Abstract
The calcineurin B-like-interacting protein kinase (CIPK) protein family plays a key role in the plant calcium ion-mediated signal transduction pathway, which regulates a plant’s response to abiotic stress. Nitraria sibirica pall. (N. sibirica) is a halophyte with a strong tolerance for high salt environments, yet how it is able to deal with salt stress on a molecular level is still unknown. Due to their function as described in other plant species, CIPK genes are prime candidates for a role in salt stress signaling in N. sibirica. In this study, we identified and analyzed the phylogenetic makeup and gene expression of the N. sibirica CIPK gene family. A total of 14 CIPKs were identified from the N. sibirica genome and were clustered into seven groups based on their phylogeny. The promoters of NsCIPK genes contained multiple elements involved in hormonal and stress response. Synteny analysis identified a total of three pairs of synteny relationships between NsCIPK genes. Each gene showed its own specific expression pattern across different tissues, with the overall expression of CIPK6 being the lowest, and that of CIPK20 being the highest. Almost all CIPK genes tended to respond to salt, drought, and cold stress, but with different sensitivity levels. In this study, we have provided a general description of the NsCIPK gene family and its expression, which will be of great significance for further understanding of the NsCIPK gene family function.
1. Introduction
Currently, the global ecological environment is progressively deteriorating. Drought and an increasing saline-alkali concentration lead to soil deterioration. More than 932.2 M ha of soil around the world have been affected by saltification and alkalization leading up to the year 2016 [1]. Most plants are sessile, and major changes in their surrounding environment will dramatically affect their growth and development. Climate change and environmental degradation lead to severe abiotic stress in plants [2,3], resulting in significant damage to plant life, both affecting their survival and limiting their global distribution.
In order to survive in harsh environments, plants have evolved a multitude of molecular mechanisms to adapt their physiology, the Ca2+ signaling pathway being one of the most important pathways involved in their response to abiotic stress [4]. When plants suffer from abiotic stress, the Ca2+ concentration in plants increases. Ca2+ signal receptors sense the change in Ca2+ concentration and activate related protective mechanisms by regulating downstream gene expression to avoid sustained cellular damage. Among the key Ca2+ receptors are Calmodulin (CaM) [5], Calmodulin-like protein (CMLs) [6], Calcium-dependent protein kinases (CDPKs) [7], and calcineurin B-like protein (CBLs) [8]. CBLs physically interact with CBL-Interacting Protein Kinases (CIPKs) to facilitate signal transduction, which is one of the Ca2+ signal transduction pathways that has been studied extensively [9]. For example, CBL4 and CBL10-CIPK24 (SOS2)-NHX7 (SOS1) interaction networks jointly regulate the salt stress response pathway [10,11]. CBL1 and CIPK7 are involved in cold stress response [12]. CIPK14 interacts with CBL1 to regulate the response to drought stress in pigeon pea [13]. In addition, CIPK genes are also involved in the regulation of responses to further stresses such as high pH [14], low potassium [15] and abscisic acid (ABA) stress [16]. Collectively, CIPKs play an important role in plant abiotic stress response pathways.
The CIPK genes encode serine/ threonine-protein kinases, which contain two key domains; the N-terminal catalytic/kinase domain and the C-terminal NAF/FISL domain. The N-terminal kinase region can be phosphorylated to enhance kinase activity, but the C-terminal NAF/FISL inhibits kinase activity [17]. However, the NAF/FISL domain of CIPK protein kinases can be converted into a protein kinase activator by physically binding with the CBL gene [18]. Because of their demonstrated importance as abiotic stress mitigators in plants, CIPK genes have been identified and studied in multiple plant species. For example, 26, 34, and 43 CIPK genes were identified in Arabidopsis, rice, and maize, respectively [19,20,21,22].
N. sibirica is a shrub plant that is placed in the Nitraria Linnaeus genus and is mainly distributed in saline-alkali arid areas as it can withstand very severe salt and drought damage. N. sibirica’s saline-alkali tolerance gives it the potential to be a pioneer species in saline-alkali soil remediation. However, the molecular mechanisms underlying the tolerance of N. sibirica to adverse environmental conditions such as drought and salinity have not yet been elucidated, and whether CIPKs participate in such abiotic stress responses is unclear as well. Therefore, in this study, we characterized the N. sibirica CIPK gene family, analyzing their expression patterns in response to salt, drought, and cold treatments. We identified a total of 14 CIPKs from the N. sibirica genome. Their expression showed distinct responses to salt, drought, and cold stress, indicating that NsCIPKs may play a role in the molecular response to these abiotic stresses in N. sibirica. These results underscore the potential of NsCIPKs as antibiotic stress regulators in N. sibirica and serve as a solid basis from which to further study their function.
