Developmental Transcriptome Analysis of Red-Spotted Apollo Butterfly, Parnassius bremeri
Abstract
1. Introduction
2. Results
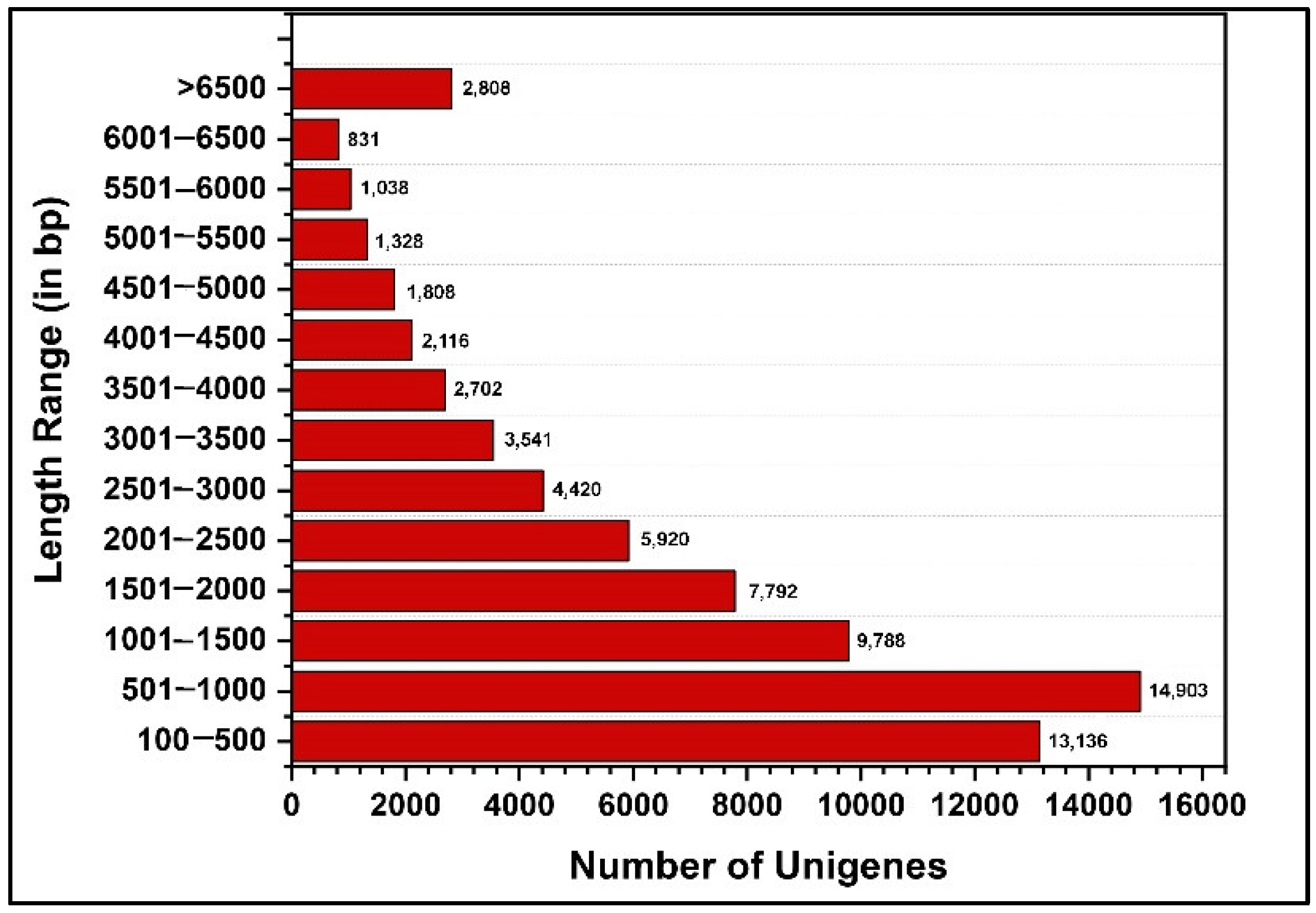
2.1. Sequencing and De Novo Assembly of the P. bremeri Transcriptome
2.2. Functional Annotation
2.3. Identification of Differently Expressed Genes (DEGs)
2.4. Differentially Expressed Genes (DEGs) across Egg versus First Instar Larval Comparisons
2.5. Differentially Expressed Genes (DEGs) across Developmental Transition Phases
