Dynamic Transcriptional Landscape of Grass Carp (Ctenopharyngodon idella) Reveals Key Transcriptional Features Involved in Fish Development
Abstract
1. Introduction
2. Results
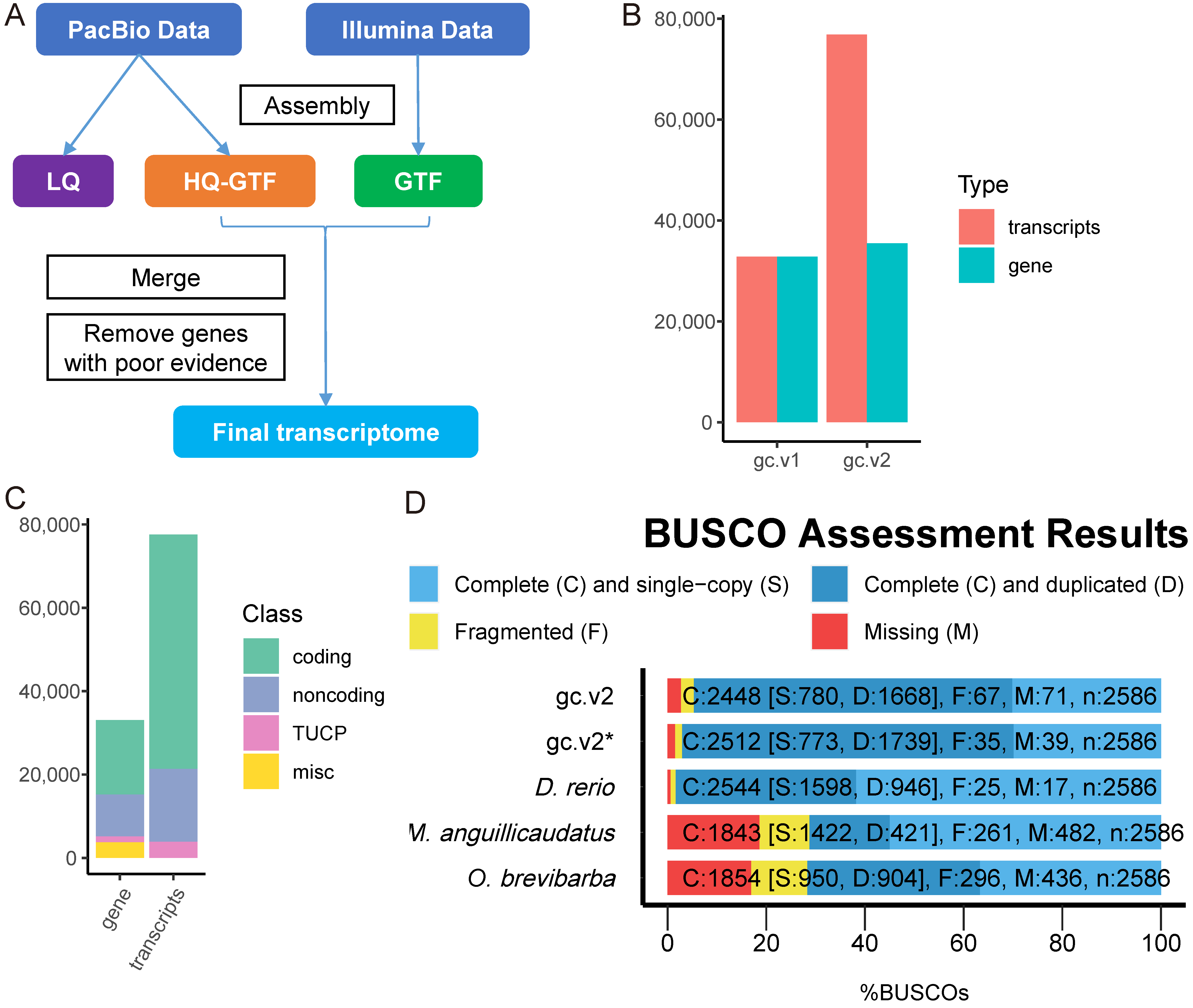
2.1. Transcriptome Sequencing and Assembly
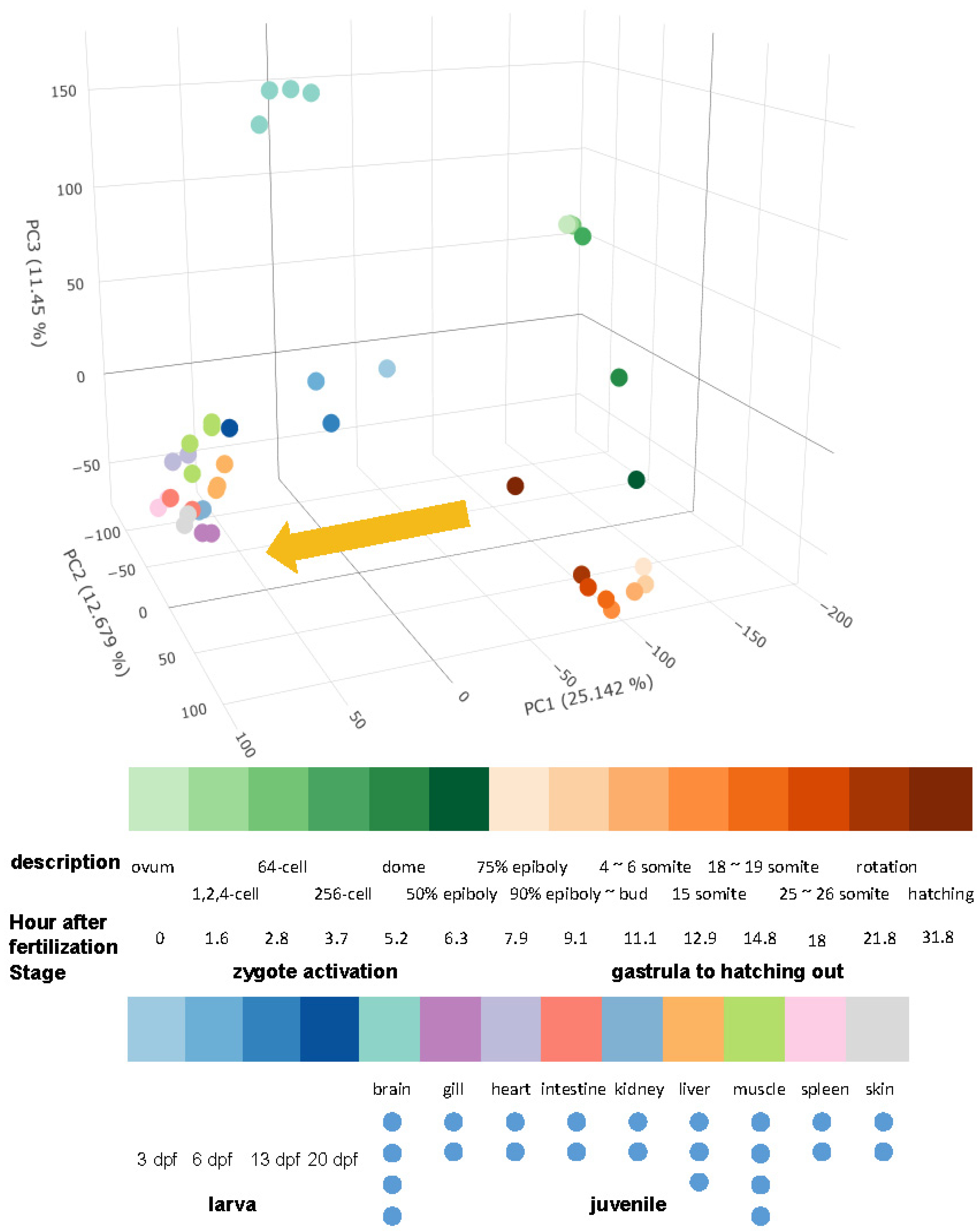
2.2. Overview of Grass Carp Development and Tissue Differentiation
2.2.1. The Developmental Trajectory of Grass Carp
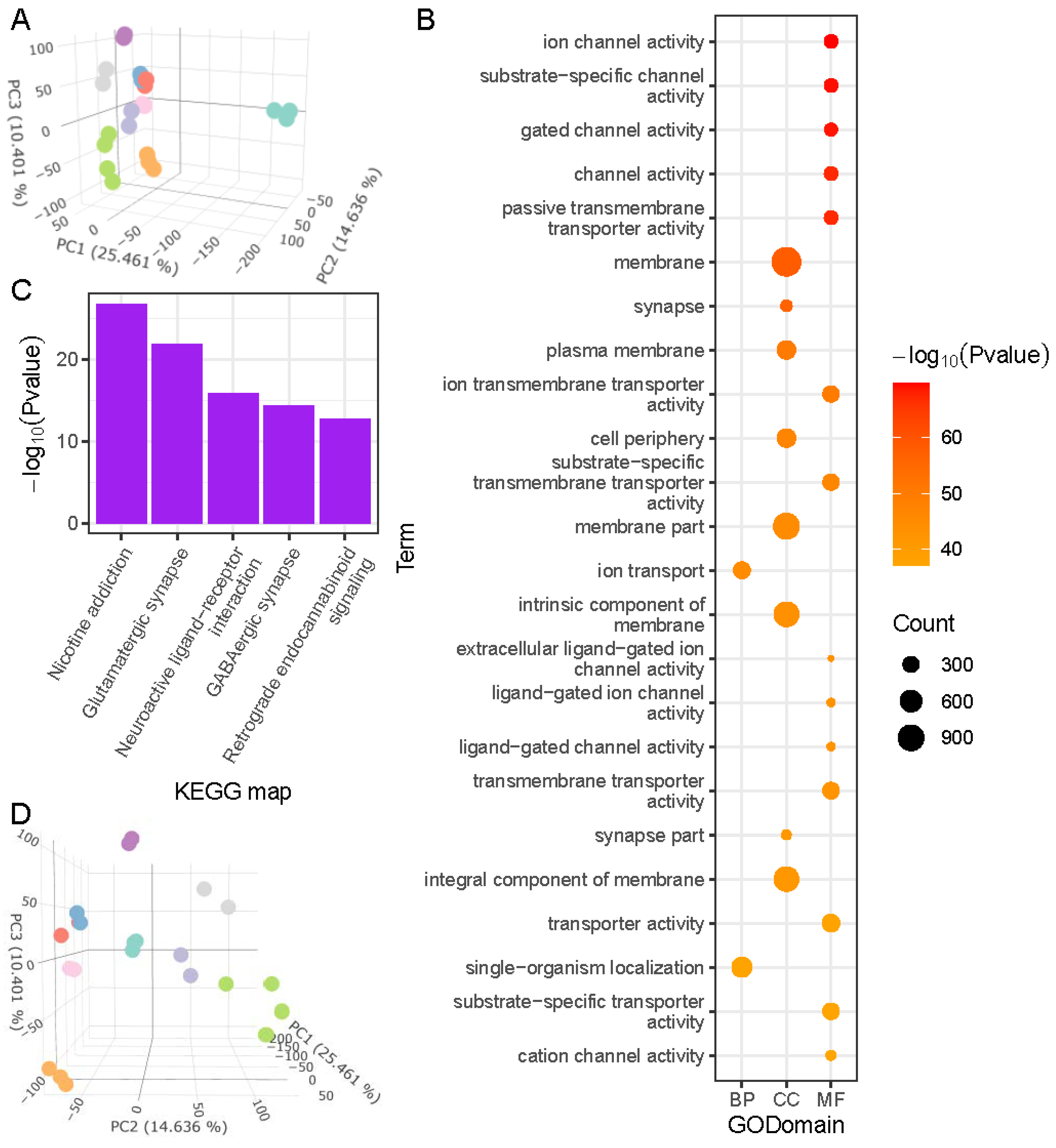
2.2.2. Fish Brain Is Highly Differentiated
2.2.3. Muscle Development and Basic Metabolic Regulation Describe Major Differences among Fish Organs