2. Result
2.1. Identification and Chromosomal Location of the N. sibirica CIPK Gene Family
In order to identify CIPK genes in N. sibirica, we used HMMER software to scan the N. sibirica genome library using both the CIPK kinase and NAF/FLSL domains. We selected as candidate genes those genes that gave a hit using either domain. In addition, we used a BLASTP algorithm to search for potential NsCIPKs, taking AtCIPKs as input sequence. The results from both the HMMER and BLASTP search were combined, and we used the SMART database and the conserved domain database to further determine whether candidate genes were indeed NsCIPKs. In total, 14 CIPK genes were identified in N. sibirica (Supplementary File S1). To identify the distribution of these identified CIPKs across the whole genome, we mapped them to the chromosome level according to their ID numbers and genome data. We were able to map 12 NsCIPK genes directly to chromosomes, which were CHR3, CHR6, CHR7, CHR9, CHR10, and CHR11, respectively, while NsCIPK1-1 and NsCIPK1-2 were mapped to sequence scaffolds (scaffold80 and scaffold97) (Figure S1). Then, we performed all against all blastp searches between the identified candidate NsCIPKs and AtCIPKs. Based on their sequence homology to AtCIPKs, the NsCIPK genes were named as NsCIPK1-1 to NsCIPK23, respectively.
We then continued to further characterize the NsCIPK proteins by determining the number of amino acids and relative molecular weight (MW) (Table 1). The number of amino acids in the N. sibirica CIPK proteins lies between 359 to 583, with the MW varying from 39.04 kDa to 64.42 kDa, and the PI index is between 5.7 to 9.33. We then performed transmembrane structure analysis and found that only NsCIPK12 has a putative transmembrane domain, while the other 13 NsCIPKs do not (Figure S2). We used Cell-PLOc2.0 software to predict subcellular localization and found that all NsCIPK proteins are potentially localized to the cytoplasm. Furthermore, NsCIPK1-2, NsCIPK11, NsCIPK15, and NsCIPK23 are predicted to be localized to both the cytoplasm and the nucleus (Table 1).

Table 1.
Physicochemical properties of NsCIPK proteins.
2.2. Phylogeny and Synteny Analysis of the CIPK Gene Family
To elucidate the evolutionary relationship of the NsCIPK gene family with other species, we used the amino acid sequences of AtCIPKs, OsCIPKs, and NsCIPKs (Table S1) to construct an unrooted phylogenetic tree. As shown in Figure 1, a total of 73 CIPKs could be divided into 7 groups. Among them, the largest is group G which contains 19 members, while groups A, D, and E have the smallest distribution with four members. The remaining groups B, C, and F have 16, 16, and 10 members, respectively (Figure 1). These results indicate that the functions of CIPKs may have diverged during evolution. We found no CIPK gene in groups A and E, which may indicate that CIPKs in this branch did not diverge or become lost during the evolution of N. sibirica.
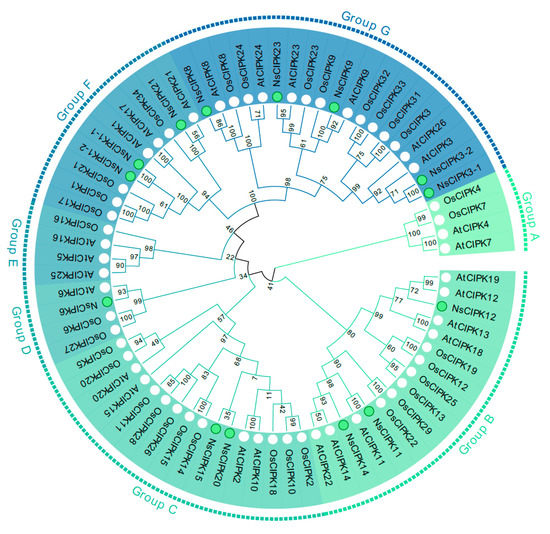
Figure 1.
