Switching of Photocatalytic Tyrosine/Histidine Labeling and Application to Photocatalytic Proximity Labeling
Abstract
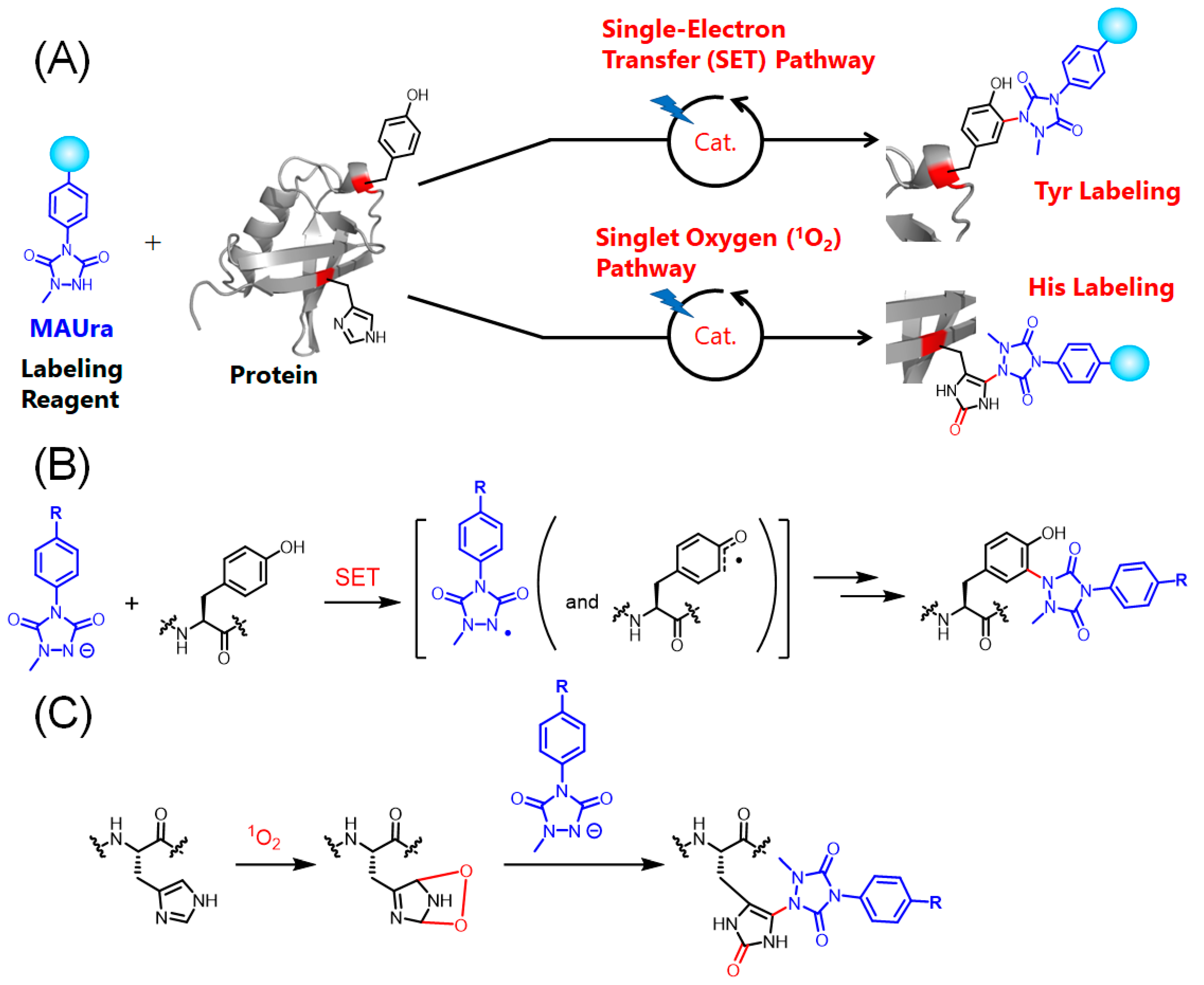
1. Introduction
2. Results
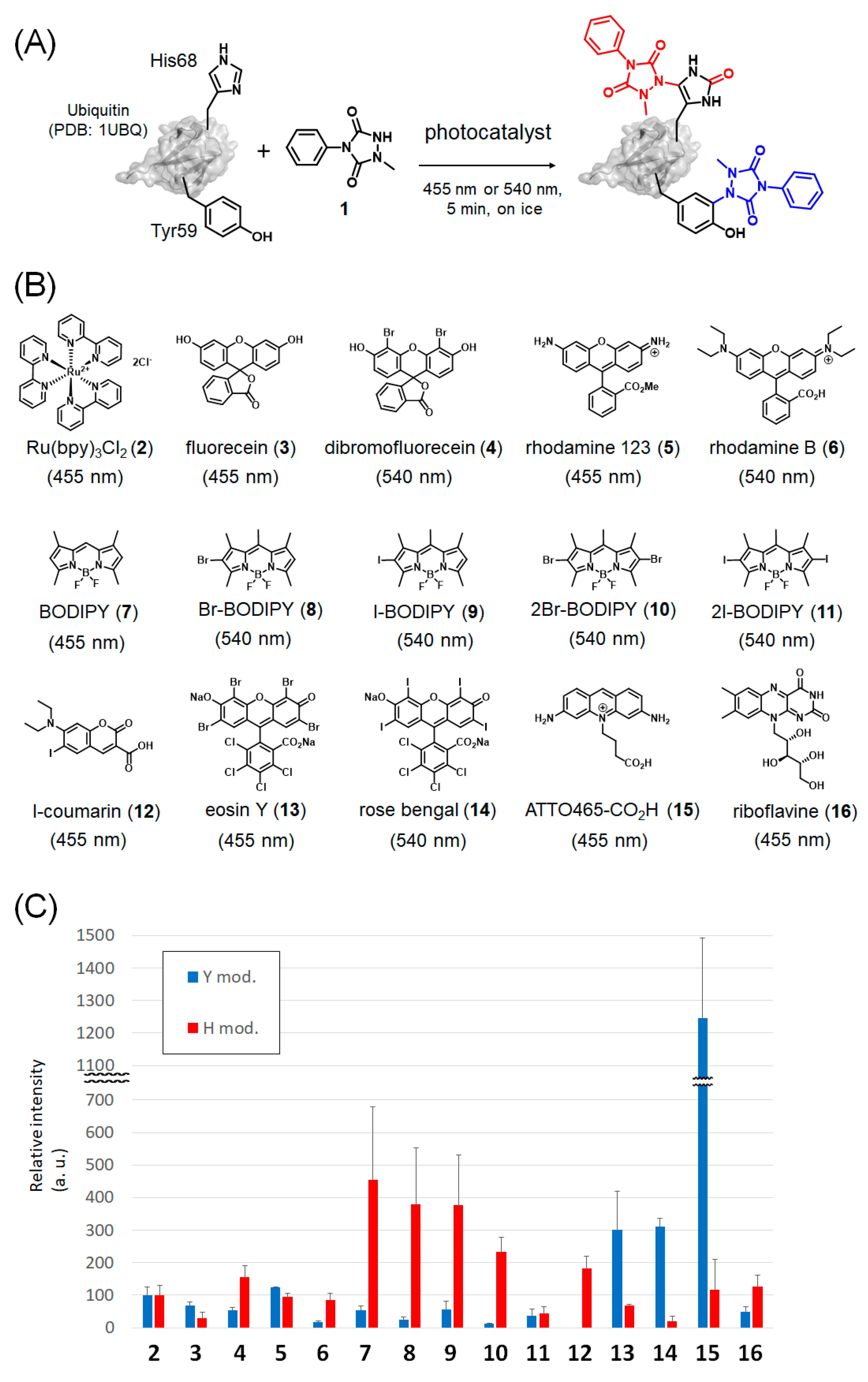
2.1. Photocatalyst Screening
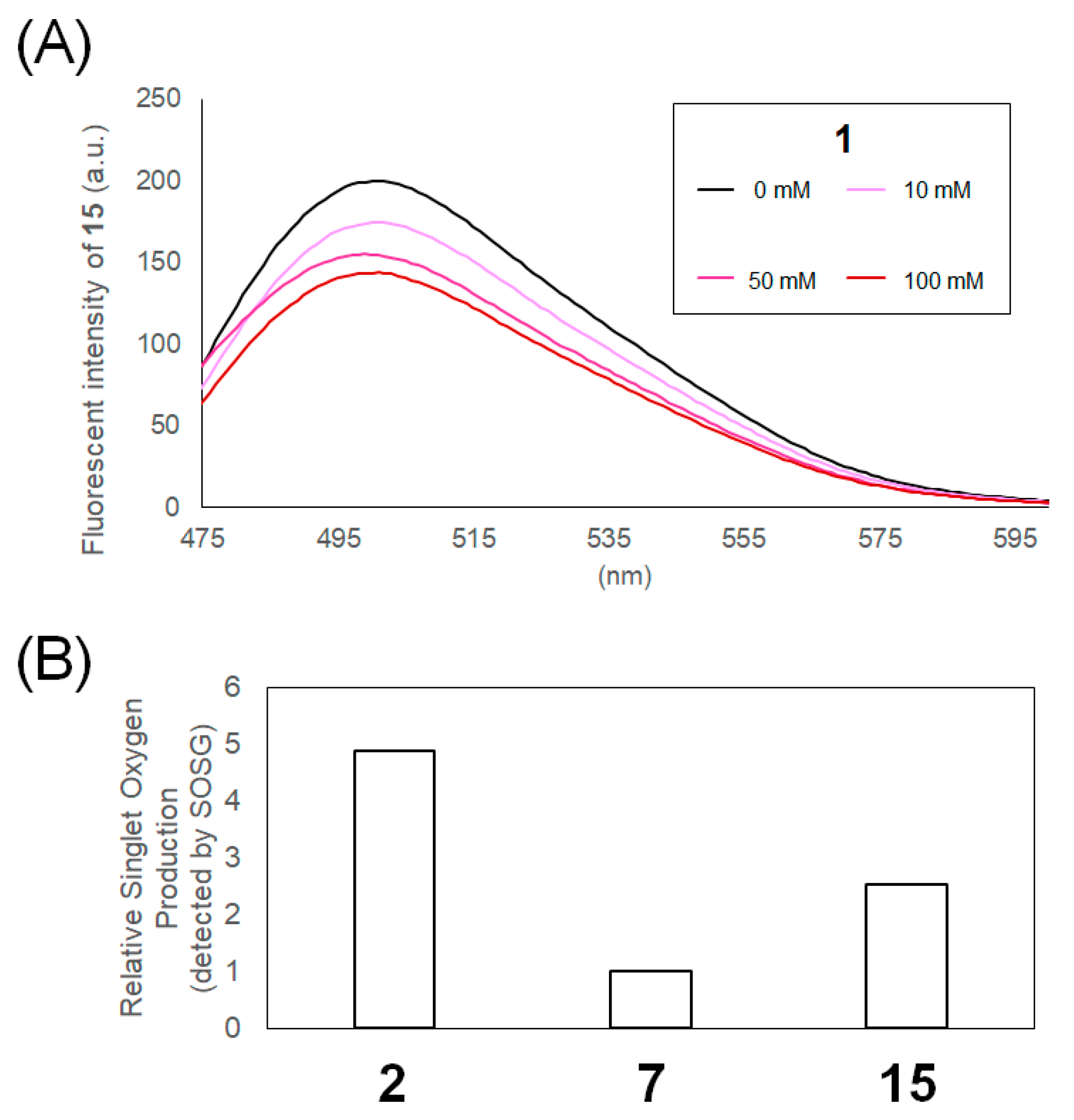
2.2. Mechanistic Analysis of Photocatalysts
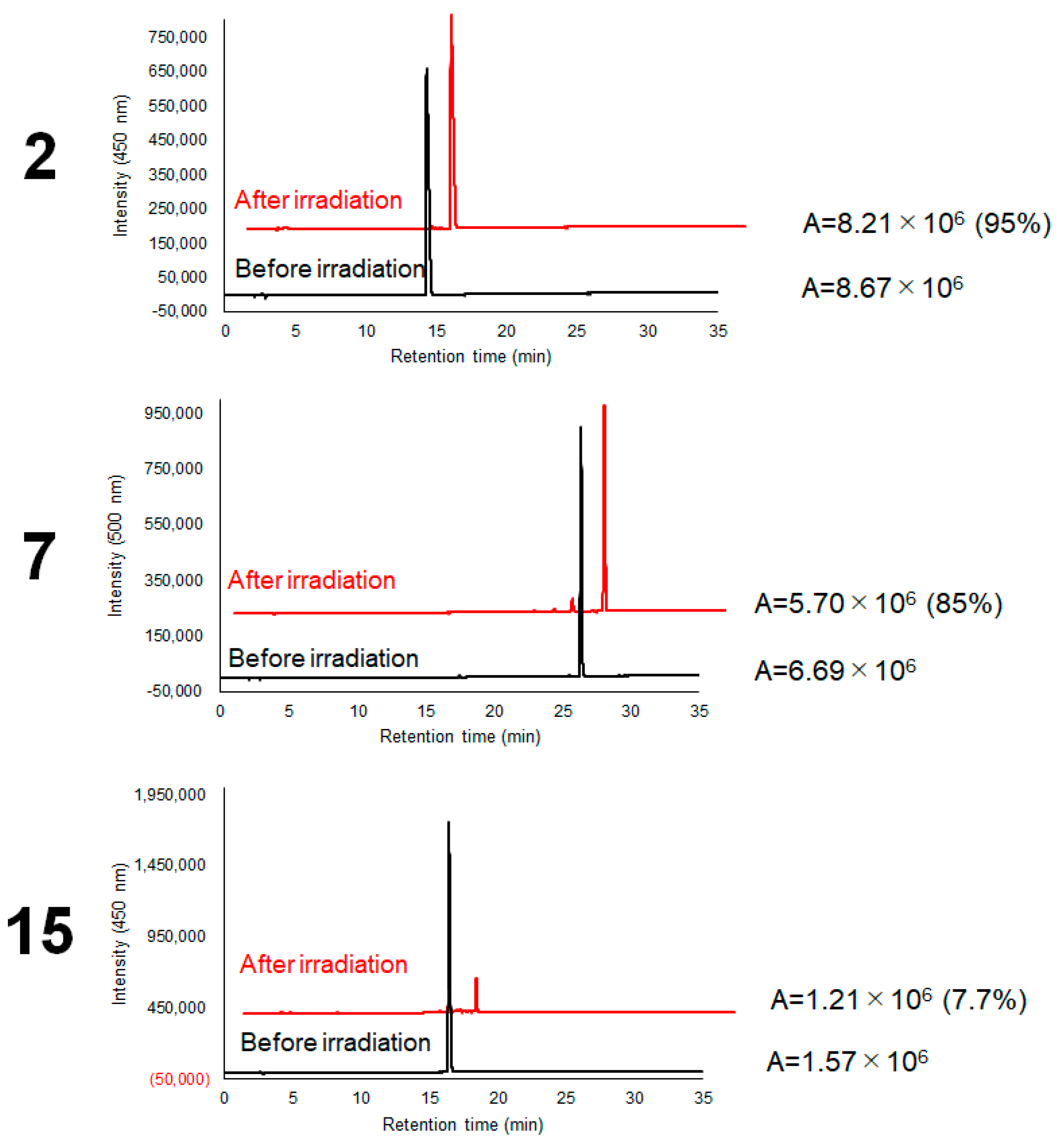
2.3. Photostability Evaluation of Photocatalysts
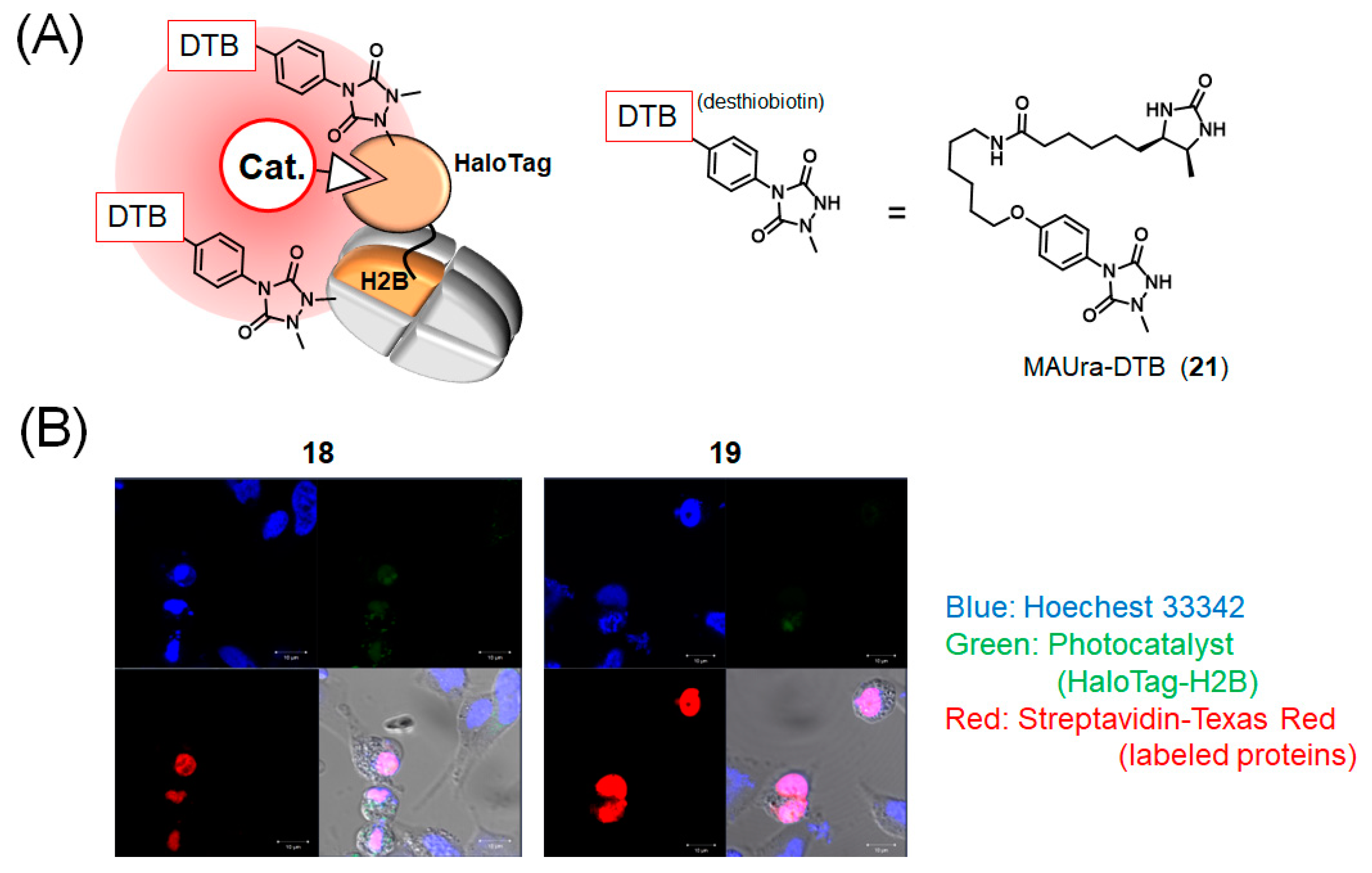
2.4. Photocatalytic Proximity Labeling using HaloTag
2.5. HaloTag-H2B Photocatalytic Proximity Labeling in Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Ubiquitin Labeling
4.3. In-Gel Digestion of Labeled Ubiquitin
4.4. NanoLC-MS/MS Analysis
4.5. Measurement of Singlet Oxygen Generation
4.6. Stern–Volmer Fluorescence Quenching Experiments
4.7. Evaluation of Photostability
4.8. GST-HaloTag Labeling in the Protein Mixture
4.9. Peptide Labeling
4.10. Transfection of HaloTag-H2B
4.11. HaloTag-H2B Proximity Labeling in Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lobingier, B.T.; Hüttenhain, R.; Eichel, K.; Miller, K.B.; Ting, A.Y.; von Zastrow, M.; Krogan, N.J. An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 2017, 169, 350–360.e12. [Google Scholar] [CrossRef]
- Li, Z.; Rodriguez, E.; Azaria, S.; Pekarek, A.; Hage, D.S. Affinity monolith chromatography: A review of general principles and applications. Electrophoresis 2017, 38, 2837–2850. [Google Scholar] [CrossRef]
