Mutation and Interaction Analysis of the Glycoprotein D and L and Thymidine Kinase of Pseudorabies Virus
Abstract
1. Introduction
2. Results
2.1. The Amino Acid Sequences of PRV Strains in China Are Different from That Reported Abroad
2.2. Mutations in PRV gD and gL Proteins May Affect the Recognition of Antigenic Epitopes
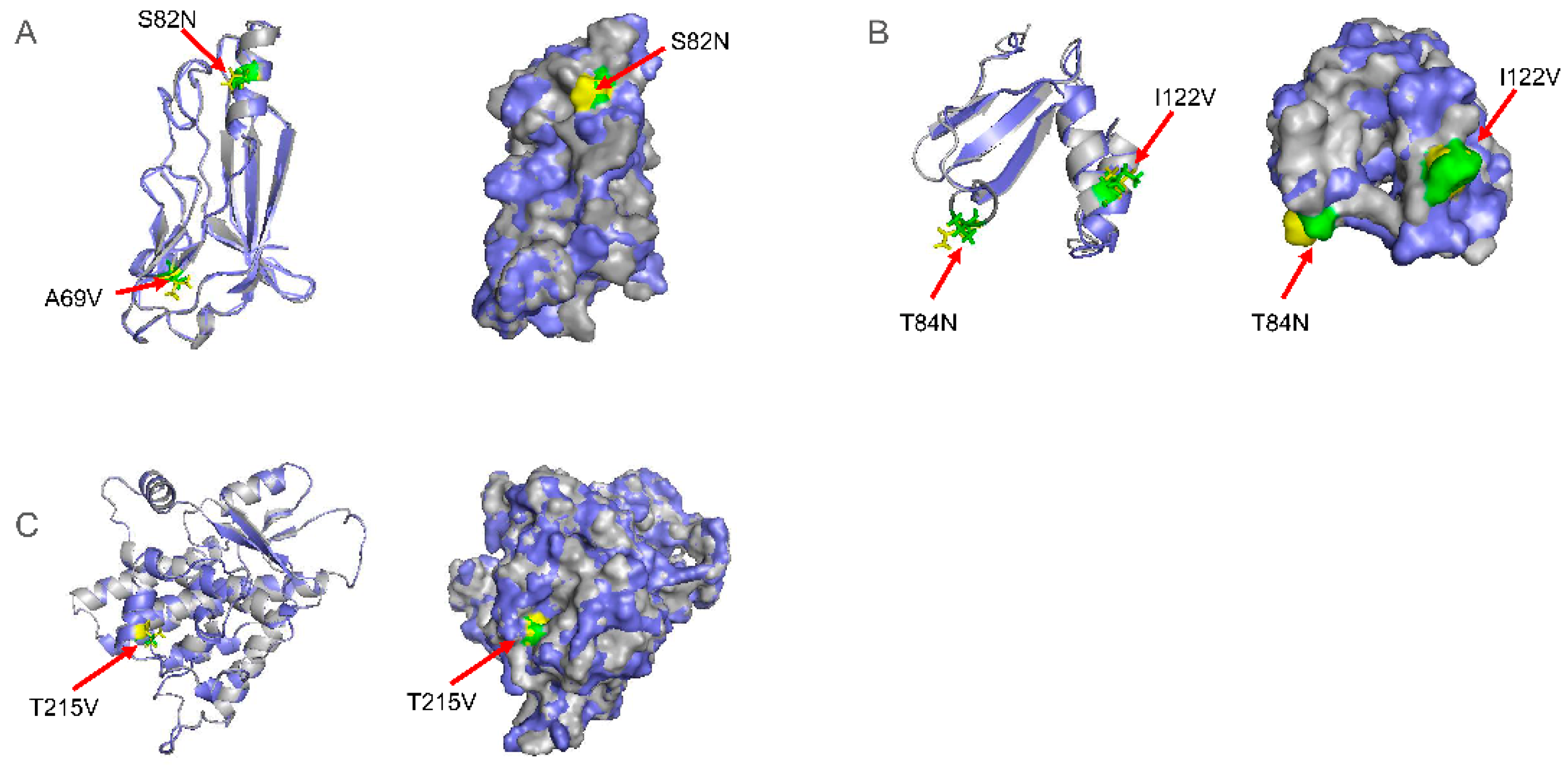
2.3. The Mutations of gD, gL, and TK Proteins Did Not Affect Their Spatial Conformation but May Affect Their Function
2.4. PRV JL-CC gD Protein Can Bind to Human Nectin-1, Nectin-2, and HS
3. Discussion
4. Materials and Methods
4.1. Virus Strain Information and Multiple Alignments
4.2. Phylogenetic Tree
4.3. Functional Domain and Epitope Analysis of gD, gL, and TK Proteins
4.4. Three-Dimensional (3D) Structures Modeling
4.5. The Mutation Analysis of the gD, gL, and TK Proteins
4.6. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peng, Z.; Ouyang, T.; Pang, D.; Ma, T.; Chen, X.; Guo, N.; Chen, F.; Yuan, L.; Ouyang, H.; Ren, L. Pseudorabies virus can escape from CRISPR-Cas9-mediated inhibition. Virus Res. 2016, 223, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kuang, Y.; Li, Y.; Guo, H.; Zhou, C.; Guo, S.; Tan, C.; Wu, B.; Chen, H.; Wang, X. The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans. Viruses 2022, 14, 1463. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Chen, J.; Li, G.; Wan, C.; Li, Y.; Peng, L.; Fang, R.; Peng, Y.; Ye, C. A Comparison of Pseudorabies Virus Latency to Other alpha-Herpesvirinae Subfamily Members. Viruses 2022, 14, 1386. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wu, Z.; Liu, J.; Ji, Q.; Ju, C. The Role of Latency-Associated Transcripts in the Latent Infection of Pseudorabies Virus. Viruses 2022, 14, 1379. [Google Scholar] [CrossRef]
- Lu, J.J.; Yuan, W.Z.; Zhu, Y.P.; Hou, S.H.; Wang, X.J. Latent pseudorabies virus infection in medulla oblongata from quarantined pigs. Transbound. Emerg. Dis. 2021, 68, 543–551. [Google Scholar] [CrossRef]
- Zheng, H.H.; Fu, P.F.; Chen, H.Y.; Wang, Z.Y. Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses 2022, 14, 1638. [Google Scholar] [CrossRef]
- Xu, L.; Wei, J.F.; Zhao, J.; Xu, S.Y.; Lee, F.Q.; Nie, M.C.; Xu, Z.W.; Zhou, Y.C.; Zhu, L. The Immunity Protection of Central Nervous System Induced by Pseudorabies Virus DelgI/gE/TK in Mice. Front. Microbiol. 2022, 13, 862907. [Google Scholar] [CrossRef]
- Kit, S.; Kit, M.; Pirtle, E.C. Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. Am. J. Vet. Res. 1985, 46, 1359–1367. [Google Scholar]
- Yin, H.; Li, Z.; Zhang, J.; Huang, J.; Kang, H.; Tian, J.; Qu, L. Construction of a US7/US8/UL23/US3-deleted recombinant pseudorabies virus and evaluation of its pathogenicity in dogs. Vet. Microbiol. 2020, 240, 108543. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Immunobiology of pseudorabies (Aujeszky’s disease). Vet. Immunol. Immunopathol. 1996, 54, 221–229. [Google Scholar] [CrossRef]
