The Giardial Arginine Deiminase Participates in Giardia-Host Immunomodulation in a Structure-Dependent Fashion via Toll-like Receptors
Abstract
1. Introduction
2. Results
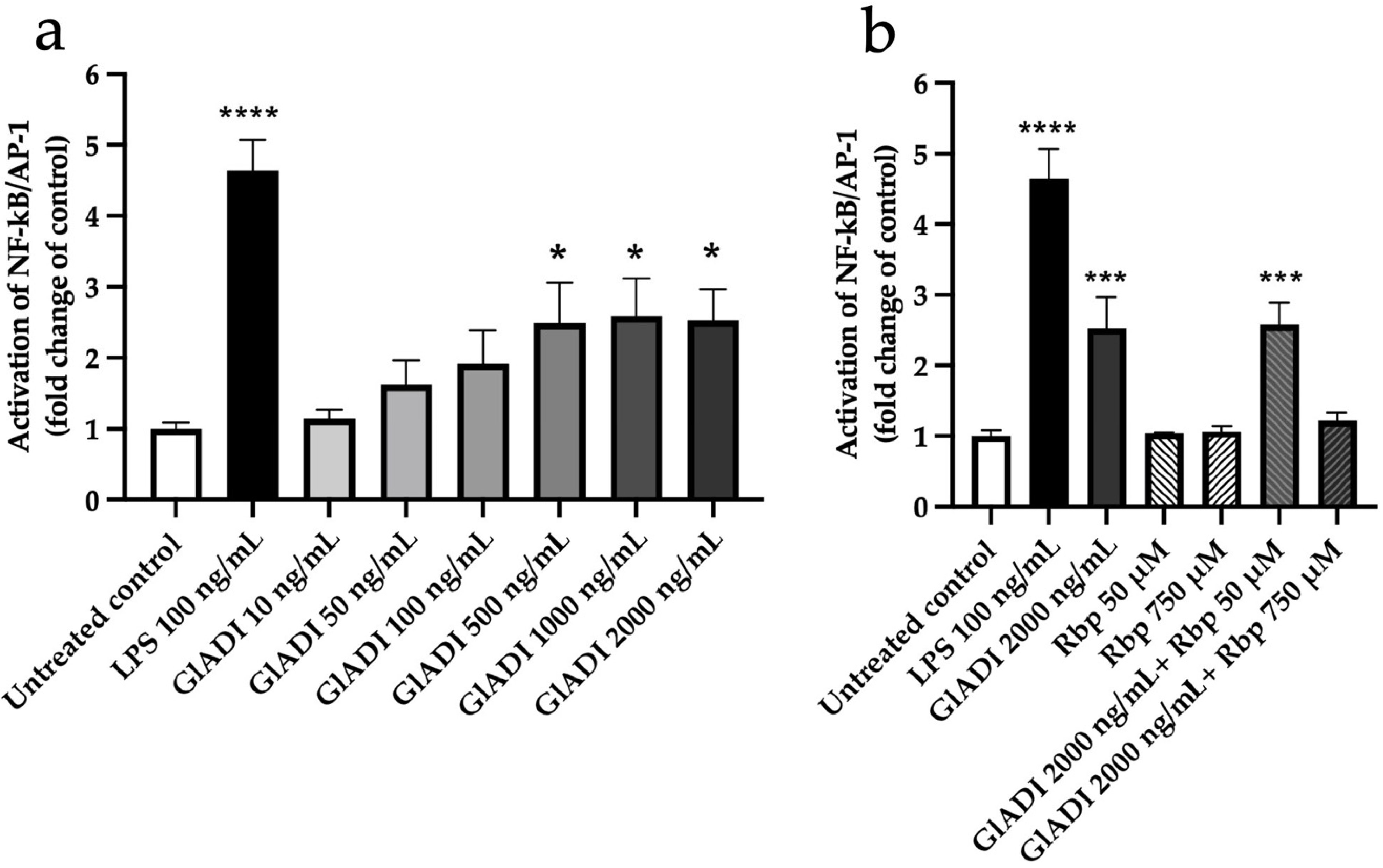
2.1. GlADI Exerts Immunomodulation by Activating TLRs
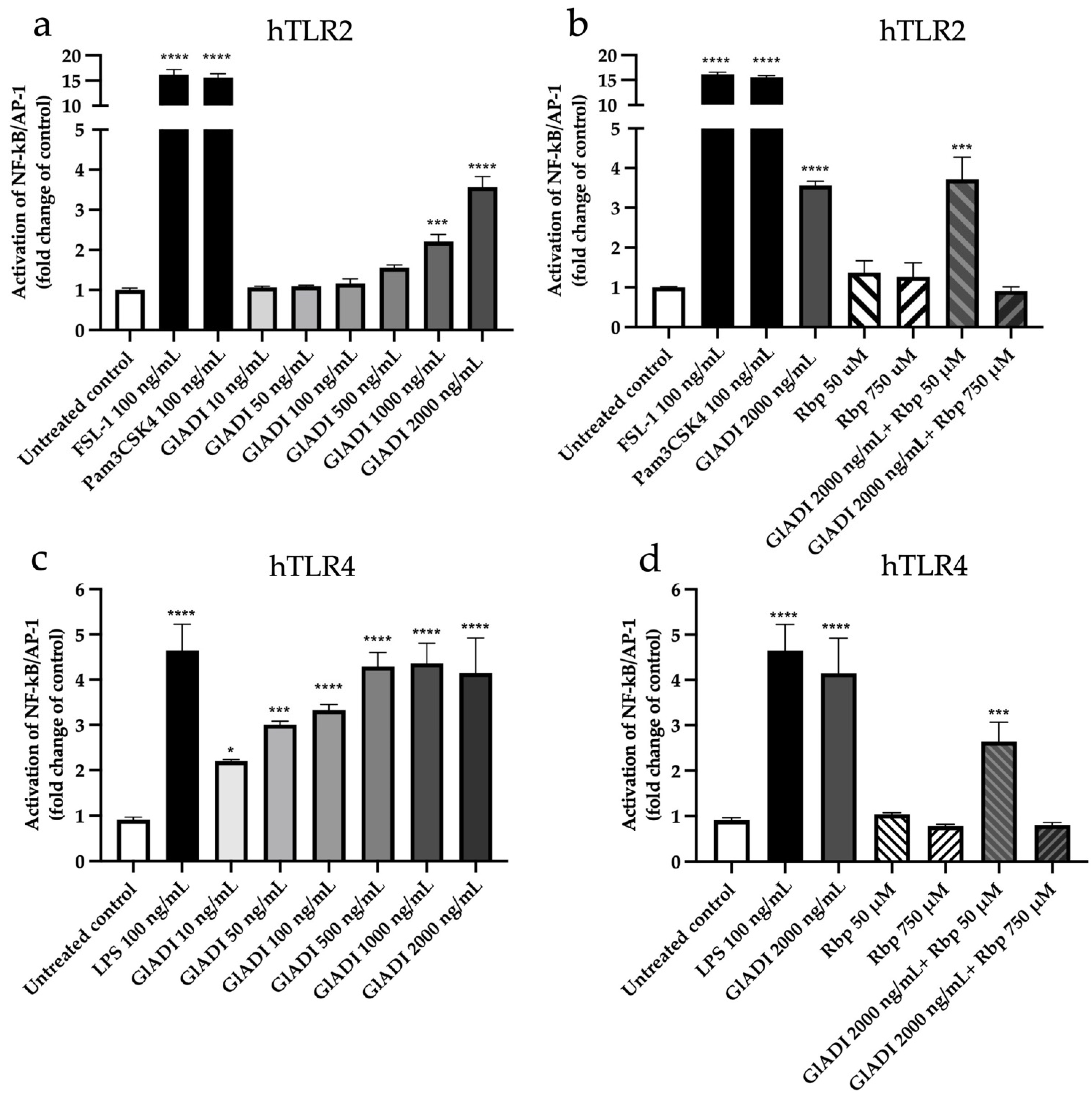
2.2. GlADI Interacts with the Host Immune System through TLR2 and TLR4
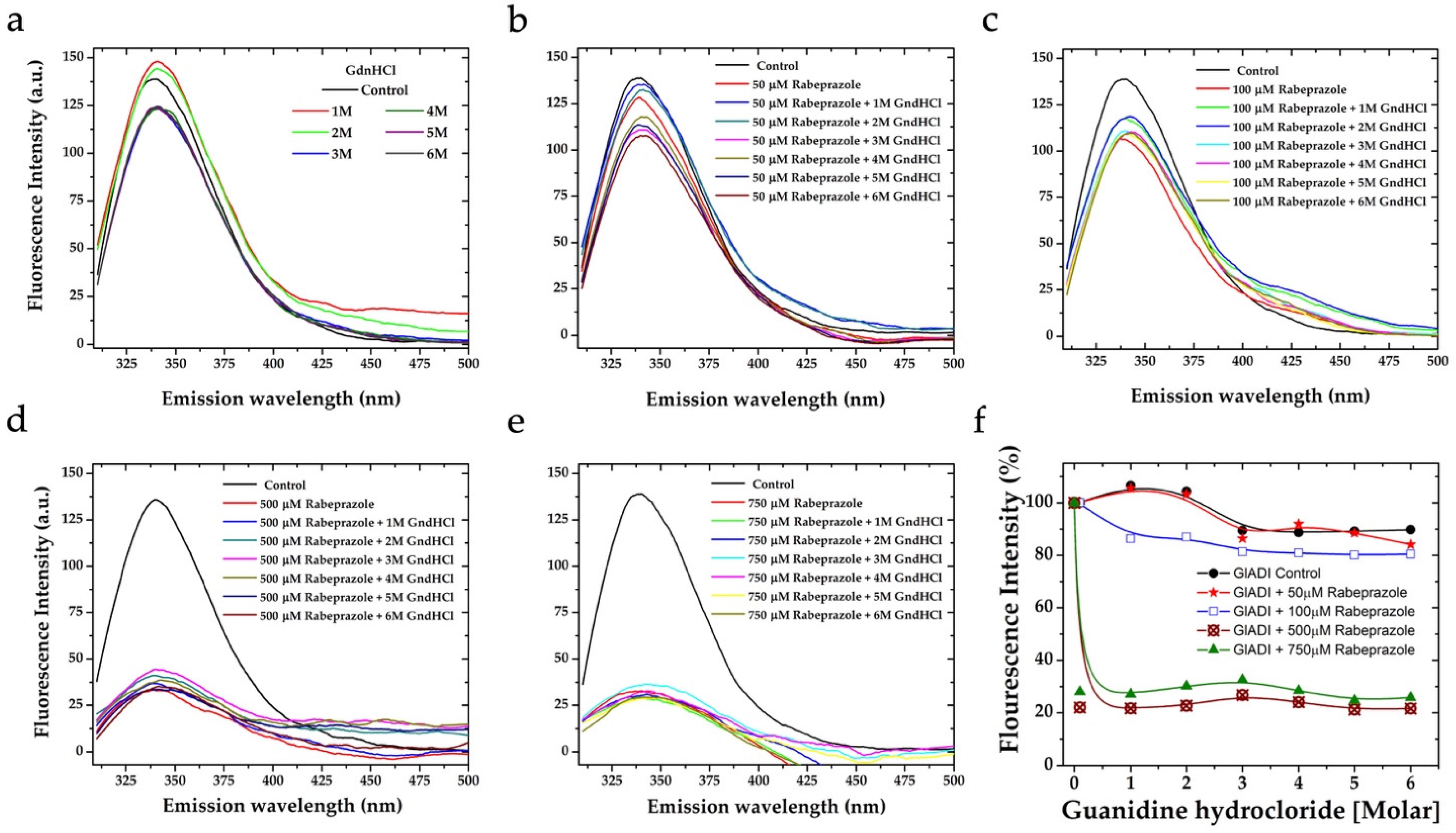
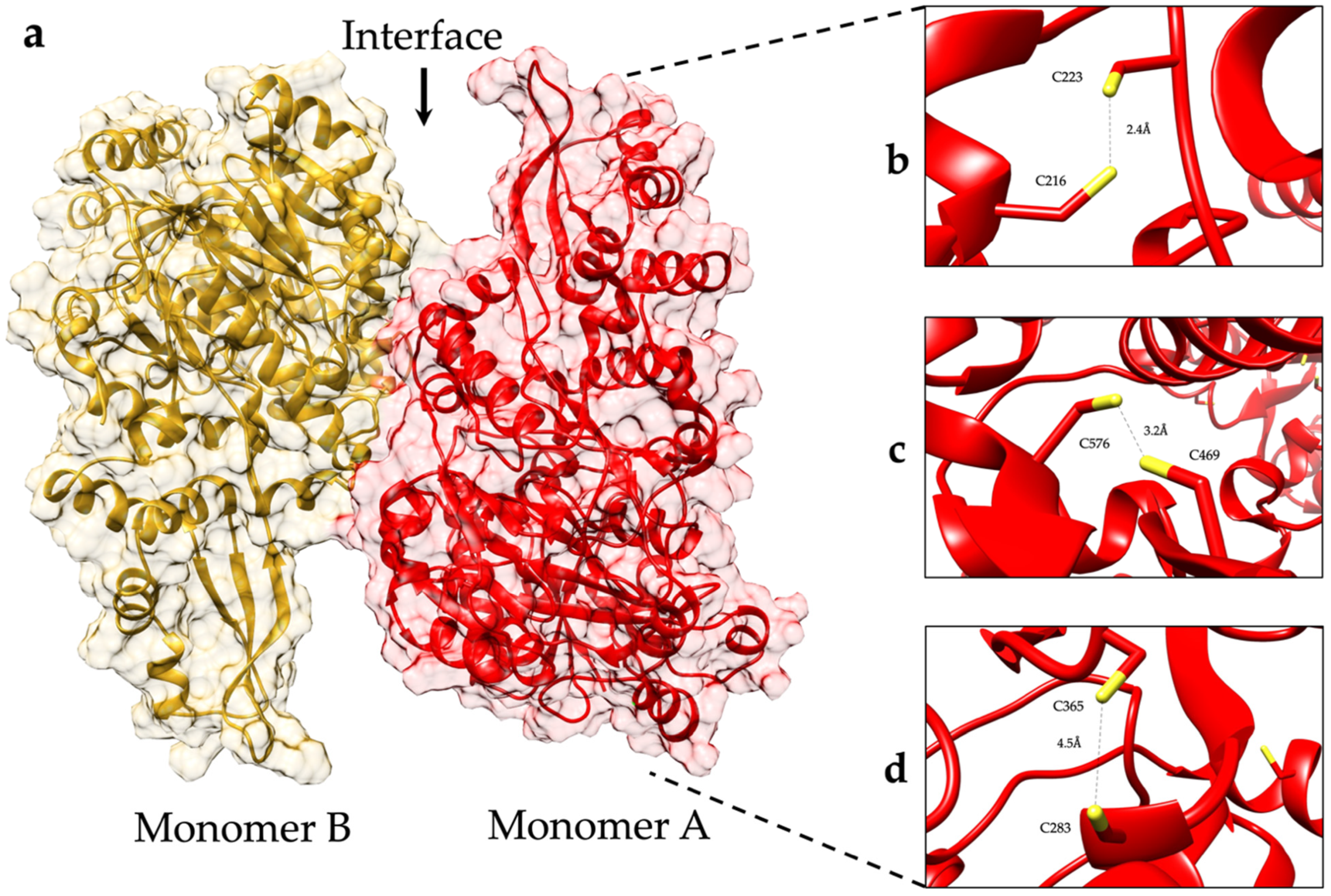
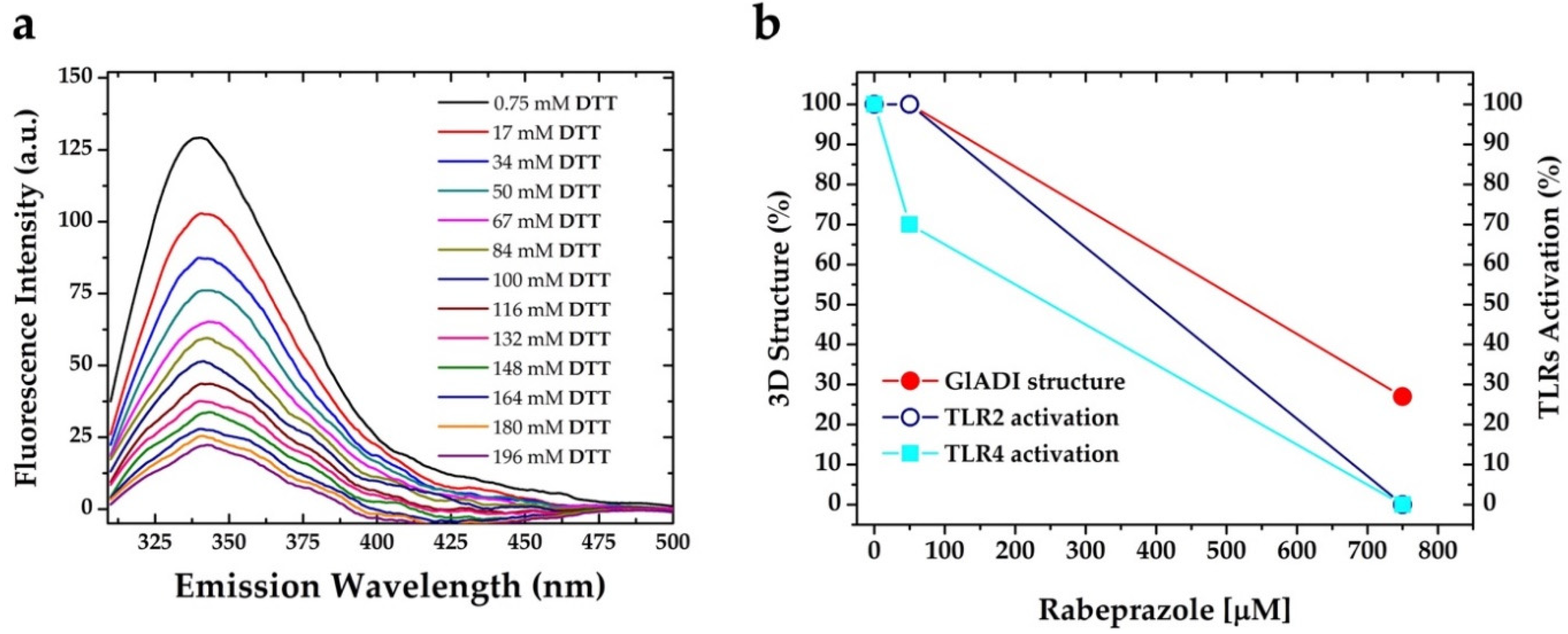
2.3. The Forces Maintaining the 3D Structure of GlADI Could Be a Pivotal Factor in Activating TLRs
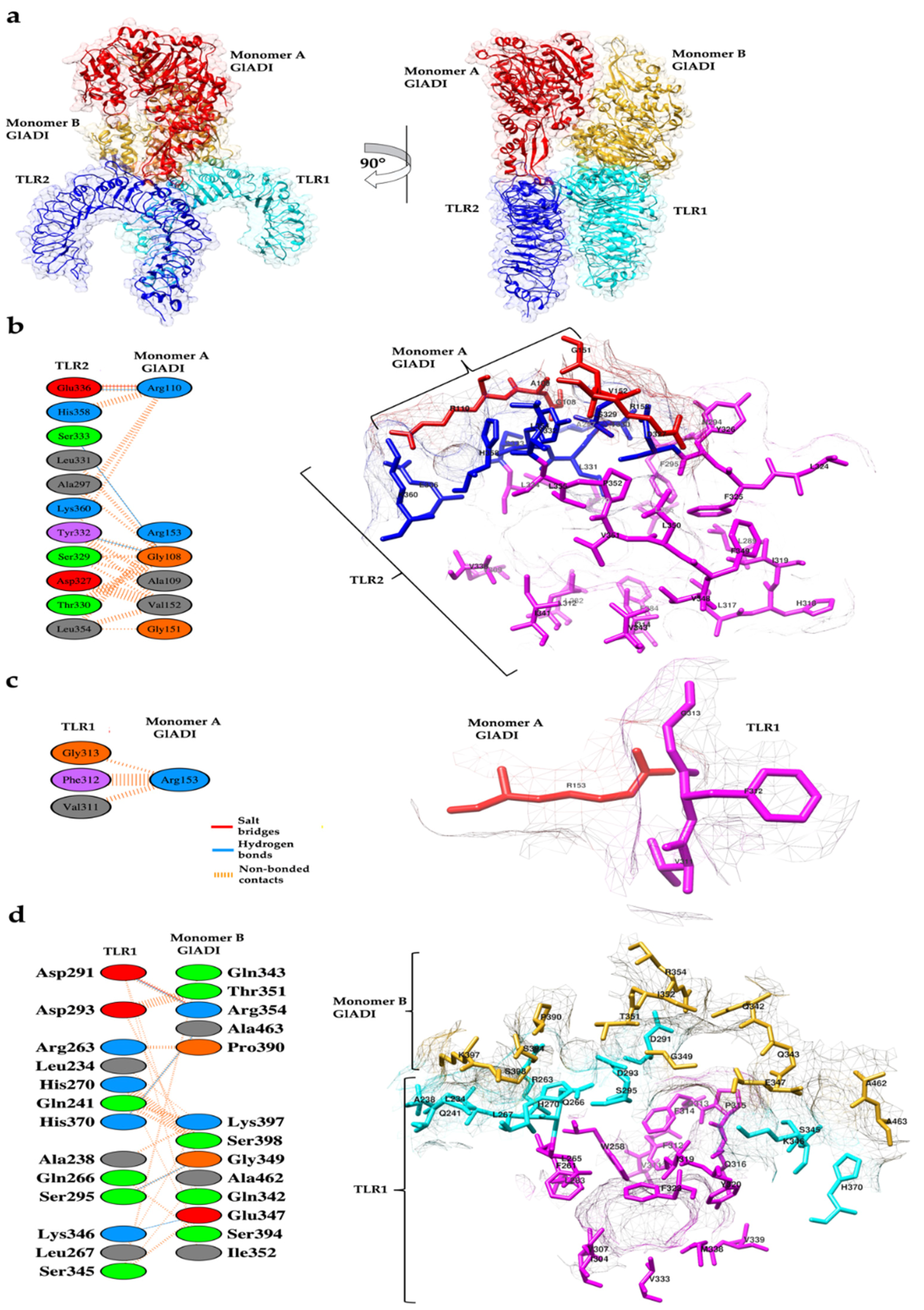
2.4. Predicted Interactions between the AlphaFold-Predicted Structure of GlADI and TLR2-TLR1
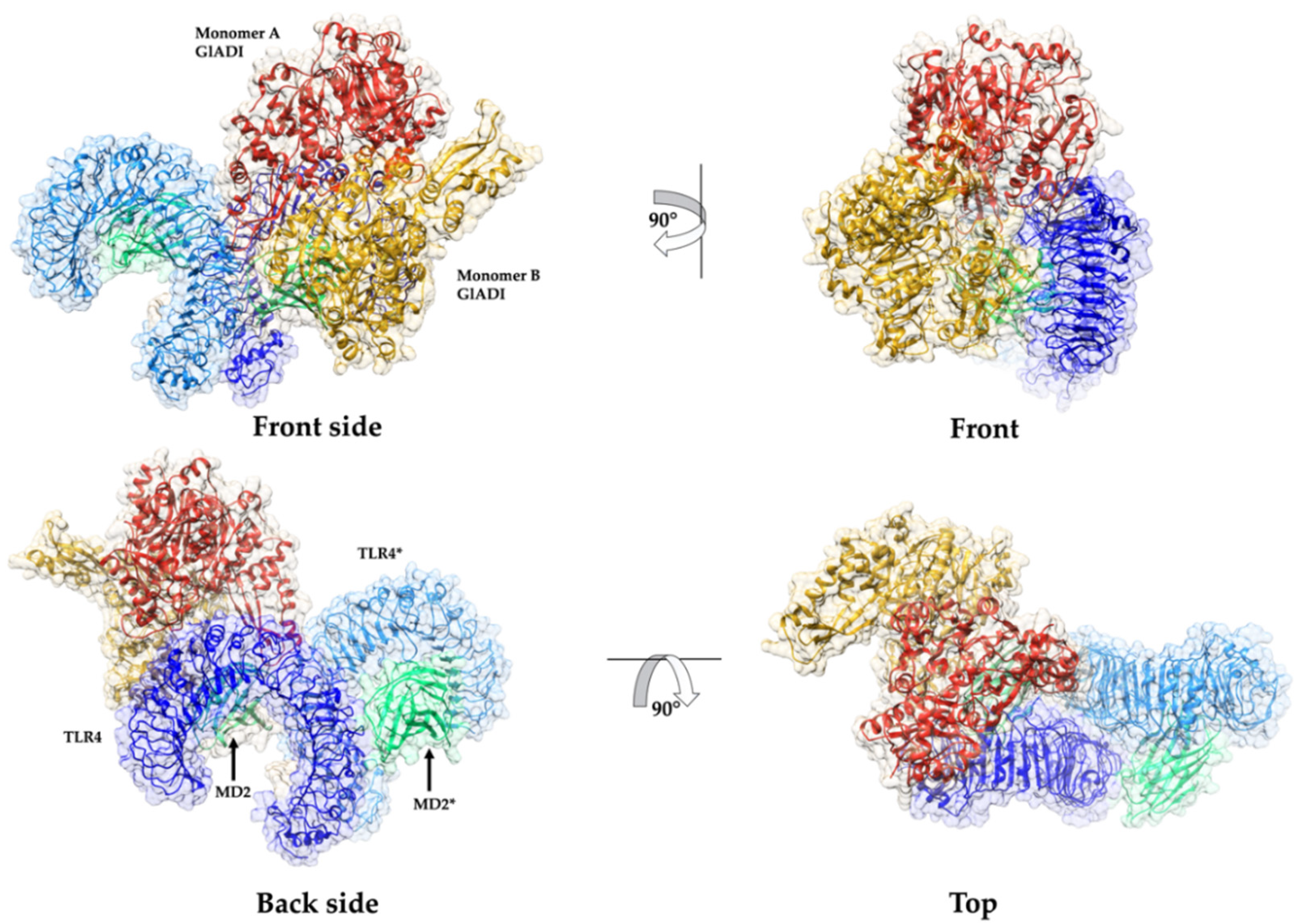
2.5. Predicted Interactions between the AlphaFold-Predicted Structure of GlADI and TLR4-MD-2
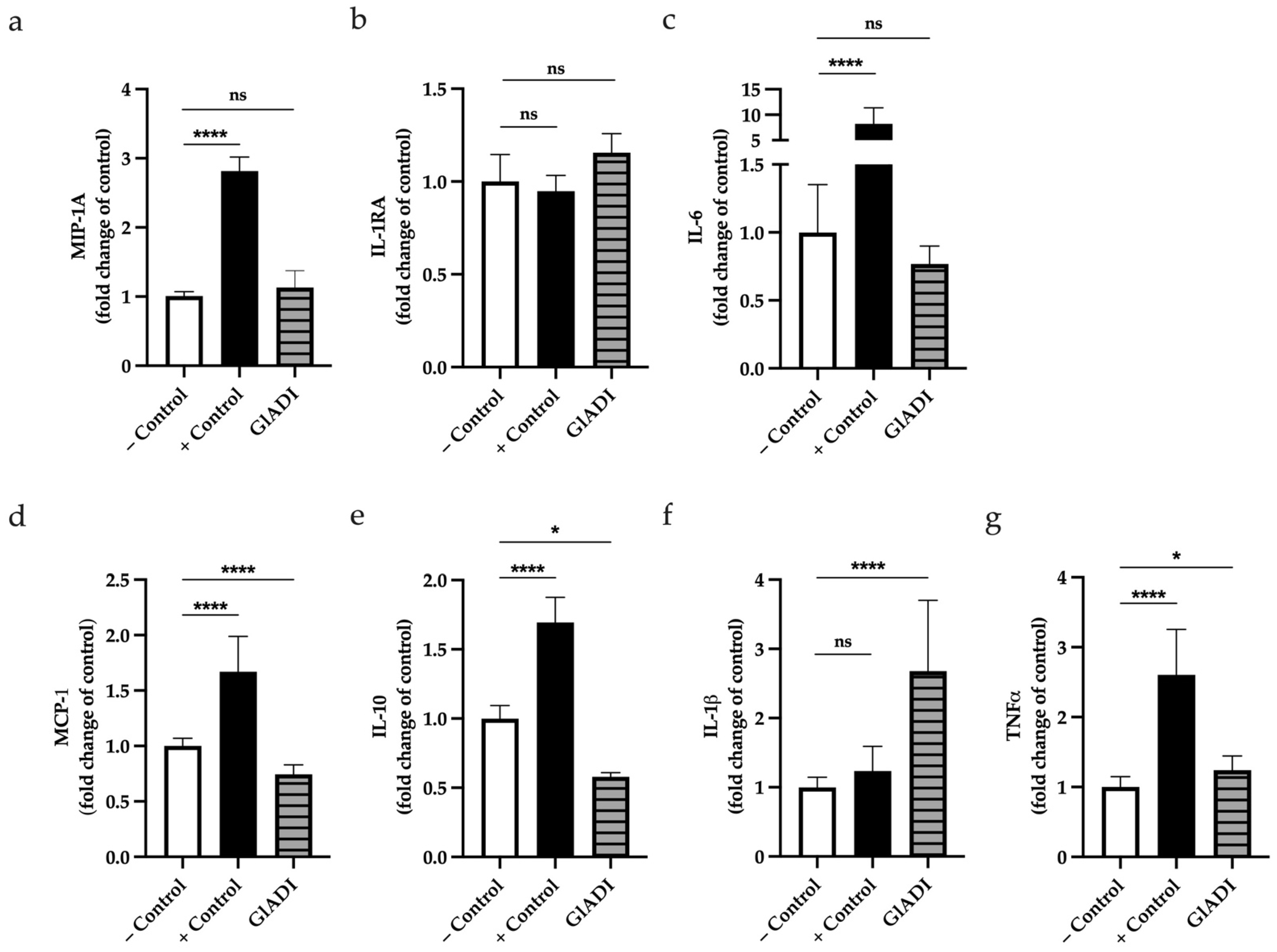
2.6. GlADI Induces Cytokine Production of DCs
3. Discussion
4. Materials and Methods
4.1. Recombinant GlADI Expression and Endotoxin Removal
4.2. Reporter Cell Lines
4.3. GlADI-TLRs Interaction Assays
4.4. Dendritic Cells Culturing and Stimulation with GlADI
4.5. Quantitation of Dendritic Cells Cytokines Production
4.6. Protein Thermal Shift Assay
4.7. Fluorescence Emission Spectra
4.8. Prediction of the Three-Dimensional (3D) Structure of GlADI
4.9. Prediction of the Probably Binding Mode of GlADI to TLRs by Using the AlphaFold-Predicted Structure and Molecular Docking
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dizdar, V.; Gilja, O.H.; Hausken, T. Increased visceral sensitivity in Giardia-induced postinfectious irritable bowel syndrome and functional dyspepsia. Effect of the 5HT3-antagonist ondansetron. Neurogastroenterol. Motil. 2007, 19, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Kamda, J.D.; Singer, S.M. Phosphoinositide 3-kinase-dependent inhibition of dendritic cell interleukin-12 production by Giardia lamblia. Infect. Immun. 2009, 77, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Buret, A.G.; Cacciò, S.M.; Favennec, L.; Svärd, S. Update on Giardia: Highlights from the seventh International Giardia and Cryptosporidium Conference. Parasite 2020, 27, 49. [Google Scholar] [CrossRef]
- Singer, S.M.; Fink, M.Y.; Angelova, V.V. Recent insights into innate and adaptive immune responses to Giardia. Adv. Parasitol. 2019, 106, 171–208. [Google Scholar] [PubMed]