- Yoo, J.; Lee, T.-S.; Choi, B.; Shon, M.J.; Yoon, T.-Y. Observing Extremely Weak Protein–Protein Interactions with Conventional Single-Molecule Fluorescence Microscopy. J. Am. Chem. Soc. 2016, 138, 14238–14241. [Google Scholar] [CrossRef]
- O’Reilly, F.J.; Rappsilber, J. Cross-linking mass spectrometry: Methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol. 2018, 25, 1000–1008. [Google Scholar] [CrossRef]
- Savitski, M.M.; Reinhard, F.B.M.; Franken, H.; Werner, T.; Savitski, M.F.; Eberhard, D.; Molina, D.M.; Jafari, R.; Dovega, R.B.; Klaeger, S.; et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 2014, 346, 1255784. [Google Scholar] [CrossRef]
- Lauc, G.; Lee, R.T.; Dumiæ, J.; Lee, Y.C. Photoaffinity glycoprobes—A new tool for the identification of lectins. Glycobiology 2000, 10, 357–364. [Google Scholar] [CrossRef][Green Version]
- Ballell, L.; van Scherpenzeel, M.; Buchalova, K.; Liskamp, R.M.J.; Pieters, R.J. A new chemical probe for the detection of the cancer-linked galectin-3. Org. Biomol. Chem. 2006, 4, 4387–4394. [Google Scholar] [CrossRef]
- Wibowo, A.; Peters, E.C.; Hsieh-Wilson, L.C. Photoactivatable Glycopolymers for the Proteome-Wide Identification of Fucose-α(1-2)-Galactose Binding Proteins. J. Am. Chem. Soc. 2014, 136, 9528–9531. [Google Scholar] [CrossRef]
- Sakurai, K.; Hatai, Y.; Okada, A. Gold nanoparticle-based multivalent carbohydrate probes: Selective photoaffinity labeling of carbohydrate-binding proteins. Chem. Sci. 2015, 7, 702–706. [Google Scholar] [CrossRef]
- Tsukiji, S.; Miyagawa, M.; Takaoka, Y.; Tamura, T.; Hamachi, I. Ligand-directed tosyl chemistry for protein labeling in vivo. Nat. Chem. Biol. 2009, 5, 341–343. [Google Scholar] [CrossRef]