2.3. Expression Analysis of Grass Carp Transposons
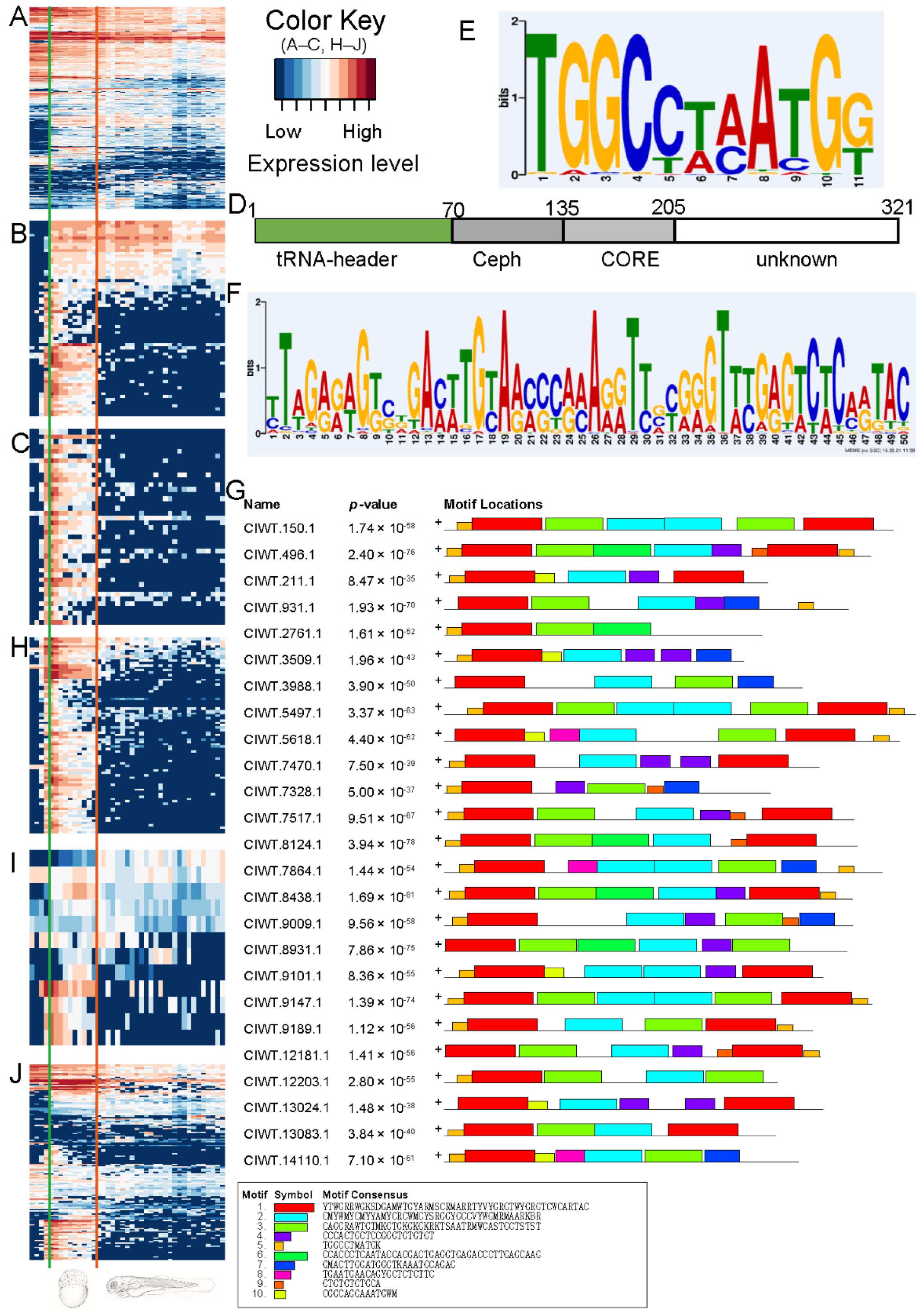
2.3.1. Expression Scenery of Grass Carp Transposon
2.3.2. SINE Element rnd-3_family-293 Is Specifically Expressed during Embryonic Development
2.3.3. Motif Analysis for Family rnd-3_family-293
2.3.4. Sequences Possessing Both Motif 1 and Motif 5 Were Specifically Expressed during Embryonic Development
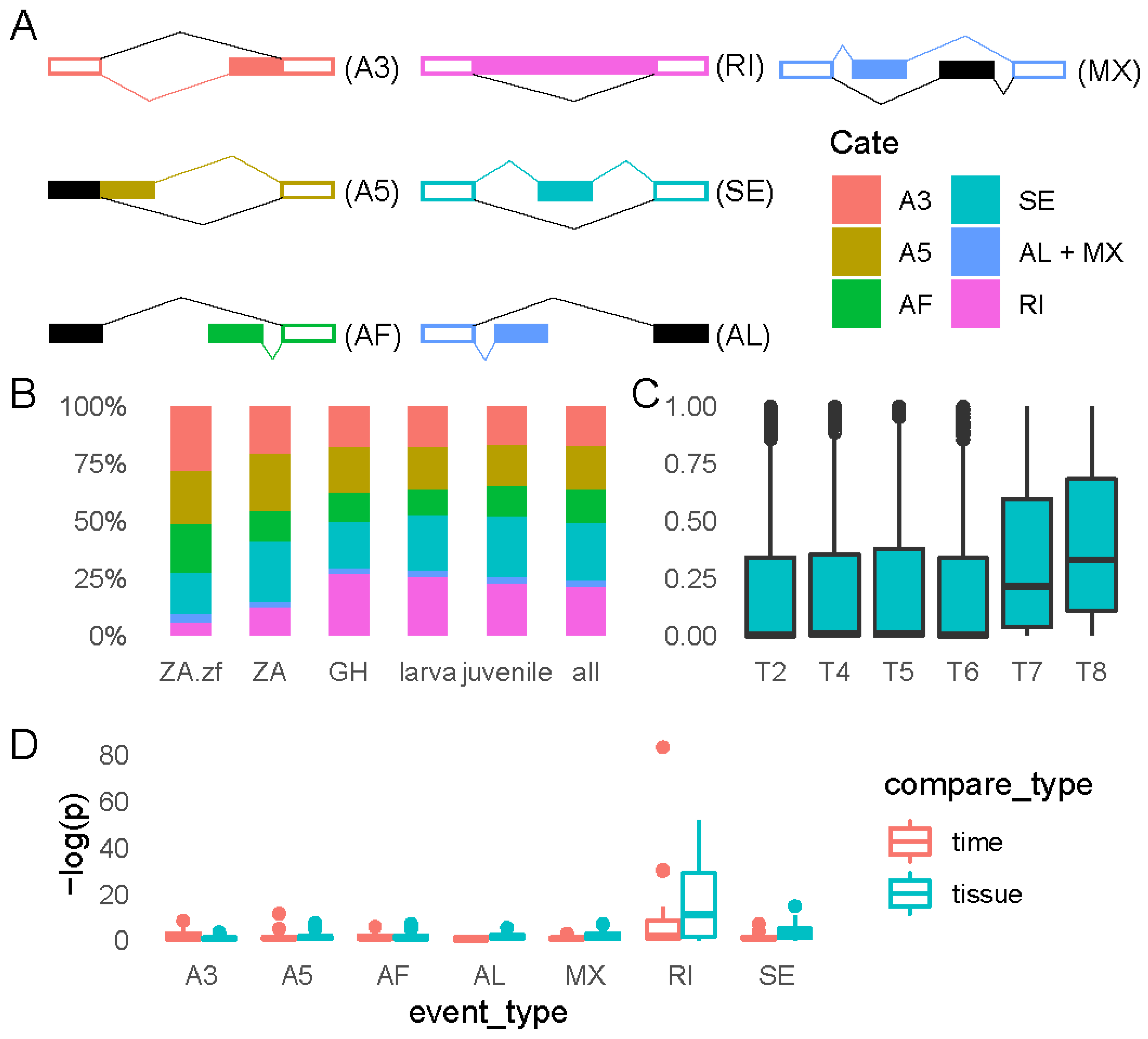
2.4. Alternative Splicing Events in Grass Carp
2.4.1. Distributions of Grass Carp Alternative Splicing Events
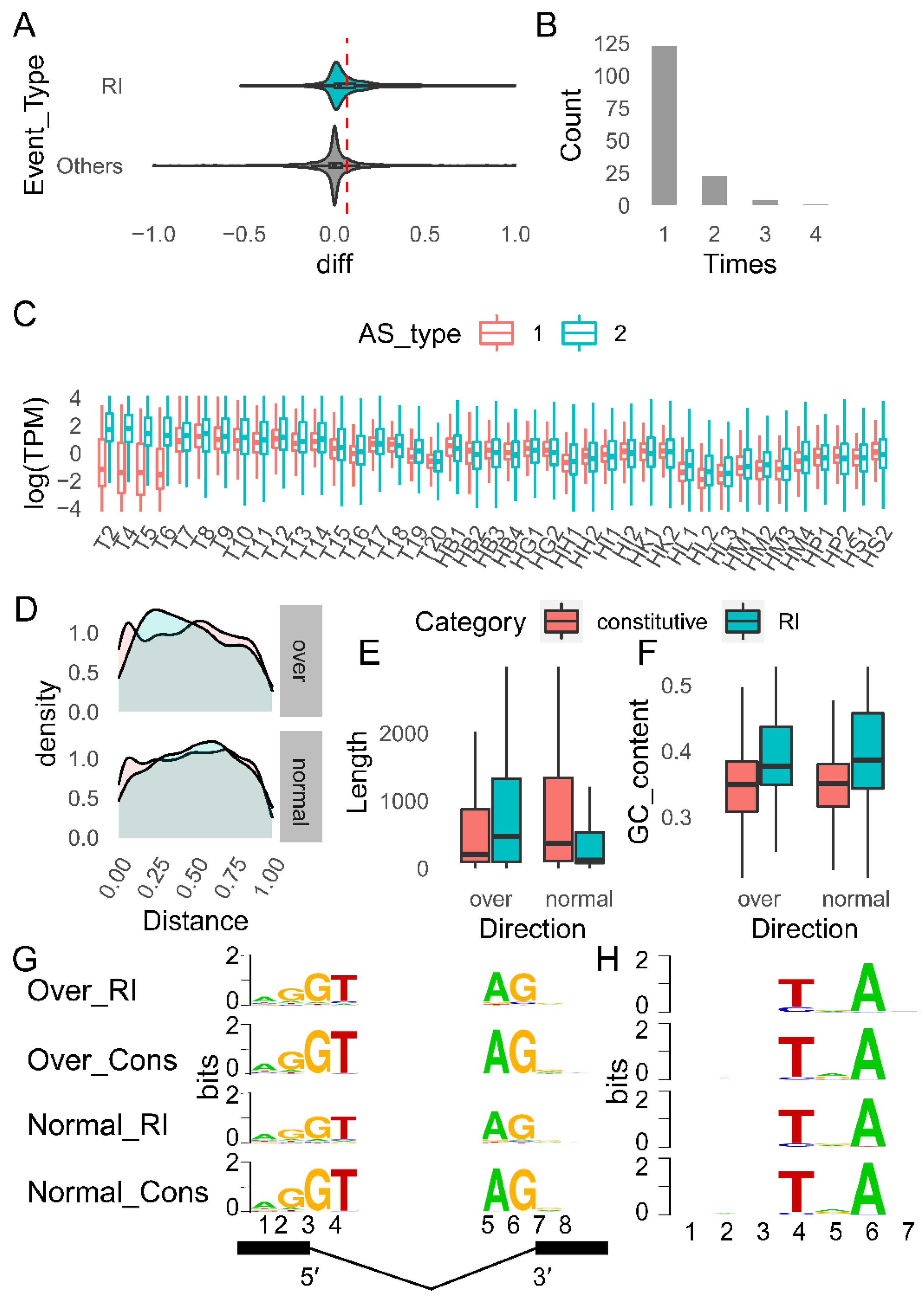
2.4.2. RI Events Changes Dramatically during T6–T7
2.4.3. Characteristics of RI Events during T6–T7
2.4.4. Distribution of Splicing Isoforms in Nine Tissues and the Tissue Specificity of RI Alternative Splicing
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Preparation of Sample for Library Construction
4.3. Library Construction and Sequencing
4.3.1. ssRNA-Seq
4.3.2. SMRT-Based RNA-Seq
4.4. Transcriptome Assembly
4.4.1. Generation Transcriptome for ssRNA-Seq Data and SMRT-Based RNA-Seq Data Separately
4.4.2. Merge ssRNA-Seq Transcriptome and SMRT Transcriptome
- Eliminate any transcripts that might have been fragmented. It is quite simple to confuse fragmented mRNA transcripts with newly assembled lncRNAs. In order to locate these transcripts, we first determined the distance between the transcript and the border of the scaffold and then checked to see if the transcript was supported by SMRT data. More than 95 percent of introns were less than 7875 base pairs, as shown in the length distribution of introns in all transcripts (Supplemental Table S12). It was decided that the transcripts whose distance from the scaffold edge was less than this value and which were not supported by SMRT data were fragmented transcripts. As a result, these transcripts were removed from the analysis.