- Balmer, E.A.; Faso, C. The Road Less Traveled? Unconventional Protein Secretion at Parasite-Host Interfaces. Front. Cell Dev. Biol. 2021, 9, 662711. [Google Scholar] [CrossRef] [PubMed]
- Rabouille, C.; Malhotra, V.; Nickel, W. Diversity in unconventional protein secretion. J. Cell Sci. 2012, 125, 5251–5255. [Google Scholar] [CrossRef] [PubMed]
- Schofield, P.; Costello, M.; Edwards, M.; O’sullivan, W. The arginine dihydrolase pathway is present in Giardia intestinalis. Int. J. Parasitol. 1990, 20, 697–699. [Google Scholar] [CrossRef]
- Schofield, P.J.; Edwards, M.R.; Matthews, J.; Wilson, J.R. The pathway of arginine catabolism in Giardia intestinalis. Mol. Biochem. Parasitol. 1992, 51, 29–36. [Google Scholar] [CrossRef]
- Eckmann, L.; Laurent, F.; Langford, T.D.; Hetsko, M.L.; Smith, J.R.; Kagnoff, M.F.; Gillin, F.D. Nitric oxide production by human intestinal epithelial cells and competition for arginine as potential determinants of host defense against the lumen-dwelling pathogen Giardia lamblia. J. Immunol. 2000, 164, 1478–1487. [Google Scholar] [CrossRef]
- Touz, M.C.; Rópolo, A.S.; Rivero, M.R.; Vranych, C.V.; Conrad, J.T.; Svard, S.G.; Nash, T.E. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 2008, 121, 2930–2938. [Google Scholar] [CrossRef]
- Ringqvist, E.; Palm, J.E.; Skarin, H.; Hehl, A.B.; Weiland, M.; Davids, B.J.; Reiner, D.S.; Griffiths, W.J.; Eckmann, L.; Gillin, F.D.; et al. Release of metabolic enzymes by Giardia in response to interaction with intestinal epithelial cells. Mol. Biochem. Parasitol. 2008, 159, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Rópolo, A.S.; Touz, M.C. A lesson in survival, by Giardia lamblia. ScientificWorldJournal 2010, 10, 2019–2031. [Google Scholar] [CrossRef] [PubMed]
- Stadelmann, B.; Merino, M.C.; Persson, L.; Svärd, S.G. Arginine consumption by the intestinal parasite Giardia intestinalis reduces proliferation of intestinal epithelial cells. PLoS ONE 2012, 7, e45325. [Google Scholar] [CrossRef]
- Bronte, V.; Zanovello, P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005, 5, 641–654. [Google Scholar] [CrossRef]