- Koshi, Y.; Nakata, E.; Miyagawa, M.; Tsukiji, S.; Ogawa, A.T.; Hamachi, I. Target-Specific Chemical Acylation of Lectins by Ligand-Tethered DMAP Catalysts. J. Am. Chem. Soc. 2007, 130, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Tsushima, M.; Sato, S.; Niwa, T.; Taguchi, H.; Nakamura, H. Catalyst-proximity protein chemical labelling on affinity beads targeting endogenous lectins. Chem. Commun. 2019, 55, 13275–13278. [Google Scholar] [CrossRef]
- Kido, K.; Yamanaka, S.; Nakano, S.; Motani, K.; Shinohara, S.; Nozawa, A.; Kosako, H.; Ito, S.; Sawasaki, T. AirID, a novel proximity biotinylation enzyme, for analysis of protein–protein interactions. Elife 2020, 9, e54983. [Google Scholar] [CrossRef]
- Trowbridge, A.D.; Seath, C.P.; Rodriguez-Rivera, F.P.; Li, B.X.; Dul, B.E.; Schwaid, A.G.; Buksh, B.F.; Geri, J.B.; Oakley, J.V.; Fadeyi, O.O.; et al. Small molecule photocatalysis enables drug target identification via energy transfer. Proc. Natl. Acad. Sci. USA 2022, 119. [Google Scholar] [CrossRef]
- Geri, J.B.; Oakley, J.V.; Reyes-Robles, T.; Wang, T.; McCarver, S.J.; White, C.H.; Rodriguez-Rivera, F.P.; Parker, D.L.; Hett, E.C.; Fadeyi, O.O.; et al. Microenvironment mapping via Dexter energy transfer on immune cells. Science 2020, 367, 1091–1097. [Google Scholar] [CrossRef]
- Li, Q.; Xie, Y.; Rice, R.; Maverakis, E.; Lebrilla, C.B. A proximity labeling method for protein–protein interactions on cell membrane. Chem. Sci. 2022, 13, 6028–6038. [Google Scholar] [CrossRef]