- Remove possible precursor mRNAs. It is anticipated that the RNA-seq technology will discover a high number of mRNA fragments that have not been spliced or are in the process of being spliced. Because these fragments, unlike mature mRNA, exactly correspond to the full continuous region of the genome, it is common practice to confuse them with lengthy single-exon transcripts; nonetheless, they are eventually discovered to be long non-coding RNA. In order to get rid of the negative effects of such transcripts, we made the assumption that if a gene contains multi-exon transcripts as well as single-exon transcripts, the single-exon transcripts are very certainly precursors and should be removed as well.
4.5. Annotation of Transcriptome
4.6. Completeness of Transcriptome
4.7. Principal Component Analysis
4.8. Annotation and Analysis of Transposons
4.8.1. Identification, Annotation, and Relationship Analysis of Transposons
4.8.2. Motif Analysis
4.9. Validation Using qPCR
4.10. Alternative Splicing Analysis
4.11. Other Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Gerdes, P.; Richardson, S.R.; Mager, D.L.; Faulkner, G.J. Transposable elements in the mammalian embryo: Pioneers surviving through stealth and service. Genome Biol. 2016, 17, 100. [Google Scholar] [CrossRef] [PubMed]
- Fadloun, A.; Le Gras, S.; Jost, B.; Ziegler-Birling, C.; Takahashi, H.; Gorab, E.; Carninci, P.; Torres-Padilla, M.E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013, 20, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Ma, H.; Liu, D. Endogenous Retroviruses Function as Gene Expression Regulatory Elements During Mammalian Pre-implantation Embryo Development. Int. J. Mol. Sci. 2019, 20, 790. [Google Scholar] [CrossRef] [PubMed]
- Bui, L.C.; Evsikov, A.V.; Khan, D.R.; Archilla, C.; Peynot, N.; Henaut, A.; Le Bourhis, D.; Vignon, X.; Renard, J.P.; Duranthon, V. Retrotransposon expression as a defining event of genome reprogramming in fertilized and cloned bovine embryos. Reproduction 2009, 138, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflug. Arch. 2018, 470, 995–1016. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, S.C.; Lopez-Oreja, I.; Valcarcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef]
- McGuire, A.M.; Pearson, M.D.; Neafsey, D.E.; Galagan, J.E. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol. 2008, 9, R50. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.; Au, A.Y.; Ritchie, W.; Rasko, J.E. Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. Bioessays 2016, 38, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Braunschweig, U.; Barbosa-Morais, N.L.; Pan, Q.; Nachman, E.N.; Alipanahi, B.; Gonatopoulos-Pournatzis, T.; Frey, B.; Irimia, M.; Blencowe, B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014, 24, 1774–1786. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Tie, W.; Fu, L.; Yan, Y.; Liu, G.; Yan, W.; Li, Y.; Wu, C.; Zhang, J.; Hu, W. Strand-specific RNA-seq based identification and functional prediction of drought-responsive lncRNAs in cassava. BMC Genom. 2019, 20, 214. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, Y.F.; Feng, Y.Z.; He, H.; Lian, J.P.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol. J. 2020, 18, 679–690. [Google Scholar] [CrossRef]
- Zhou, Q.; Wan, Q.; Jiang, Y.; Liu, J.; Qiang, L.; Sun, L. A Landscape of Murine Long Non-Coding RNAs Reveals the Leading Transcriptome Alterations in Adipose Tissue during Aging. Cell Rep. 2020, 31, 107694. [Google Scholar] [CrossRef]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Wang, B.; Kumar, V.; Olson, A.; Ware, D. Reviving the Transcriptome Studies: An Insight Into the Emergence of Single-Molecule Transcriptome Sequencing. Front. Genet. 2019, 10, 384. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef]
- Tan, M.H.; Au, K.F.; Yablonovitch, A.L.; Wills, A.E.; Chuang, J.; Baker, J.C.; Wong, W.H.; Li, J.B. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013, 23, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.B.; Boley, N.; Eisman, R.; May, G.E.; Stoiber, M.H.; Duff, M.O.; Booth, B.W.; Wen, J.; Park, S.; Suzuki, A.M.; et al. Diversity and dynamics of the Drosophila transcriptome. Nature 2014, 512, 393–399. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Rozowsky, J.; Yan, K.K.; Wang, D.; Cheng, C.; Brown, J.B.; Davis, C.A.; Hillier, L.; Sisu, C.; Li, J.J.; et al. Comparative analysis of the transcriptome across distant species. Nature 2014, 512, 445–448. [Google Scholar] [CrossRef]
- Hashimshony, T.; Feder, M.; Levin, M.; Hall, B.K.; Yanai, I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature 2015, 519, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Boeck, M.E.; Huynh, C.; Gevirtzman, L.; Thompson, O.A.; Wang, G.; Kasper, D.M.; Reinke, V.; Hillier, L.W.; Waterston, R.H. The time-resolved transcriptome of C. elegans. Genome Res. 2016, 26, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Anavy, L.; Cole, A.G.; Winter, E.; Mostov, N.; Khair, S.; Senderovich, N.; Kovalev, E.; Silver, D.H.; Feder, M.; et al. The mid-developmental transition and the evolution of animal body plans. Nature 2016, 531, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Owens, N.D.L.; Blitz, I.L.; Lane, M.A.; Patrushev, I.; Overton, J.D.; Gilchrist, M.J.; Cho, K.W.Y.; Khokha, M.K. Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cell Rep. 2016, 14, 632–647. [Google Scholar] [CrossRef]