Phylogenetic relationships between CIPKs from Arabidopsis, rice, and N. sibirica. Genes marked with a green dot are NsCIPKs; those marked with a white dot are AtCIPK and OsCIPK.
We found that the number of CIPKs in Arabidopsis and rice is higher than that in N. sibirica. In order to verify whether some of the CIPKs have been lost during N. sibirica evolution, we used CIPKs from additional species (Table S2) to draw a more extensive phylogenetic tree (Figure S3). This tree showed that, although N. sibirica has only 14 CIPKs, they are distributed on almost every branch of the phylogenetic tree. There is no clustering of NsCIPK on CIPK4 or 7, consistent with a phylogenetic tree with only three species (Figure 1). In addition, the synteny analysis between NsCIPKs also shows that there are only three collinear gene pairs (Figure 2), which indicates that the internal collinearity of CIPK is not strong. These results may indicate that the CIPK gene family is not expanded in N. sibirica.
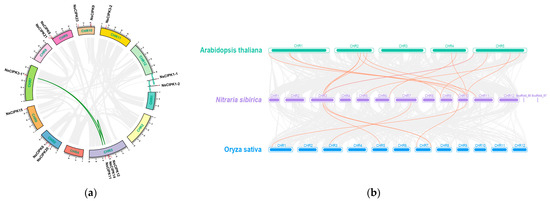
Figure 2.
Genome-wide synteny analysis of CIPK gene family among N. sibirica and other three species: (a) inter-chromosomal relationships of CIPKs in N. sibirica (the links on the green curve indicate synteny relationships between genes); (b) synteny analyses between the CIPKs of N. sibirica, Arabidopsis, and Rice. The green, purple and blue chromosome level genomes belong to Arabidopsis, N. sibirica, and rice, respectively. The orange lines represent a syntenic relationship between two genes.
The presence of collinearity between genes in different species often indicates the similarity of gene functions. Therefore, we performed synteny analysis on the CIPK gene families of N. sibirica, Arabidopsis, and rice. A total of 15 pairs of syntenic genes were found between Arabidopsis and N. sibirica. However, there were only six syntenic genes between N. sibirica and rice. We found more syntenic gene pairs between NsCIPKs and AtCIPKs than between NsCIPKs and OsCIPKs, indicating that NsCIPKs and AtCIPKs are more closely related to each other than they are to OsCIPKs (Figure 2).
To analyze whether the CIPK gene family had duplication events over a long evolutionary history, we searched for duplications using McScanX software [23]. However, we could not detect any segmental duplications in NsCIPKs. Next, we performed a synteny analysis to look for synteny between NsCIPKs. A total of three pairs of synteny relationships were identified in these NsCIPKs, namely, NsCIPK14 and NsCIPK11, NsCIPK11 and NsCIPK3-1, and NsCIPK14 and NsCIPK3-1, respectively (Figure 2). This suggests that segment duplications may be the main expansion mode of the CIPK gene family.
2.3. NsCIPKs Contain Multiple Conserved Motifs
In order to further study the structure of these CIPK genes, we analyzed their conservative motifs, domain distribution and gene structure. Via domain analysis, we found that the kinase domains of NsCIPKs are all located near the N-terminus, while the NAF domains are all located at the C-terminus of the protein (Figure 3B and Figure S4). A total of 11 major motifs were identified in these NsCIPKs (Figure 3A and Figure S5). Among them, motif 1, motif 7, and motif 8 exist in all NsCIPK genes. The remaining motifs were present in most CIPK genes, which suggest that these CIPKs may have similar functions. Analysis of the NsCIPK gene structure showed that they can be divided into two categories, depending on whether introns are present or not (Figure 3C). NsCIPK11, NsCIPK14, and NsCIPK15 have no intron structure, while the remaining NsCIPKs have one or multiple introns, with the total gene length remaining under 9000 bp.
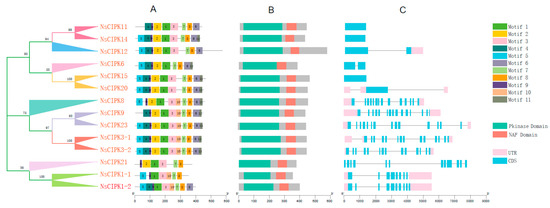
Figure 3.