- Joeh, E.; O’Leary, T.; Li, W.; Hawkins, R.; Hung, J.R.; Parker, C.G.; Huang, M.L. Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc. Natl. Acad. Sci. USA 2020, 117, 27329–27338. [Google Scholar] [CrossRef]
- Müller, M.; Gräbnitz, F.; Barandun, N.; Shen, Y.; Wendt, F.; Steiner, S.N.; Severin, Y.; Vetterli, S.U.; Mondal, M.; Prudent, J.R.; et al. Light-mediated discovery of surfaceome nanoscale organization and intercellular receptor interaction networks. Nat. Commun. 2021, 12, 7036. [Google Scholar] [CrossRef]
- Oslund, R.C.; Reyes-Robles, T.; White, C.H.; Tomlinson, J.H.; Crotty, K.A.; Bowman, E.P.; Chang, D.; Peterson, V.M.; Li, L.; Frutos, S.; et al. Detection of cell–cell interactions via photocatalytic cell tagging. Nat. Chem. Biol. 2022, 18, 850–858. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Shiraiwa, K.; Cheng, R.; Nonaka, H.; Tamura, T.; Hamachi, I. Chemical Tools for Endogenous Protein Labeling and Profiling. Cell Chem. Biol. 2020, 27, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Lechner, V.M.; Nappi, M.; Deneny, P.J.; Folliet, S.; Chu, J.C.K.; Gaunt, M.J. Visible-Light-Mediated Modification and Manipulation of Biomacromolecules. Chem. Rev. 2021, 122, 1752–1829. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-G.; Rhee, H.-W. Molecular Spatiomics by Proximity Labeling. Accounts Chem. Res. 2022, 55, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Hatano, K.; Tsushima, M.; Nakamura, H. 1-Methyl-4-aryl-urazole (MAUra) labels tyrosine in proximity to ruthenium photocatalysts. Chem. Commun. 2018, 54, 5871–5874. [Google Scholar] [CrossRef]