- Yi, S.; Zhou, X.; Li, J.; Zhang, M.; Luo, S. Full-length transcriptome of Misgurnus anguillicaudatus provides insights into evolution of genus Misgurnus. Sci. Rep. 2018, 8, 11699. [Google Scholar] [CrossRef]
- Hu, F.; Fan, J.; Wu, C.; Zhu, M.; Zhou, Y.; Wang, S.; Zhang, C.; Tao, M.; Zhao, R.; Tang, C.; et al. Analysis of Chromosomal Numbers, Mitochondrial Genome, and Full-Length Transcriptome of Onychostoma brevibarba. Mar. Biotechnol. 2019, 21, 515–525. [Google Scholar] [CrossRef]
- Luo, H.; Liu, H.; Zhang, J.; Hu, B.; Zhou, C.; Xiang, M.; Yang, Y.; Zhou, M.; Jing, T.; Li, Z.; et al. Full-length transcript sequencing accelerates the transcriptome research of Gymnocypris namensis, an iconic fish of the Tibetan Plateau. Sci. Rep. 2020, 10, 9668. [Google Scholar] [CrossRef]
- Ren, L.; Yan, X.; Gao, X.; Cui, J.; Yan, P.; Wu, C.; Li, W.; Liu, S. Maternal effects shape the alternative splicing of parental alleles in reciprocal cross hybrids of Megalobrama amblycephala x Culter alburnus. BMC Genom. 2020, 21, 457. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, G.; Frasca, A.; Kent, B.; Sadler, K.C.; Sealfon, S.C.; Walsh, M.J.; Zaslavsky, E. High resolution annotation of zebrafish transcriptome using long-read sequencing. Genome Res. 2018, 28, 1415–1425. [Google Scholar] [CrossRef]
- Mehjabin, R.; Xiong, L.; Huang, R.; Yang, C.; Chen, G.; He, L.; Liao, L.; Zhu, Z.; Wang, Y. Full-Length Transcriptome Sequencing and the Discovery of New Transcripts in the Unfertilized Eggs of Zebrafish (Danio rerio). G3 2019, 9, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Mohindra, V.; Dangi, T.; Chowdhury, L.M.; Jena, J.K. Tissue specific alpha-2-Macroglobulin (A2M) splice isoform diversity in Hilsa shad, Tenualosa ilisha (Hamilton, 1822). PLoS ONE 2019, 14, e0216144. [Google Scholar] [CrossRef]
- Diao, J.; Yu, X.; Wang, X.; Fan, Y.; Wang, S.; Li, L.; Wang, Y.; Gai, C.; Ye, H.; Liu, H.; et al. Full-length transcriptome sequencing combined with RNA-seq analysis revealed the immune response of fat greenling (Hexagrammos otakii) to Vibrio harveyi in early infection. Microb. Pathog. 2020, 149, 104527. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, J.; Li, L.; Huang, W.; Ahmad, H.I.; Li, H.; Jiang, H.; Chen, J. Full-length transcriptome sequencing and comparative transcriptomic analysis to uncover genes involved in early gametogenesis in the gonads of Amur sturgeon (Acipenser schrenckii). Front. Zool 2020, 17, 11. [Google Scholar] [CrossRef]
- Tian, Y.; Wen, H.; Qi, X.; Zhang, X.; Liu, S.; Li, B.; Sun, Y.; Li, J.; He, F.; Yang, W.; et al. Characterization of Full-Length Transcriptome Sequences and Splice Variants of Lateolabrax maculatus by Single-Molecule Long-Read Sequencing and Their Involvement in Salinity Regulation. Front. Genet. 2019, 10, 1126. [Google Scholar] [PubMed]
- Feng, X.; Jia, Y.; Zhu, R.; Chen, K.; Chen, Y. Characterization and analysis of the transcriptome in Gymnocypris selincuoensis on the Qinghai-Tibetan Plateau using single-molecule long-read sequencing and RNA-seq. DNA Res. 2019, 26, 353–363. [Google Scholar] [CrossRef]
- Mathavan, S.; Lee, S.G.; Mak, A.; Miller, L.D.; Murthy, K.R.; Govindarajan, K.R.; Tong, Y.; Wu, Y.L.; Lam, S.H.; Yang, H.; et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005, 1, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Rauwerda, H.; Pagano, J.F.; de Leeuw, W.C.; Ensink, W.; Nehrdich, U.; de Jong, M.; Jonker, M.; Spaink, H.P.; Breit, T.M. Transcriptome dynamics in early zebrafish embryogenesis determined by high-resolution time course analysis of 180 successive, individual zebrafish embryos. BMC Genom. 2017, 18, 287. [Google Scholar] [CrossRef] [PubMed]
- Nepal, C.; Hadzhiev, Y.; Previti, C.; Haberle, V.; Li, N.; Takahashi, H.; Suzuki, A.M.; Sheng, Y.; Abdelhamid, R.F.; Anand, S.; et al. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res. 2013, 23, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.A.; Sealy, I.; Kettleborough, R.; Fenyes, F.; White, R.; Stemple, D.; Smith, J.C. Identification of the zebrafish maternal and paternal transcriptomes. Development 2013, 140, 2703–2710. [Google Scholar] [CrossRef] [PubMed]
- Aanes, H.; Winata, C.L.; Lin, C.H.; Chen, J.P.; Srinivasan, K.G.; Lee, S.G.; Lim, A.Y.; Hajan, H.S.; Collas, P.; Bourque, G.; et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011, 21, 1328–1338. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 2017, 6, e30860. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Jiang, K.; Zhang, F.; Lin, Y.; Ma, L. RNA-sequencing of the sturgeon Acipenser baeri provides insights into expression dynamics of morphogenic differentiation and developmental regulatory genes in early versus late developmental stages. BMC Genom. 2016, 17, 564. [Google Scholar] [CrossRef]
- Singh, P.; Borger, C.; More, H.; Sturmbauer, C. The Role of Alternative Splicing and Differential Gene Expression in Cichlid Adaptive Radiation. Genome Biol. Evol. 2017, 9, 2764–2781. [Google Scholar] [CrossRef]