- Das, P.; Lahiri, A.; Lahiri, A.; Chakravortty, D. Modulation of the arginase pathway in the context of microbial pathogenesis: A metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 2010, 6, e1000899. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lainez, C.; de la Mora-de la Mora, I.; García-Torres, I.; Enríquez-Flores, S.; Flores-López, L.A.; Gutiérrez-Castrellón, P.; Yépez-Mulia, L.; Matadamas-Martínez, F.; de Vos, P.; López-Velázquez, G. Multilevel Approach for the Treatment of Giardiasis by Targeting Arginine Deiminase. Int. J. Mol. Sci. 2021, 22, 9491. [Google Scholar] [CrossRef]
- Palm, J.E.; Weiland, M.E.; Griffiths, W.J.; Ljungström, I.; Svärd, S.G. Identification of immunoreactive proteins during acute human giardiasis. J. Infect. Dis. 2003, 187, 1849–1859. [Google Scholar] [CrossRef]
- Banik, S.; Renner Viveros, P.; Seeber, F.; Klotz, C.; Ignatius, R.; Aebischer, T. Giardia duodenalis arginine deiminase modulates the phenotype and cytokine secretion of human dendritic cells by depletion of arginine and formation of ammonia. Infect. Immun. 2013, 81, 2309–2317. [Google Scholar] [CrossRef]
- Muñoz-Cruz, S.; Gomez-García, A.; Matadamas-Martínez, F.; Alvarado-Torres, J.A.; Meza-Cervantez, P.; Arriaga-Pizano, L.; Yépez-Mulia, L. Giardia lamblia: Identification of molecules that contribute to direct mast cell activation. Parasitol. Res. 2018, 117, 2555–2567. [Google Scholar] [CrossRef]
- Bryant, C.E.; Gay, N.J.; Heymans, S.; Sacre, S.; Schaefer, L.; Midwood, K.S. Advances in Toll-like receptor biology: Modes of activation by diverse stimuli. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 359–379. [Google Scholar] [CrossRef]
- Fernández-Lainez, C.; Akkerman, R.; Oerlemans, M.; Logtenberg, M.; Schols, H.; Silva-Lagos, L.; López-Velázquez, G.; de Vos, P. β (2→6)-Type fructans attenuate proinflammatory responses in a structure dependent fashion via Toll-like receptors. Carbohydr. Polym. 2022, 277, 118893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, X.; Bao, X.; Xiao, W.; Chen, G. Toll-like receptor 4 (TLR4) inhibitors: Current research and prospective. Eur. J. Med. Chem. 2022, 235, 114291. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, F.; Clote, P. DiANNA: A web server for disulfide connectivity prediction. Nucleic Acids Res. 2005, 33 (Suppl. S2), W230–W232. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.G.; Lee, H.; Lee, J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Karmakar, S.; Babu, S.P. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016, 20, 193–204. [Google Scholar] [CrossRef]