- Nakane, K.; Sato, S.; Niwa, T.; Tsushima, M.; Tomoshige, S.; Taguchi, H.; Ishikawa, M.; Nakamura, H. Proximity Histidine Labeling by Umpolung Strategy Using Singlet Oxygen. J. Am. Chem. Soc. 2021, 143, 7726–7731. [Google Scholar] [CrossRef]
- Tsushima, M.; Sato, S.; Miura, K.; Niwa, T.; Taguchi, H.; Nakamura, H. Intracellular photocatalytic-proximity labeling for profiling protein–protein interactions in microenvironments. Chem. Commun. 2022, 58, 1926–1929. [Google Scholar] [CrossRef]
- Romero, N.; Nicewicz, D.A. Organic Photoredox Catalysis. Chem. Rev. 2016, 116, 10075–10166. [Google Scholar] [CrossRef]
- Tamura, T.; Takato, M.; Shiono, K.; Hamachi, I. Development of a Photoactivatable Proximity Labeling Method for the Identification of Nuclear Proteins. Chem. Lett. 2020, 49, 145–148. [Google Scholar] [CrossRef]
- Li, Y.; Aggarwal, M.B.; Ke, K.; Nguyen, K.; Spitale, R.C. Improved Analysis of RNA Localization by Spatially Restricted Oxidation of RNA–Protein Complexes. Biochemistry 2018, 57, 1577–1581. [Google Scholar] [CrossRef]
- Liu, H.; Luo, H.; Xue, Q.; Qin, S.; Qiu, S.; Liu, S.; Lin, J.; Li, J.P.; Chen, P.R. Antigen-Specific T Cell Detection via Photocatalytic Proximity Cell Labeling (PhoXCELL). J. Am. Chem. Soc. 2022, 144, 5517–5526. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Zeng, K.; Qiang, J.; Cao, Y.; Li, Y.; Fang, Y.; Zhang, Y.; Chen, Y. Selective Mitochondrial Protein Labeling Enabled by Biocompatible Photocatalytic Reactions inside Live Cells. JACS Au 2021, 1, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Nakane, K.; Niwa, T.; Tsushima, M.; Tomoshige, S.; Taguchi, H.; Nakamura, H.; Ishikawa, M.; Sato, S. BODIPY Catalyzes Proximity-Dependent Histidine Labelling. ChemCatChem 2022, 14, e202200077. [Google Scholar] [CrossRef]