Conserved motifs, domain distribution and gene structure of N.sibirica CIPK genes. (A) Conserved motif distribution of NsCIPK genes. (B) Domain distribution of NsCIPK genes. (C) Gene structure of NsCIPK gene families.
2.4. NsCIPK Promoters Contain Multiple Stress Responsive cis Regulatory Elements
A cis regulatory element is a target nucleotide sequence that can be bound by different trans-acting factors, which are involved in the regulation of gene expression. We used PlantCare online tools to identify cis regulatory elements within the sequence region 2500 bp upstream of the start codon of each NsCIPK gene. On average, we could identify ten cis regulatory elements within these regions (Figure 4). These cis regulatory elements are mainly related to hormone response, salt, low temperature, and drought stress, which suggests that NsCIPKs may respond to hormones and abiotic stress.
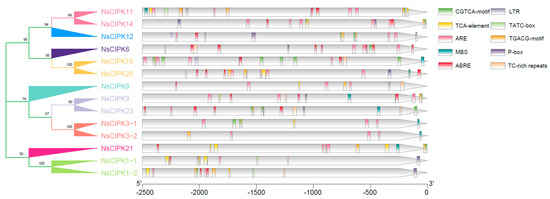
Figure 4.
cis regulatory elements analysis of N. sibirica NsCIPK gene upstream regions. The colored squares represent the different cis regulatory elements.
2.5. NsCIPK Gene Expression Analysis across Different N. sibirica Tissues
To explore the expression of NsCIPKs in different tissues, we collected the root, stem, and leaves of two-month-old seedlings of N. sibirica, isolated RNA, and performed qRT-PCR. Since the mRNA sequence of NsCIPK1-1 is highly similar to NsCIPK1-2, no specific primers could be designed. Therefore, NsCIPK1-1 and NsCIPK1-2 are collectively referred as NsCIPK1. We designed customized qRT-PCR primers for each gene (Table S3), and the result showed that there were clear differences between the expression levels of individual NsCIPK genes in different tissues (Figure 5); NsCIPK12 shows the highest expression in the root, while NsCIPK20 is the most abundant in the stem and leaf. In general, CIPK3-2, CIPK12, CIPK20, CIPK21, and CIPK23 show comparatively high expression levels in all tissues analyzed, while the rest of the genes showed a lower expression level. Notably, the expression of CIPK6 in roots, stems, and leaves was at an extremely low level compared with other CIPK genes (about 1/1000 of the CIPK1 expression level). These results suggest that there are dedicated CIPK genes functioning in different tissues while some CIPK genes may only act in the event of special environmental conditions.
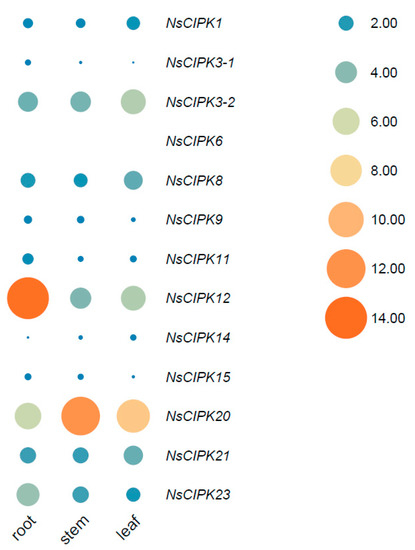
Figure 5.
Comparative analysis of the relative expression of CIPK genes in different tissues of N. sibirica. Different circle sizes and colors indicate the relative expression size. The expression of CIPK1 in the root was used as a reference for all relative quantification.