- Zuckermann, F.A.; Zsak, L.; Mettenleiter, T.C.; Ben-Porat, T. Pseudorabies virus glycoprotein gIII is a major target antigen for murine and swine virus-specific cytotoxic T lymphocytes. J. Virol. 1990, 64, 802–812. [Google Scholar] [CrossRef]
- Spear, P.G.; Longnecker, R. Herpesvirus entry: An update. J. Virol. 2003, 77, 10179–10185. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Chen, Z.; Yang, F.; Ye, F.; Lin, S.; He, B.; Cheng, Y.; Wang, J.; Chen, Z.; Lin, X.; et al. Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor. Sci. Adv. 2020, 6, eaba5147. [Google Scholar] [CrossRef] [PubMed]
- Vallbracht, M.; Backovic, M.; Klupp, B.G.; Rey, F.A.; Mettenleiter, T.C. Common characteristics and unique features: A comparison of the fusion machinery of the alphaherpesviruses Pseudorabies virus and Herpes simplex virus. Adv. Virus Res. 2019, 104, 225–281. [Google Scholar]
- Li, A.; Lu, G.; Qi, J.; Wu, L.; Tian, K.; Luo, T.; Shi, Y.; Yan, J.; Gao, G.F. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. PLoS Pathog. 2017, 13, e1006314. [Google Scholar] [CrossRef]
- Sharthiya, H.; Seng, C.; Van Kuppevelt, T.H.; Tiwari, V.; Fornaro, M. HSV-1 interaction to 3-O-sulfated heparan sulfate in mouse-derived DRG explant and profiles of inflammatory markers during virus infection. J. Neurovirol. 2017, 23, 483–491. [Google Scholar] [CrossRef]
- Choudhary, S.; Marquez, M.; Alencastro, F.; Spors, F.; Zhao, Y.; Tiwari, V. Herpes simplex virus type-1 (HSV-1) entry into human mesenchymal stem cells is heavily dependent on heparan sulfate. J. Biomed. Biotechnol. 2011, 2011, 264350. [Google Scholar] [CrossRef]
- Klupp, B.G.; Lomniczi, B.; Visser, N.; Fuchs, W.; Mettenleiter, T.C. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology 1995, 212, 466–473. [Google Scholar] [CrossRef][Green Version]
- Lomniczi, B.; Watanabe, S.; Ben-Porat, T.; Kaplan, A.S. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. J. Virol. 1987, 61, 796–801. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, Z.; Hu, D.; Zhang, Q.; Han, T.; Li, X.; Gu, X.; Yuan, L.; Zhang, S.; Wang, B.; et al. Pathogenic pseudorabies virus, China, 2012. Emerg. Infect. Dis. 2014, 20, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.J.; Jiang, Y.F.; Shan, T.L.; Gao, F.; Li, G.X.; Tong, G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cui, X.; Wang, X.; Wang, W.; Gao, S.; Liu, X.; Kai, Y.; Chen, C. Efficacy of the Bartha-K61 vaccine and a gE−/gI−/TK− prototype vaccine against variant porcine pseudorabies virus (vPRV) in piglets with sublethal challenge of vPRV. Res. Vet. Sci. 2020, 128, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zhang, Q.Z.; Tian, Z.J.; Zheng, H.; Zhao, K.; Liu, F.; Guo, J.C.; Tong, W.; Jiang, C.G.; Wang, S.J.; et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 2015, 483, 32–43. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; Zhang, Y.; Yu, H.; Zhao, Y.; Yi, A. Human Encephalitis Caused by Pseudorabies Virus in China: A Case Report and Systematic Review. Vector Borne Zoonotic Dis. 2022, 22, 391–396. [Google Scholar] [CrossRef]