- Prieto-Montero, R.; Prieto-Castaneda, A.; Sola-Llano, R.; Agarrabeitia, A.R.; Garcia-Fresnadillo, D.; Lopez-Arbeloa, I.; Villanueva, A.; Ortiz, M.J.; de la Moya, S.; Martinez-Martinez, V. Exploring BODIPY Derivatives as Singlet Oxygen Photosensitizers for PDT. Photochem. Photobiol. 2020, 96, 458–477. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Kim, D.K.; Bloom, S.; Huang, R.Y.-C.; Qiao, J.X.; Ewing, W.R.; Oblinsky, D.G.; Scholes, G.D.; MacMillan, D.W.C. Site-selective tyrosine bioconjugation via photoredox catalysis for native-to-bioorthogonal protein transformation. Nat. Chem. 2021, 13, 902–908. [Google Scholar] [CrossRef]
- Arias-Rotondo, D.M.; McCusker, J.K. The photophysics of photoredox catalysis: A roadmap for catalyst design. Chem. Soc. Rev. 2016, 45, 5803–5820. [Google Scholar] [CrossRef]
- Turksoy, A.; Yildiz, D.; Akkaya, E.U. Photosensitization and controlled photosensitization with BODIPY dyes. Coordin. Chem. Rev. 2019, 379, 47–64. [Google Scholar] [CrossRef]
- Yogo, T.; Urano, Y.; Ishitsuka, Y.; Maniwa, A.F.; Nagano, T. Highly Efficient and Photostable Photosensitizer Based on BODIPY Chromophore. J. Am. Chem. Soc. 2005, 127, 12162–12163. [Google Scholar] [CrossRef]