2.6. Expression Patterns of NsCIPK Genes in Response to Different Abiotic Stresses
Studies in other species have found that CIPK families generally respond to abiotic stress [24]. To explore whether NsCIPKs respond to abiotic stress as well, we used treatments with 300 mmol/L NaCl, 20% PEG6000 and 4 °C to simulate salt, drought and cold stress, respectively, after which we detected the expression level of the NsCIPK gene family by quantitative real-time PCR. As shown in Figure 6, for salt stress, we found that most of the CIPK genes were induced to express, although the period of expression is different. However, NsCIPK3-1 and NsCIPK12 were down-regulated in some periods, and only NsCIPK15 did not show an obvious upward or downward trend. For drought stress, the relative expression levels of NsCIPK1-1, NsCIPK3-1, NsCIPK3-2, NsCIPK6, NsCIPK11, NsCIPK12, NsCIPK14, and NsCIPK20 increased in different degrees, indicating that they may play a role in drought stress. In contrast, the rest of the CIPK genes showed a downward trend. In cold stress, most NsCIPKs increased and changed little, but only NsCIPK8 expression tended to decrease during stress. These analyses indicated that different NsCIPK had different sensitivities to salt, drought, or cold stress environments, but in general, NsCIPKs responded to abiotic stress processes such as salt, drought, and cold stress, suggesting that they may play a key role in these abiotic stress processes.
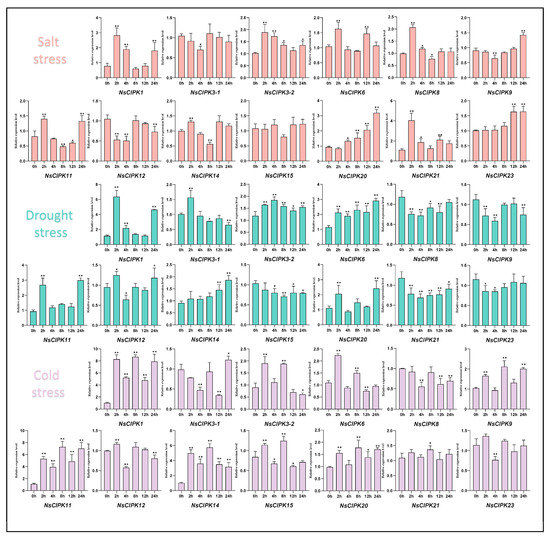
Figure 6.
Expression analysis of NsCIPKs in response to salt, drought, and cold stress. A student’s t-test was used to analyze statistically significant differences (* p < 0.05, ** p < 0.01).
3. Discussion
Salinization and aridification are becoming more and more severe, limiting the further development of global agriculture and affecting food output [25]. Phytoremediation has always been an effective measure for soil remediation because it is a fully natural and low-cost approach. Salt-alkali tolerant and drought-tolerant plants are especially suited for this purpose. N. sibirica has excellent tolerance for drought and saline-alkali environments and is widely distributed in the east of Eurasia, making it an ideal plant for the treatment of salinization and alkalinity.
The calcium signaling pathway is one of the key molecular pathways involved in the biological response to an adverse environment and includes multiple typical response pathways such as the CAM, CDPK, and CBL-CIPK pathways. CIPK genes regulate a plant’s response to Ca2+ by binding the CBL gene. CIPK binds CBL through its own C-terminal NAF domain, making the NAF domain indispensable for CIPK function [17]. A NAF domain was found in all the CIPK genes identified in this study (Figure S4), indicating that all these identified CIPK genes have the potential to interact with the CBL gene. CBL-CIPK is a unique pathway in plants and plays an important role in plant stress resistance, such as resistance to cold, drought, and salt stress. For example, ScCBL-ScCIPK signaling network regulates the response process of Solanum commersonii to cold stress [26]; ScCIPKs were involved in the response of cotton to drought stress [27]; TaCIPKs were responsive to salt stress in Triticum aestivum [28]. In this study, the NsCIPK genes were systematically analyzed at a genome-wide level in N. sibirica, which will be beneficial for further understanding the function of the CIPKs and the molecular mechanisms underlying salt tolerance and drought resistance in N. sibirica.
The number of CIPK gene family members often varies between plant species; in this study, fourteen NsCIPK genes have been identified, which is relatively low compared to the number of CIPK genes in Arabidopsis (26), Oryza sativa (34), Vitis vinifera (20), and Zea mays (36). From the phylogenetic tree showing the interrelatedness between NsCIPK, AtCIPKs, and OsCIPKs, we found there was no clustering of NsCIPKs on CIPK4 or 7. This may indicate that these CIPK genes were lost during N. sibirica evolution. To further investigate whether CIPKs were lost during N. sibirica’s long evolutionary history, we constructed phylogenetic trees with more species added (Figure S3) and found that NsCIPKs were distributed in almost every branch, which may indicate that a lack of extensive expansion of NsCIPKs may be the reason for the low number of CIPKs in N. sibirica.