- Kucera, K.; Koblansky, A.A.; Saunders, L.P.; Frederick, K.B.; De La Cruz, E.M.; Ghosh, S.; Modis, Y. Structure-based analysis of Toxoplasma gondii profilin: A parasite-specific motif is required for recognition by Toll-like receptor 11. J. Mol. Biol. 2010, 403, 616–629. [Google Scholar] [CrossRef]
- Rana, A.; Akhter, Y. A multi-subunit based, thermodynamically stable model vaccine using combined immunoinformatics and protein structure based approach. Immunobiology 2016, 221, 544–557. [Google Scholar] [CrossRef]
- Safavi, A.; Kefayat, A.; Mahdevar, E.; Abiri, A.; Ghahremani, F. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine 2020, 38, 7612–7628. [Google Scholar] [CrossRef]
- Cheng, L.; Kiewiet, M.B.; Groeneveld, A.; Nauta, A.; de Vos, P. Human milk oligosaccharides and its acid hydrolysate LNT2 show immunomodulatory effects via TLRs in a dose and structure-dependent way. J. Funct. Foods 2019, 59, 174–184. [Google Scholar] [CrossRef]
- Serradell, M.C.; Rupil, L.L.; Martino, R.A.; Prucca, C.G.; Carranza, P.G.; Saura, A.; Fernández, E.A.; Gargantini, P.R.; Tenaglia, A.H.; Petiti, J.P.; et al. Efficient oral vaccination by bioengineering virus-like particles with protozoan surface proteins. Nat. Commun. 2019, 10, 361. [Google Scholar] [CrossRef]
- Emery-Corbin, S.J.; Grüttner, J.; Svärd, S. Transcriptomic and proteomic analyses of Giardia intestinalis: Intestinal epithelial cell interactions. Adv. Parasitol. 2020, 107, 139–171. [Google Scholar] [PubMed]
- Heras, B.; Kurz, M.; Shouldice, S.R.; Martin, J.L. The name’s bond......disulfide bond. Curr. Opin. Struct. Biol. 2007, 17, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Goemans, C.; Denoncin, K.; Collet, J.F. Folding mechanisms of periplasmic proteins. Biochim. Biophys. Acta 2014, 1843, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Buwitt-Beckmann, U.; Heine, H.; Wiesmüller, K.H.; Jung, G.; Brock, R.; Akira, S.; Ulmer, A.J. TLR1- and TLR6-independent recognition of bacterial lipopeptides. J. Biol. Chem. 2006, 281, 9049–9057. [Google Scholar] [CrossRef]
- Van Bergenhenegouwen, J.; Plantinga, T.S.; Joosten, L.A.; Netea, M.G.; Folkerts, G.; Kraneveld, A.D.; Garssen, J.; Vos, A.P. TLR2 & Co: A critical analysis of the complex interactions between TLR2 and coreceptors. J. Leukoc. Biol 2013, 94, 885–902. [Google Scholar]