- Gorman, A.; Killoran, J.; O’Shea, C.; Kenna, T.; Gallagher, A.W.M.; O’Shea, D.F. In Vitro Demonstration of the Heavy-Atom Effect for Photodynamic Therapy. J. Am. Chem. Soc. 2004, 126, 10619–10631. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.-W.; Cui, A.-J.; Cai, X.-X.; Wan, Y.; Chen, Q.; He, M.-Y.; Zhang, W. Regioselective 2,6-dihalogenation of BODIPYs in 1,1,1,3,3,3-hexafluoro-2-propanol and preparation of novel meso-alkyl polymeric BODIPY dyes. RSC Adv. 2013, 3, 9219–9222. [Google Scholar] [CrossRef]
- Yan, D.; Zhao, Q.; Rao, L.; Chen, J.; Xiao, W. Eosin Y as a Redox Catalyst and Photosensitizer for Sequential Benzylic C−H Amination and Oxidation. Chem. A Eur. J. 2018, 24, 16895–16901. [Google Scholar] [CrossRef]
- Yu, Z.-Y.; Zhao, J.-N.; Yang, F.; Tang, X.-F.; Wu, Y.-F.; Ma, C.-F.; Song, B.; Yun, L.; Meng, Q.-W. Rose bengal as photocatalyst: Visible light-mediated Friedel–Crafts alkylation of indoles with nitroalkenes in water. RSC Adv. 2020, 10, 4825–4831. [Google Scholar] [CrossRef]
- Chu, X.-Q.; Ge, D.; Wang, M.; Rao, W.; Loh, T.; Shen, Z.-L. Chemo- and Regioselective Ring Construction Driven by Visible-Light Photoredox Catalysis: An Access to Fluoroalkylated Oxazolidines Featuring an All-Substituted Carbon Stereocenter. Adv. Synth. Catal. 2019, 361, 4082–4090. [Google Scholar] [CrossRef]
- Sato, S.; Matsumura, M.; Kadonosono, T.; Abe, S.; Ueno, T.; Ueda, H.; Nakamura, H. Site-Selective Protein Chemical Modification of Exposed Tyrosine Residues Using Tyrosine Click Reaction. Bioconjugate Chem. 2020, 31, 1417–1424. [Google Scholar] [CrossRef]
- Geren, L.; Durham, B.; Millett, F. Chapter 28 Use of Ruthenium Photoreduction Techniques to Study Electron Transfer in Cytochrome Oxidase. Methods Enzymol. 2009, 456, 507–520. [Google Scholar] [CrossRef]