Genes tend to have tissue-specific expression, related to their function. In our expression analysis, we found that the genes NsCIPK3-2, NsCIPK12, NsCIPK14, and NsCIPK20 are expressed in a tissue specific manner. By comparison, NsCIPK3-1, NsCIPK6, NsCIPK9, and NsCIPK15 are always lowly expressed (Figure 5). The level of basal gene expression does not necessarily indicate whether a gene is functional or not; rather, it is the rise or fall of expression in a given environment that may determine the potential function of a gene. Some CIPKs, such as NsCIPK1 and NsCIPK6, are expressed at a very low level during normal growth, but are strongly induced during stress, suggesting that these genes are likely transducing Ca2+ signals when plants are stressed.
We also noticed that some CIPK genes respond to multiple abiotic stresses simultaneously, while others only respond to one specific abiotic stress. These results suggest that there may be functional differentiation between CIPK genes.
ABA has been shown to play an important role in plant tolerance to abiotic stress [29,30,31]. Several studies have found that CIPK genes play an important role in the ABA signaling response. For example, AtCIPK1 plays a role in the ABA response pathways in Arabidopsis, as mutation of AtCIPK1 results in an abnormal ABA response and affects plant tolerance to abiotic stress [32]. There are also studies showing that AtCIPK3 is involved in cold stress and ABA response. AtCIPK6 plays a role in plant tolerance to salt or osmotic stress [33,34]. In this study, we found that NsCIPK1, NsCIPK3-1, and NsCIPK3-2 expression was significantly induced by salt, drought, and cold stress, suggesting that a similar response mechanism may exist in N. sibirica. NsCIPK6 has similar expression trends to NsCIPK3, although CIPK6 has not been reported to regulate cold stress, even though it has been reported that CIPK6 can be induced by cold stress [35], which is consistent with what we found in this study. This suggests that NsCIPK6 may have a function in plant cold stress response, motivating further functional studies. In addition, even NsCIPK genes with less pronounced differential expression during abiotic stress did contain ABA responsive elements (ABRE) in their promoter region, which may indicate that ABA activates these CIPKs during abiotic stress in N. sibirica. It is important for us to further study the relationship of NsCIPKs and ABA in the process of abiotic stress response. The function of NsCIPK genes cannot be predicted only by their expression level and still needs to be verified by further experiments. It is of great significance to further study the function of CIPK genes in N. sibirica as they may have an important function in the molecular pathways underlying its profound abiotic stress resistance.
4. Materials and Methods
4.1. Plant Materials and Abiotic Stress Treatments
N. sibirica seeds were collected in Dengkou County, Inner Mongolia, China. Seeds were mixed with wet sand and placed at 4 °C to vernalize for two months. After the vernalized seeds germinated, they were planted in a soil mix of peat soil: perlite, with a ratio of 4:1. Plants were cultivated in the green-house under a 16 h-light/8 h dark- light cycle and 60% air humidity at 23 °C. After a growth period of around two months, the plant materials were used for expression analysis. For abiotic stress treatments, a 300 mM NaCl solution, 20% PEG 6000, and 4 °C were used to simulate salt, drought and cold stress, respectively. Plant materials were collected at 0, 2, 4, 8, 12, and 24 h after corresponding treatments and were quickly put into liquid nitrogen and transferred to −80 °C refrigerator for storage until RNA was extracted.
4.2. Identification of CIPK Genes in N. sibirica
N. sibirica genomic data were obtained from the unpublished complete genomic sequence that we obtained previously in our laboratory. CIPK gene sequences from Arabidopsis were extracted from the Arabidopsis genome (TAIR.10) according to their annotation ID. Subsequently, the Protein Kinase domain (PF00069) and NAF/FLSL (PF03822) domain from the Pfam database [36] (http://pfam.xfam.org/ (accessed on 21 May 2022)) were used to scan the putative protein sequence libraries of N. sibirica using HMMER software (v3.0) with an e-value of 10−5. Blastp software (v2.9) and SMART [37] (http://smart.embl-heidelberg.de/ (accessed on 21 May 2022)), and CDD search tools [38] (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi/ (accessed on 21 May 2022)) were then used to manually confirm the extracted sequences. Finally, these NsCIPKs were named according to their homology with AtCIPK proteins. The expasy tool [39] (https://prosite.expasy.org/ (accessed on 21 May 2022)) was used for the identification of the basic physical properties including MWs (molecular weights). The transmembrane structure was analyzed using the online analysis tool TMHMM2.0 [40] (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 (accessed on 21 May 2022)). The subcellular localization of NsCIPK proteins was predicted by using the online tool Cell-PLoc 2.0 [41] (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/ (accessed on 21 May 2022)).