- Mukherjee, S.; Karnam, A.; Das, M.; Babu, S.P.S.; Bayry, J. Wuchereria bancrofti filaria activates human dendritic cells and polarizes T helper 1 and regulatory T cells via toll-like receptor 4. Commun. Biol. 2019, 2, 169. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mukherjee, S.; Bhattacharya, S.; Sinha Babu, S.P. Surface proteins of Setaria cervi induce inflammation in macrophage through Toll-like receptor 4 (TLR4)-mediated signalling pathway. Parasite Immunol. 2017, 39, e12389. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mukherjee, S.; Maiti, T.K.; Bhattacharya, S.; Sinha Babu, S.P. A Novel Ligand of Toll-like Receptor 4 From the Sheath of Wuchereria bancrofti Microfilaria Induces Proinflammatory Response in Macrophages. J. Infect. Dis. 2017, 215, 954–965. [Google Scholar] [CrossRef]
- Bryant, C.E.; Spring, D.R.; Gangloff, M.; Gay, N.J. The molecular basis of the host response to lipopolysaccharide. Nat. Rev. Microbiol. 2010, 8, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Bulut, Y.; Faure, E.; Thomas, L.; Karahashi, H.; Michelsen, K.S.; Equils, O.; Morrison, S.G.; Morrison, R.P.; Arditi, M. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J. Immunol. 2002, 168, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhou, Y.; Fan, S.; Wen, Q. The Multiple Roles and Therapeutic Potential of HSP60 in Cancer. Biochem. Pharmacol. 2022, 201, 115096. [Google Scholar] [CrossRef]
- Asea, A.; Rehli, M.; Kabingu, E.; Boch, J.A.; Baré, O.; Auron, P.E.; Stevenson, M.A.; Calderwood, S.K. Novel signal transduction pathway utilized by extracellular HSP70: Role of toll-like receptor (TLR) 2 and TLR4. J. Biol. Chem. 2002, 277, 15028–15034. [Google Scholar] [CrossRef]
- Lee, J.H.; Jeon, J.; Bai, F.; Jin, S.; Wu, W.; Ha, U.H. The Pseudomonas aeruginosa HSP70-like protein DnaK induces IL-1β expression via TLR4-dependent activation of the NF-κB and JNK signaling pathways. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101373. [Google Scholar] [CrossRef] [PubMed]
- Smiley, S.T.; King, J.A.; Hancock, W.W. Fibrinogen stimulates macrophage chemokine secretion through toll-like receptor 4. J. Immunol. 2001, 167, 2887–2894. [Google Scholar] [CrossRef]