- Rozhkov, R.V.; Davisson, V.J.; Bergstrom, D.E. Fluorogenic Transformations Based on Formation of C-C Bonds Catalyzed by Palladium: An Efficient Approach for High Throughput Optimizations and Kinetic Studies. Adv. Synth. Catal. 2008, 350, 71–75. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakane, K.; Nagasawa, H.; Fujimura, C.; Koyanagi, E.; Tomoshige, S.; Ishikawa, M.; Sato, S. Switching of Photocatalytic Tyrosine/Histidine Labeling and Application to Photocatalytic Proximity Labeling. Int. J. Mol. Sci. 2022, 23, 11622. https://doi.org/10.3390/ijms231911622
Nakane K, Nagasawa H, Fujimura C, Koyanagi E, Tomoshige S, Ishikawa M, Sato S. Switching of Photocatalytic Tyrosine/Histidine Labeling and Application to Photocatalytic Proximity Labeling. International Journal of Molecular Sciences. 2022; 23(19):11622. https://doi.org/10.3390/ijms231911622
Chicago/Turabian StyleNakane, Keita, Haruto Nagasawa, Chizu Fujimura, Eri Koyanagi, Shusuke Tomoshige, Minoru Ishikawa, and Shinichi Sato. 2022. "Switching of Photocatalytic Tyrosine/Histidine Labeling and Application to Photocatalytic Proximity Labeling" International Journal of Molecular Sciences 23, no. 19: 11622. https://doi.org/10.3390/ijms231911622
APA StyleNakane, K., Nagasawa, H., Fujimura, C., Koyanagi, E., Tomoshige, S., Ishikawa, M., & Sato, S. (2022). Switching of Photocatalytic Tyrosine/Histidine Labeling and Application to Photocatalytic Proximity Labeling. International Journal of Molecular Sciences, 23(19), 11622. https://doi.org/10.3390/ijms231911622