- Yang, X.; Guan, H.; Li, C.; Li, Y.; Wang, S.; Zhao, X.; Zhao, Y.; Liu, Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int. J. Infect. Dis. 2019, 87, 92–99. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, X.X.; Zhang, G. Human PRV Infection in China: An Alarm to Accelerate Eradication of PRV in Domestic Pigs. Virol. Sin. 2021, 36, 823–828. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef]
- Wong, G.; Lu, J.; Zhang, W.; Gao, G.F. Pseudorabies virus: A neglected zoonotic pathogen in humans? Emerg. Microbes Infect. 2019, 8, 150–154. [Google Scholar] [CrossRef]
- Masot, A.J.; Gil, M.; Risco, D.; Jimenez, O.M.; Nunez, J.I.; Redondo, E. Pseudorabies virus infection (Aujeszky’s disease) in an Iberian lynx (Lynx pardinus) in Spain: A case report. BMC Vet. Res. 2017, 13, 6. [Google Scholar] [CrossRef]
- Denner, J.; Bigley, T.M.; Phan, T.L.; Zimmermann, C.; Zhou, X.; Kaufer, B.B. Comparative Analysis of Roseoloviruses in Humans, Pigs, Mice, and Other Species. Viruses 2019, 11, 1108. [Google Scholar] [CrossRef] [PubMed]
- Skinner, G.R.; Ahmad, A.; Davies, J.A. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp. Immunol. Microbiol. Infect. Dis. 2001, 24, 255–269. [Google Scholar] [CrossRef]
- Hurst, E.W. Studies on Pseudorabies (Infectious Bulbar Paralysis, Mad Itch: III. The Disease in the Rhesus Monkey, Macaca Mulatta. J. Exp. Med. 1936, 63, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhu, L.; Cai, Y.; Yan, J.; Fan, Y.; Lv, W.; Gong, S.; Yin, X.; Yang, X.; Sun, X.; et al. Immunogenicity and protective efficacy induced by an mRNA vaccine encoding gD antigen against pseudorabies virus infection. Vet. Microbiol. 2020, 251, 108886. [Google Scholar] [CrossRef]
- Wang, J.; Song, Z.; Ge, A.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Zheng, Y.; Fan, H.; et al. Safety and immunogenicity of an attenuated Chinese pseudorabies variant by dual deletion of TK&gE genes. BMC Vet. Res. 2018, 14, 287. [Google Scholar]
- Zhang, L.; Ruan, K.; Sang, G.; Xu, Z.; Tong, W.; Yu, H.; Shan, T.; Gao, F.; Li, L.; Kong, N.; et al. Tk-deleted Pseudorabies Virus Retains High Pathogenicity in Rats. J. Vet. Res. 2021, 65, 401–405. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, L.; Fu, Z.F. Effective cross-protection of a lyophilized live gE/gI/TK-deleted pseudorabies virus (PRV) vaccine against classical and variant PRV challenges. Vet. Microbiol. 2022, 267, 109387. [Google Scholar] [CrossRef]