- Guillot, L.; Balloy, V.; McCormack, F.X.; Golenbock, D.T.; Chignard, M.; Si-Tahar, M. Cutting edge: The immunostimulatory activity of the lung surfactant protein-A involves Toll-like receptor 4. J. Immunol. 2002, 168, 5989–5992. [Google Scholar] [CrossRef]
- Okamura, Y.; Watari, M.; Jerud, E.S.; Young, D.W.; Ishizaka, S.T.; Rose, J.; Chow, J.C.; Strauss, J.F., 3rd. The extra domain A of fibronectin activates Toll-like receptor 4. J. Biol. Chem. 2001, 276, 10229–10233. [Google Scholar] [CrossRef]
- Vabulas, R.M.; Braedel, S.; Hilf, N.; Singh-Jasuja, H.; Herter, S.; Ahmad-Nejad, P.; Kirschning, C.J.; Da Costa, C.; Rammensee, H.G.; Wagner, H.; et al. The endoplasmic reticulum-resident heat shock protein Gp96 activates dendritic cells via the Toll-like receptor 2/4 pathway. J. Biol. Chem. 2002, 277, 20847–208453. [Google Scholar] [CrossRef]
- Park, J.S.; Svetkauskaite, D.; He, Q.; Kim, J.Y.; Strassheim, D.; Ishizaka, A.; Abraham, E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 2004, 279, 7370–7377. [Google Scholar] [CrossRef]
- Zhao, Y.; Kuang, M.; Li, J.; Zhu, L.; Jia, Z.; Guo, X.; Hu, Y.; Kong, J.; Yin, H.; Wang, X.; et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021, 31, 818–820. [Google Scholar] [CrossRef]
- Choudhury, A.; Mukherjee, S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020, 92, 2105–2113. [Google Scholar] [CrossRef]
- Stagg, A.J. Intestinal Dendritic Cells in Health and Gut Inflammation. Front. Immunol. 2018, 9, 2883. [Google Scholar] [CrossRef]
- Tezuka, H.; Ohteki, T. Regulation of IgA Production by Intestinal Dendritic Cells and Related Cells. Front. Immunol. 2019, 10, 1891. [Google Scholar] [CrossRef]
- Mowat, A.M. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 2003, 3, 331–341. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Abo-Zaid, M.A.; Hamdi, A.A. Evaluation of immune response and haematological parameters in infected male albino rats by giardiasis. Parasite Immunol. 2022, 44, e12908. [Google Scholar] [CrossRef]
- Zhou, P.; Li, E.; Shea-Donohue, T.; Singer, S.M. Tumour necrosis factor alpha contributes to protection against Giardia lamblia infection in mice. Parasite Immunol. 2007, 29, 367–374. [Google Scholar] [CrossRef]
- Troeger, H.; Epple, H.J.; Schneider, T.; Wahnschaffe, U.; Ullrich, R.; Burchard, G.D.; Jelinek, T.; Zeitz, M.; Fromm, M.; Schulzke, J.D. Effect of chronic Giardia lamblia infection on epithelial transport and barrier function in human duodenum. Gut 2007, 56, 328–335. [Google Scholar] [CrossRef]