4.3. Motifs and Gene Structure Analysis
Conserved gene motifs were analyzed using MEME online tools [42] (https://meme-suite.org/meme/tools/meme/ (accessed on 21 May 2022)). Pfam sequence search (http://pfam.xfam.org/ (accessed on 21 May 2022)) was used to analyze the protein kinase domain and NAF/FLSL domain of predicted genes, and gene structure was analyzed by the GSDS [43] (http://gsds.cbi.pku.edu.cn/ (accessed on 21 May 2022)) online analysis tool.
4.4. Promoter cis Regulatory Element Prediction
Sequences 2500 bp in length located upstream of the coding region of putative genes were extracted for cis regulatory element analysis using PlantCare online tool [44] (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 21 May 2022)) and visualized by Tbtools software [45].
4.5. Chromosomal Location and Synteny Analysis
Chromosomal location was analyzed and visualized by Tbtools [45]. Criteria for gene duplication refer to the methods previously reported [23], and the result was visualized by Tbtools [42].
4.6. Phylogenetic and Multiple Alignment Analysis
The phylogeny of CIPKs from N. sibirica, rice, and Arabidopsis was analyzed using MEGA X v10.1.8 software (Temple, Philadelphia, PA, USA). The Muscle method was used to align the CIPK amino acid sequences [46], and the NJ (Neighbor-joining) method was used to construct the phylogenetic tree. The bootstrap was repeated 1000 times. The phylogenetic tree was visualized using Evolview software [47]. DNAMAN v9.0 (Lynnon Corporation, San Ramon, CA, USA) software was used for multi-fragment alignment of amino acid sequences.
4.7. Expression Analysis of CIPK Genes in N. sibirica
Total RNA was extracted from plant materials using an RNA extraction kit (Promega, Shanghai, China). cDNA was synthesized using a reverse transcription kit (Vazyme, Nanjing, China). Quantitative real-time PCR (qRT-PCR) was performed using a Lightcyler 480II (Roche, Basel, Switzerland) and the AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China). Primers for qRT-PCR were designed using the NCBI online primer design tool [48] (https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 21 May 2022)). The primers used in this experiment are shown in Table S3. Three biological replicates of each sample were used for quantitative real-time PCR. The relative expression level was calculated using the 2−ΔΔCT method [49], and the graph was finally plotted using GraphPad Prism (v8.01).
5. Conclusions
In this study, we identified and analyzed the phylogenetic makeup and gene expression of the N. sibirica CIPK gene family. A total of 14 CIPKs were identified from the N. sibirica genome and were clustered into seven groups based on their phylogeny. The promoters of NsCIPK genes contained multiple elements involved in hormonal and stress response. Synteny analysis identified a total of three pairs of synteny relationships between NsCIPK genes. Each gene showed its own specific expression pattern across different tissues, with the overall expression of CIPK6 being the lowest, and that of CIPK20 being the highest. Almost all CIPK genes tended to respond to salt, drought, and cold stress, but with different sensitivity levels. In this study, we have provided a general description of the NsCIPK gene family and its expression, which will be of great significance for further understanding of the NsCIPK gene family function.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911599/s1.