- Yokoyama, N.; Maeda, K.; Tohya, Y.; Kawaguchi, Y.; Shin, Y.S.; Ono, M.; Ishiguro, S.; Fujikawa, Y.; Mikami, T. Pathogenicity and vaccine efficacy of a thymidine kinase-deficient mutant of feline herpesvirus type 1 in cats. Arch. Virol. 1996, 141, 481–494. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Zhang, C.; Freddolino, P.L.; Zhang, Y. COFACTOR: Improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res. 2017, 45, W291–W299. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef]


| Protein | Domain/Family | T Cell Epitopes | B Cell Epitopes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Bartha | hSD-1/2019 | JL-CC | Bartha | hSD-1/2019 | JL-CC | Bartha | hSD-1/2019 | JL-CC | |
| gD | 56–181 | A69V S82N | A69V S82N | Y35TESWQLTL43, G358TAMGALLV356, H230REVVNYWY238 | Q76VDRLLSEAV86, R165RLVSVDGVN174, I122FGRCRRRTT131, A70LISDPQVDR79, S169VDGVNILTD178 | S82N | S82N | ||
| gL | 70–138 | T84N I122V | T84N I122V | V86PSVVVKPY94, M83TAVTSVVV91 | H58PLLGLEPPV67 R44APRREELEW53 | A45H G62D | A45H G62D | ||
| TK | 10–278 | T215V | T215V | ||||||
| Clash | Buried Hydrophilic Introduced | Buried Charge Introduced and Switch | Secondary Structure Altered | Disallowed phi/psi | Buried Charge Replaced | Buried H-Bond Breakage | Cavity Altered | Buried/Exposed Switch | |
|---|---|---|---|---|---|---|---|---|---|
| gD A59V | +++ | +++ | +++ | NA | −−− | −−− | −−− | ++− | +−− |
| gD S82N | +−− | ++− | ++− | NA | −−− | ++− | ++− | −−− | ++− |
| gL T84N | +−− | +−− | +−− | NA | −−− | −−− | −−− | −−− | ++− |
| gL I122V | +−− | −−− | −−− | NA | +−− | −−− | −−− | −−− | −−− |
| TK T215V | −−− | +++ | ++− | NA | −−− | ++− | −−− | −−− | ++− |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Chen, S.; Zhang, L.; Zheng, J.; Niu, G.; Yang, L.; Zhang, X.; Ren, L. Mutation and Interaction Analysis of the Glycoprotein D and L and Thymidine Kinase of Pseudorabies Virus. Int. J. Mol. Sci. 2022, 23, 11597. https://doi.org/10.3390/ijms231911597
Li X, Chen S, Zhang L, Zheng J, Niu G, Yang L, Zhang X, Ren L. Mutation and Interaction Analysis of the Glycoprotein D and L and Thymidine Kinase of Pseudorabies Virus. International Journal of Molecular Sciences. 2022; 23(19):11597. https://doi.org/10.3390/ijms231911597
Chicago/Turabian StyleLi, Xue, Si Chen, Liying Zhang, Jiawei Zheng, Guyu Niu, Lin Yang, Xinwei Zhang, and Linzhu Ren. 2022. "Mutation and Interaction Analysis of the Glycoprotein D and L and Thymidine Kinase of Pseudorabies Virus" International Journal of Molecular Sciences 23, no. 19: 11597. https://doi.org/10.3390/ijms231911597
APA StyleLi, X., Chen, S., Zhang, L., Zheng, J., Niu, G., Yang, L., Zhang, X., & Ren, L. (2022). Mutation and Interaction Analysis of the Glycoprotein D and L and Thymidine Kinase of Pseudorabies Virus. International Journal of Molecular Sciences, 23(19), 11597. https://doi.org/10.3390/ijms231911597