- Schmitz, H.; Fromm, M.; Bentzel, C.J.; Scholz, P.; Detjen, K.; Mankertz, J.; Bode, H.; Epple, H.J.; Riecken, E.O.; Schulzke, J.D. Tumor necrosis factor-alpha (TNFalpha) regulates the epithelial barrier in the human intestinal cell line HT-29/B6. J. Cell Sci. 1999, 112, 137–146. [Google Scholar] [CrossRef]
- Maciorkowska, E.; Kaczmarski, M.; Kemona, A. The role of cytokines in giardiasis in children. Med. Wieku Rozw. 2005, 9, 665–673. [Google Scholar]
- Annunziato, F.; Romagnani, C.; Romagnani, S. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol. 2015, 135, 626–635. [Google Scholar] [CrossRef] [PubMed]
- Saghaug, C.S.; Sørnes, S.; Peirasmaki, D.; Svärd, S.; Langeland, N.; Hanevik, K. Human memory CD4+ T cell immune responses against Giardia lamblia. Clin. Vaccine Immunol. 2016, 23, 11–18. [Google Scholar] [CrossRef]
- Lépine, A.; de Vos, P. Synbiotic Effects of the Dietary Fiber Long-Chain Inulin and Probiotic Lactobacillus acidophilus W37 Can be Caused by Direct, Synergistic Stimulation of Immune Toll-Like Receptors and Dendritic Cells. Mol. Nutr. Food Res. 2018, 62, 1800251. [Google Scholar] [CrossRef]
- Vogt, L.; Ramasamy, U.; Meyer, D.; Pullens, G.; Venema, K.; Faas, M.M.; Schols, H.A.; de Vos, P. Immune modulation by different types of β2→ 1-fructans is toll-like receptor dependent. PLoS ONE 2013, 8, e68367. [Google Scholar] [CrossRef]
- Sahasrabudhe, N.M.; Beukema, M.; Tian, L.; Troost, B.; Scholte, J.; Bruininx, E.; Bruggeman, G.; van den Berg, M.; Scheurink, A.; Schols, H.A. Dietary fiber pectin directly blocks toll-like receptor 2–1 and prevents doxorubicin-induced ileitis. Front. Immunol. 2018, 9, 383. [Google Scholar] [CrossRef]
- Li, Z.; Kulakova, L.; Li, L.; Galkin, A.; Zhao, Z.; Nash, T.E.; Mariano, P.S.; Herzberg, O.; Dunaway-Mariano, D. Mechanisms of catalysis and inhibition operative in the arginine deiminase from the human pathogen Giardia lamblia. Bioorganic Chem. 2009, 37, 149–161. [Google Scholar] [CrossRef][Green Version]
- Trejo-Soto, P.J.; Aguayo-Ortiz, R.; Yépez-Mulia, L.; Hernández-Campos, A.; Medina-Franco, J.L.; Castillo, R. Insights into the structure and inhibition of Giardia intestinalis arginine deiminase: Homology modeling, docking, and molecular dynamics studies. J. Biomol. Struct. Dyn. 2016, 34, 732–748. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. PyMOL. In The PyMOL Molecular Graphics System; Version 1.2r3pre; Schrödinger, LLC: New York, NY, USA, 2010. [Google Scholar]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 2009, 77, 114–122. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]