Author Contributions
Conceptualization, Z.L.; methodology and validation, H.F., L.Z. and J.Z.; formal analysis, X.L.; investigation, L.L., Y.L.; data curation; writing—review and editing, Z.L.; visualization, Z.L.; supervision, J.S.; project administration, J.C.; funding acquisition, T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Nature Science Foundation of (32071784, 31770715) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data in this study can be requested from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef]
- Etesami, H.; Jeong, B.R. Silicon (Si): Review and future prospects on the action mechanisms in alleviating biotic and abiotic stresses in plants. Ecotoxicol. Environ. Saf. 2018, 147, 881–896. [Google Scholar] [CrossRef]
- Kumar, M.; Kour, D.; Yadav, A.N.; Saxena, R.; Rai, P.K.; Jyoti, A.; Tomar, R.S. Biodiversity of methylotrophic microbial communities and their potential role in mitigation of abiotic stresses in plants. Biologia 2019, 74, 287–308. [Google Scholar] [CrossRef]
- Steinhorst, L.; Kudla, J. Calcium—A central regulator of pollen germination and tube growth. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 1573–1581. [Google Scholar] [CrossRef]
- Chin, D.; Means, A.R. Calmodulin: A Prototypical Calcium Sensor; Elsevier Ltd.: London, UK, 2000; Volume 10, pp. 322–328. [Google Scholar]
- Perochon, A.; Aldon, D.; Galaud, J.; Ranty, B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 2011, 93, 2048–2053. [Google Scholar] [CrossRef]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B–like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef] [PubMed]
- Batistič, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta (BBA) Mol.Cell Res. 2009, 1793, 985–992. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Huang, C.; Ding, S.; Zhang, H.; Du, H.; An, L. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 2011, 181, 57–64. [Google Scholar] [CrossRef]
- Meng, D.; Dong, B.; Niu, L.; Song, Z.; Wang, L.; Amin, R.; Cao, H.; Li, H.; Yang, Q.; Fu, Y. The pigeon pea CcCIPK14-CcCBL1 pair positively modulates drought tolerance by enhancing flavonoid biosynthesis. Plant J. 2021, 106, 1278–1297. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef]
- Li, L.; Kim, B.; Cheong, Y.H.; Pandey, G.K.; Luan, S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 12625–12630. [Google Scholar] [CrossRef]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.; Luan, S. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 2008, 1, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063. [Google Scholar] [CrossRef]
- Tang, R.; Wang, C.; Li, K.; Luan, S. The CBL–CIPK calcium signaling network: Unified paradigm from 20 years of discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef]
- Chen, X.; Gu, Z.; Xin, D.; Hao, L.; Liu, C.; Huang, J.; Ma, B.; Zhang, H. Identification and characterization of putative CIPK genes in maize. J. Genet. Genom. 2011, 38, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, P.; Sanyal, S.K.; Tokas, I.; Yadav, A.K.; Pandey, A.; Kapoor, S.; Pandey, G.K. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 2014, 56, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Liu, H.; Schofield, A.; Muise-Hennessey, A.; Stone, S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin–proteasome system. J. Exp. Bot. 2013, 64, 2779–2791. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.; Bouman, P.; Li, W. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, X.N.; Liu, S.; Yu, A.; Yang, C.; Chen, X.; Liu, J.; Wang, A. Identification and functional analysis of tomato CIPK gene family. Int. J. Mol. Sci. 2020, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W. Eco-environmental impact of inter-basin water transfer projects: A review. Environ. Sci. Pollut. Res. 2016, 23, 12867–12879. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; D’Amelia, V.; Carputo, D.; Aversano, R. Genes involved in stress signals: The CBLs-CIPKs network in cold tolerant Solanum commersonii. Biol. Plant. 2019, 63, 699–709. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, B.; Deng, J.; Chen, L.; Ullah, A.; Yang, X. Genome-wide analysis of CBL and CIPK family genes in cotton: Conserved structures with divergent interactions and expression. Physiol. Mol. Biol. Plants 2021, 27, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Paul, S.; Basu, S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013, 32, 985–1006. [Google Scholar] [CrossRef]
- D’Angelo, C.; Weinl, S.; Batistic, O.; Pandey, G.K.; Cheong, Y.H.; Schültke, S.; Albrecht, V.; Ehlert, B.; Schulz, B.; Harter, K. Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006, 48, 857–872. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Q.; Zhou, L.; Ren, F.; Li, D.; Li, X. Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 2013, 40, 4759–4767. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ren, F.; Zhou, L.; Wang, Q.; Zhong, H.; Li, X. The Brassica napus calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 2012, 63, 6211–6222. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Parasuraman, B.; Laxmi, A.; Chattopadhyay, D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009, 58, 778–790. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Heijne, G.V.; Sonnhammer, E. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes-ScienceDirect. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Chou, K.; Shen, H. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).