- Fu, J.; Zhu, W.; Wang, L.; Luo, M.; Song, F.; Dong, Z. Dynamic transcriptome sequencing and analysis during early development in the bighead carp (Hypophthalmichthys nobilis). BMC Genom. 2019, 20, 781. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Su, B.; Tian, Y.; Backenstose, N.J.C.; Ye, Z.; Moss, A.; Duong, T.Y.; Wang, X.; Dunham, R.A. Deep Transcriptomic Analysis Reveals the Dynamic Developmental Progression during Early Development of Channel Catfish (Ictalurus punctatus). Int. J. Mol. Sci. 2020, 21, 5535. [Google Scholar] [CrossRef]
- FAO. FAO Yearbook. Fishery and Aquaculture Statistics 2018; Food and Agricultural Organization (FAO): Rome, Italy, 2020. [Google Scholar]
- Gan, L.; Wang, Y.-z.; Chen, S.-j.; Lin, Z.-h.; Sun, J.-j.; He, Y.-h.; Tang, H.-j.; Peng, J.; Guo, H.-h. Identification and characterization of long non-coding RNAs in muscle sclerosis of grass carp, Ctenopharyngodon idellus fed with faba bean meal. Aquaculture 2020, 516, 734521. [Google Scholar] [CrossRef]
- Fan, K.; Shen, Y.; Xu, X.; Tao, L.; Bao, T.; Li, J. LncRNA-WAS and lncRNA-C8807 interact with miR-142a-3p to regulate the inflammatory response in grass carp. Fish Shellfish. Immunol. 2021, 111, 201–207. [Google Scholar] [CrossRef]
- He, Y.; Yu, H.; Zhao, H.; Zhu, H.; Zhang, Q.; Wang, A.; Shen, Y.; Xu, X.; Li, J. Transcriptomic analysis to elucidate the effects of high stocking density on grass carp (Ctenopharyngodon idella). BMC Genom. 2021, 22, 620. [Google Scholar] [CrossRef]
- Li, L.; Jia, X.; Liu, Y.; He, Y.; Pang, Y.; Shen, Y.; Xu, X.; Li, J. lncRNA-SUMO3 and lncRNA-HDMO13 modulate the inflammatory response by binding miR-21 and miR-142a-3p in grass carp. Dev. Comp. Immunol. 2021, 121, 104082. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Y.; Zhang, Y.; Ning, Z.; Li, Y.; Zhao, Q.; Lu, H.; Huang, R.; Xia, X.; Feng, Q.; et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015, 47, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wei, X.; Liao, Z.; Gao, Y.; Liu, X.; Su, J.; Yuan, G. Transcriptome Analysis Provides Insights into the Markers of Resting and LPS-Activated Macrophages in Grass Carp (Ctenopharyngodon idella). Int. J. Mol. Sci. 2018, 19, 3562. [Google Scholar] [CrossRef]
- Huang, X.; Jiang, Y.; Zhang, W.; Cheng, Y.; Wang, Y.; Ma, X.; Duan, Y.; Xia, L.; Chen, Y.; Wu, N.; et al. Construction of a high-density genetic map and mapping of growth related QTLs in the grass carp (Ctenopharyngodon idellus). BMC Genom. 2020, 21, 313. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, H.-M.; Qian, X.-Q.; Gui, J.-F. Transcriptome analysis of grass carp (Ctenopharyngodon idella) between fast- and slow-growing fish. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100688. [Google Scholar] [CrossRef]
- Akasaki, T.; Nikaido, M.; Nishihara, H.; Tsuchiya, K.; Segawa, S.; Okada, N. Characterization of a novel SINE superfamily from invertebrates: “Ceph-SINEs” from the genomes of squids and cuttlefish. Gene 2010, 454, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.; Labuda, D. Evolutionary inventions and continuity of CORE-SINEs in mammals. J. Mol. Biol. 2000, 298, 365–377. [Google Scholar] [CrossRef]
- Eng, L.; Coutinho, G.; Nahas, S.; Yeo, G.; Tanouye, R.; Babaei, M.; Dörk, T.; Burge, C.; Gatti, R.A. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: Maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004, 23, 67–76. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef] [PubMed]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef]
- Irie, N.; Kuratani, S. The developmental hourglass model: A predictor of the basic body plan? Development 2014, 141, 4649–4655. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, V.N. The Ground Zero of Organismal Life and Aging. Trends Mol. Med. 2021, 27, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Rugh, R. The Frog Its Reproduction and Development; Blakiston: Philadelphia, PA, USA, 1951. [Google Scholar]
- Kellicott, W.E. The Development of the Vascular and Respiratory Systems of Ceratodus; New York Academy of Sciences: New York, NY, USA, 1905; Volume 2. [Google Scholar]
- Gillis, J.A.; Tidswell, O.R. The Origin of Vertebrate Gills. Curr. Biol. 2017, 27, 729–732. [Google Scholar] [CrossRef] [PubMed]
- de Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef]
- Rebollo, R.; Romanish, M.T.; Mager, D.L. Transposable elements: An abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 2012, 46, 21–42. [Google Scholar] [CrossRef]
- Alexander, R.P.; Fang, G.; Rozowsky, J.; Snyder, M.; Gerstein, M.B. Annotating non-coding regions of the genome. Nat. Rev. Genet. 2010, 11, 559–571. [Google Scholar] [CrossRef]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef] [PubMed]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Arensburger, P.; Hice, R.H.; Zhou, L.; Smith, R.C.; Tom, A.C.; Wright, J.A.; Knapp, J.; O’Brochta, D.A.; Craig, N.L.; Atkinson, P.W. Phylogenetic and functional characterization of the hAT transposon superfamily. Genetics 2011, 188, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, C.; Liu, X.; Gao, S. Insights into epigenetic patterns in mammalian early embryos. Protein Cell 2020, 12, 7–28. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X. Exploratory bioinformatics investigation reveals importance of “junk” DNA in early embryo development. BMC Genom. 2017, 18, 200. [Google Scholar] [CrossRef]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sun, C.; Clinton, M.; Yang, N. Dynamic Transcriptional Landscape of the Early Chick Embryo. Front. Cell Dev. Biol. 2019, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Monteuuis, G.; Wong, J.J.L.; Bailey, C.G.; Schmitz, U.; Rasko, J.E.J. The changing paradigm of intron retention: Regulation, ramifications and recipes. Nucleic Acids Res. 2019, 47, 11497–11513. [Google Scholar] [CrossRef]
- Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Pertea, M. GFF Utilities: GffRead and GffCompare [version 1; peer review: 3 approved]. F1000Research 2020, 9, 304. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, W.; Cheng, Y.; Shi, M.; Xia, X.-Q. A systematic evaluation of bioinformatics tools for identification of long noncoding RNAs. Rna 2021, 27, 80–98. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Vassetzky, N.S.; Kramerov, D.A. SINEBase: A database and tool for SINE analysis. Nucleic Acids Res. 2013, 41, D83–D89. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef]
- Bailey, T.L.; Gribskov, M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef]
- Zhong, Z.; Ao, L.; Zhao, L.; Zhang, Z.; Jiang, Y. Screening and validation of reference genes for qPCR analysis in gonads and embryos of Takifugu bimaculatus. Aquac. Fish. 2020, 7, 278–286. [Google Scholar] [CrossRef]
- Caracausi, M.; Piovesan, A.; Antonaros, F.; Strippoli, P.; Vitale, L.; Pelleri, M.C. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol. Med. Rep. 2017, 16, 2397–2410. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Fan, X.; Wang, Y.; Sun, M.A.; Shao, J.; Guo, D. BPP: A sequence-based algorithm for branch point prediction. Bioinformatics 2017, 33, 3166–3172. [Google Scholar] [CrossRef] [PubMed]
- Beißbarth, T.; Speed, T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 2004, 20, 1464–1465. [Google Scholar] [CrossRef] [PubMed]

| Sample ID | Time after Fertilization | Tissue | Characteristics |
|---|---|---|---|
| T2 | 0 hpf | ovum | ovum |
| T4 | 1.6 hpf | embryo | 1, 2, 4 cells |
| T5 | 2.8 hpf | embryo | 64 cells |
| T6 | 3.7 hpf | embryo | 256 cells |
| T7 | 5.2 hpf | embryo | dome |
| T8 | 6.3 hpf | embryo | 50% epiboly |
| T9 | 7.9 hpf | embryo | 75% epiboly |
| T10 | 9.1 hpf | embryo | 90% epiboly ~ bud |
| T11 | 11.1 hpf | embryo | 4~6 somite |
| T12 | 12.9 hpf | embryo | 15 somite |
| T13 | 14.8 hpf | embryo | 18~19 somite |
| T14 | 18.0 hpf | embryo | 25~26 somite, starting to shrink |
| T15 | 21.8 hpf | embryo | rotation |
| T16 | 31.8 hpf | full fish | hatching |
| T17 | 3 dpf | full fish | yolk exhausted, appearance of eyes, start to swim |
| T18 | 6 dpf | full fish | first feeding |
| T19 | 13 dpf | full fish | NA |
| T20 | 20 dpf | full fish | NA |
| B1, M1 | 39 dpf | B, M | NA |
| B2, M2, L1 | 69 dpf | B, M, L | NA |
| B3, M3, L2, G1, Sk1, Sp1, K1, H1, I1 | 134 dpf | B, M, L, G, Sk, Sp, K, H, I | NA |
| B4, M4, L3, G2, Sk2, Sp2, K2, H2, I2 | 197 dpf | B, M, L, G, Sk, Sp, K, H, I | NA |
| Class | Count | Ratio in Expressed TE | Count in Genome | Fraction in Total Transposon Elements |
|---|---|---|---|---|
| unclassified LTR | 2 | 3.13% | 14,757 | 0.88% |
| DNA/TcMar | 8 | 12.50% | 106,014 | 6.29% |
| LINE/L2 | 13 | 20.31% | 44,915 | 2.66% |
| unclassified SINE | 41 | 64.06% | 10,123 | 0.60% |
| Category | Over Events | Normal Events | Classic Features [80] |
|---|---|---|---|
| Relative position | 5′ (not significant) | 3′ (not significant) | 3′ |
| Length | Long | Short | Short |
| GC content | High | High | High |
| Strength of splice site | Weak | Weak | Weak |
| Strength of BPS | Weak | Weak | Weak |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Zhang, Q.; Jiang, Y.; Zhang, W.; Cheng, Y.; Shi, M.; Xia, X.-Q. Dynamic Transcriptional Landscape of Grass Carp (Ctenopharyngodon idella) Reveals Key Transcriptional Features Involved in Fish Development. Int. J. Mol. Sci. 2022, 23, 11547. https://doi.org/10.3390/ijms231911547
Duan Y, Zhang Q, Jiang Y, Zhang W, Cheng Y, Shi M, Xia X-Q. Dynamic Transcriptional Landscape of Grass Carp (Ctenopharyngodon idella) Reveals Key Transcriptional Features Involved in Fish Development. International Journal of Molecular Sciences. 2022; 23(19):11547. https://doi.org/10.3390/ijms231911547
Chicago/Turabian StyleDuan, You, Qiangxiang Zhang, Yanxin Jiang, Wanting Zhang, Yingyin Cheng, Mijuan Shi, and Xiao-Qin Xia. 2022. "Dynamic Transcriptional Landscape of Grass Carp (Ctenopharyngodon idella) Reveals Key Transcriptional Features Involved in Fish Development" International Journal of Molecular Sciences 23, no. 19: 11547. https://doi.org/10.3390/ijms231911547
APA StyleDuan, Y., Zhang, Q., Jiang, Y., Zhang, W., Cheng, Y., Shi, M., & Xia, X.-Q. (2022). Dynamic Transcriptional Landscape of Grass Carp (Ctenopharyngodon idella) Reveals Key Transcriptional Features Involved in Fish Development. International Journal of Molecular Sciences, 23(19), 11547. https://doi.org/10.3390/ijms231911547