| Melting Temperatures of GlADI (°C) | ||||||||
|---|---|---|---|---|---|---|---|---|
| [NaCl] mM | pH 5.0 | pH 6.0 | pH 6.5 | pH 7.0 | pH 7.5 | pH 8.0 | pH 9.0 | pH 9.5 |
| 0 | 55.98 | 59.14 | 55.88 | 55.64 | 60.34 | 58.96 | 50.04 | 56.99 |
| 200 | 49.82 | 60.94 | 60.97 | 63.01 | 70.56 | 56.4 | 59.74 | 56.24 |
| 400 | 57.04 | 50.59 | 47.17 | 48.12 | 53.57 | 52.05 | 51.93 | 56.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Lainez, C.; de la Mora-de la Mora, I.; Enríquez-Flores, S.; García-Torres, I.; Flores-López, L.A.; Gutiérrez-Castrellón, P.; de Vos, P.; López-Velázquez, G. The Giardial Arginine Deiminase Participates in Giardia-Host Immunomodulation in a Structure-Dependent Fashion via Toll-like Receptors. Int. J. Mol. Sci. 2022, 23, 11552. https://doi.org/10.3390/ijms231911552
Fernández-Lainez C, de la Mora-de la Mora I, Enríquez-Flores S, García-Torres I, Flores-López LA, Gutiérrez-Castrellón P, de Vos P, López-Velázquez G. The Giardial Arginine Deiminase Participates in Giardia-Host Immunomodulation in a Structure-Dependent Fashion via Toll-like Receptors. International Journal of Molecular Sciences. 2022; 23(19):11552. https://doi.org/10.3390/ijms231911552
Chicago/Turabian StyleFernández-Lainez, Cynthia, Ignacio de la Mora-de la Mora, Sergio Enríquez-Flores, Itzhel García-Torres, Luis A. Flores-López, Pedro Gutiérrez-Castrellón, Paul de Vos, and Gabriel López-Velázquez. 2022. "The Giardial Arginine Deiminase Participates in Giardia-Host Immunomodulation in a Structure-Dependent Fashion via Toll-like Receptors" International Journal of Molecular Sciences 23, no. 19: 11552. https://doi.org/10.3390/ijms231911552
APA StyleFernández-Lainez, C., de la Mora-de la Mora, I., Enríquez-Flores, S., García-Torres, I., Flores-López, L. A., Gutiérrez-Castrellón, P., de Vos, P., & López-Velázquez, G. (2022). The Giardial Arginine Deiminase Participates in Giardia-Host Immunomodulation in a Structure-Dependent Fashion via Toll-like Receptors. International Journal of Molecular Sciences, 23(19), 11552. https://doi.org/10.3390/ijms231911552









