; Vejmelkova, D.; Zwolsman, E.; Hurkova, K.; Navratilova, K.; Laureni, M.; Vodickova, P.; Podzimek, T.; Hajslova, J.; Pabst, M. Adaptation of anammox bacteria to low temperature via gradual acclimation and cold shocks: Distinctions in protein expression, membrane composition and activities. Water Res. 2022, 209, 117822. [Google Scholar]
- Pecankova, K.; Pecherkova, P.; Gasova, Z.; Sovova, Z.; Riedel, T.; Jäger, E.; Cermak, J.; Majek, P. Proteome changes of plasma-derived extracellular vesicles in patients with myelodysplastic syndrome. PLoS ONE 2022, 17, e0262484. [Google Scholar]
- Yaeshima, C.; Murata, N.; Ishino, S.; Sagawa, I.; Ito, K.; Uchiumi, T. A novel ribosome-dimerization protein found in the hyperthermophilic archaeon Pyrococcus furiosus using ribosome-associated proteomics. Biochem. Biophys. Res. Commun. 2022, 593, 116–121. [Google Scholar] [CrossRef]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom. 2012, 11, M111.010587. [Google Scholar] [CrossRef]
- Polasky, D.A.; Yu, F.; Teo, G.C.; Nesvizhskii, A.I. Fast and comprehensive N-and O-glycoproteomics analysis with MSFragger-Glyco. Nat. Methods 2020, 17, 1125–1132. [Google Scholar] [CrossRef]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef]
- Yu, F.; Teo, G.C.; Kong, A.T.; Haynes, S.E.; Avtonomov, D.M.; Geiszler, D.J.; Nesvizhskii, A.I. Identification of modified peptides using localization-aware open search. Nat. Commun. 2020, 11, 4065. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. Clustvis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Paulo, J.A. Practical and efficient searching in proteomics: A cross engine comparison. Webmedcentral 2013, 4, WMCPLS0052. [Google Scholar] [PubMed]
- Fernández-Costa, C.; Martínez-Bartolomé, S.; McClatchy, D.B.; Saviola, A.J.; Yu, N.-K.; Yates, J.R., III. Impact of the Identification Strategy on the Reproducibility of the DDA and DIA Results. J. Proteome Res. 2020, 19, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Lanckmans, K.; Clinckers, R.; Van Eeckhaut, A.; Sarre, S.; Smolders, I.; Michotte, Y. Use of microbore LC–MS/MS for the quantification of oxcarbazepine and its active metabolite in rat brain microdialysis samples. J. Chromatogr. B 2006, 831, 205–212. [Google Scholar] [CrossRef]
- Peaks Team. PEAKS Studio User Manual, version 5.3; Bioinformatics Solutions, Inc.: Waterloo, ON, Canada, 2011. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Liu, P.N.; Mooney, B.P.; Nguyen, T.T.; Greenlief, C.M. A Comprehensive Study of Gradient Conditions for Deep Proteome Discovery in a Complex Protein Matrix. Int. J. Mol. Sci. 2022, 23, 11714. https://doi.org/10.3390/ijms231911714
Wei X, Liu PN, Mooney BP, Nguyen TT, Greenlief CM. A Comprehensive Study of Gradient Conditions for Deep Proteome Discovery in a Complex Protein Matrix. International Journal of Molecular Sciences. 2022; 23(19):11714. https://doi.org/10.3390/ijms231911714
Chicago/Turabian StyleWei, Xing, Pei N. Liu, Brian P. Mooney, Thao Thi Nguyen, and C. Michael Greenlief. 2022. "A Comprehensive Study of Gradient Conditions for Deep Proteome Discovery in a Complex Protein Matrix" International Journal of Molecular Sciences 23, no. 19: 11714. https://doi.org/10.3390/ijms231911714
APA StyleWei, X., Liu, P. N., Mooney, B. P., Nguyen, T. T., & Greenlief, C. M. (2022). A Comprehensive Study of Gradient Conditions for Deep Proteome Discovery in a Complex Protein Matrix. International Journal of Molecular Sciences, 23(19), 11714. https://doi.org/10.3390/ijms231911714