2.5.1. Initial Transition Phase: First Instar Larval to Second Instar Larval
2.5.2. Final Transition Phase: Fifth Instar Larval to Male Adults and Female Adults
2.6. Differential Expressed Genes across the Mid-Larval Developmental Stages (L2 to L5)
2.7. DEGs in Adult Male (AM) vs. Adult Female (AF)
2.8. Differential Expression Genes in the Butterfly Tissues of Both the Sexes
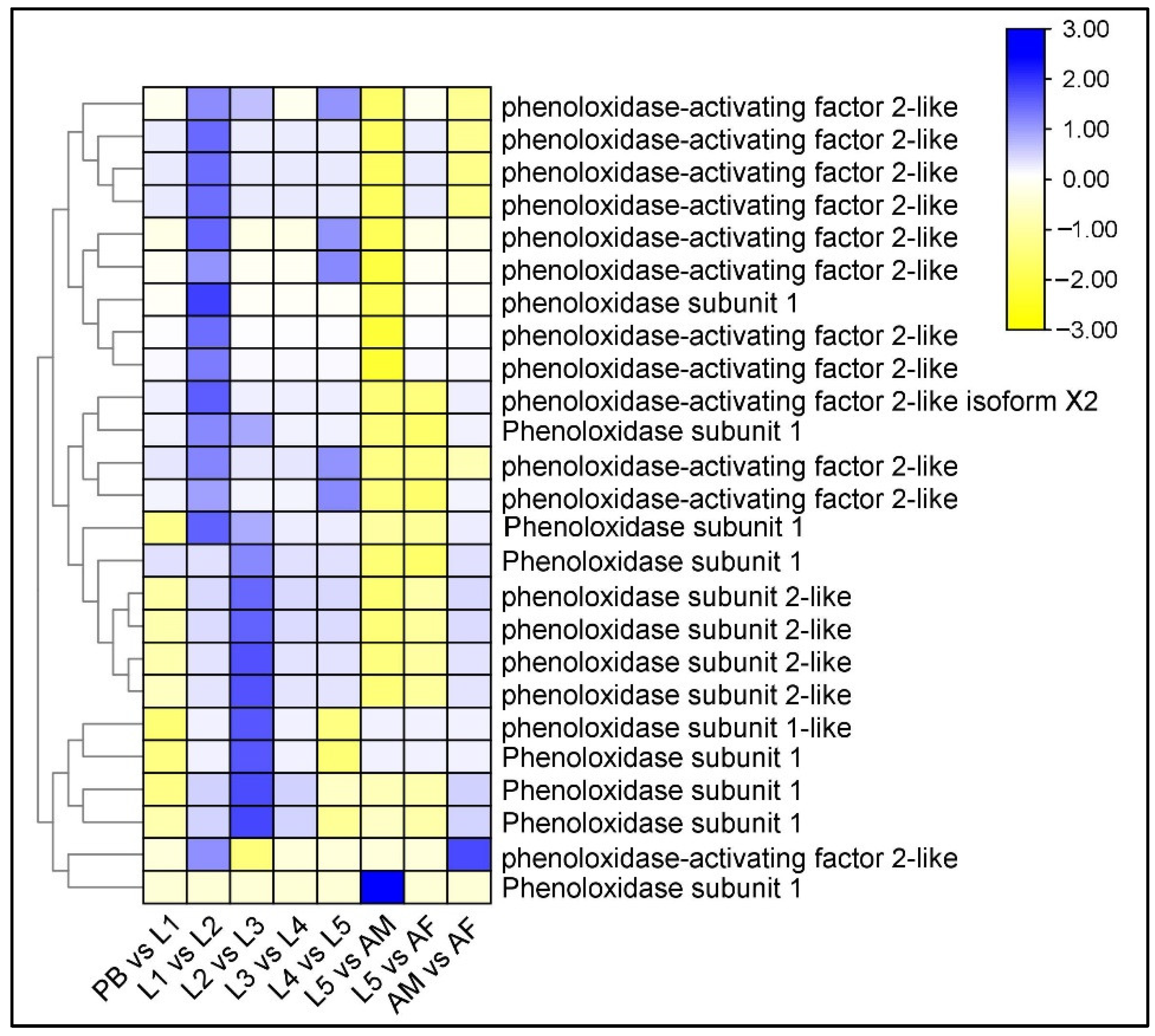
2.9. Differential Regulation of Larval Cuticle and Its Related Protein
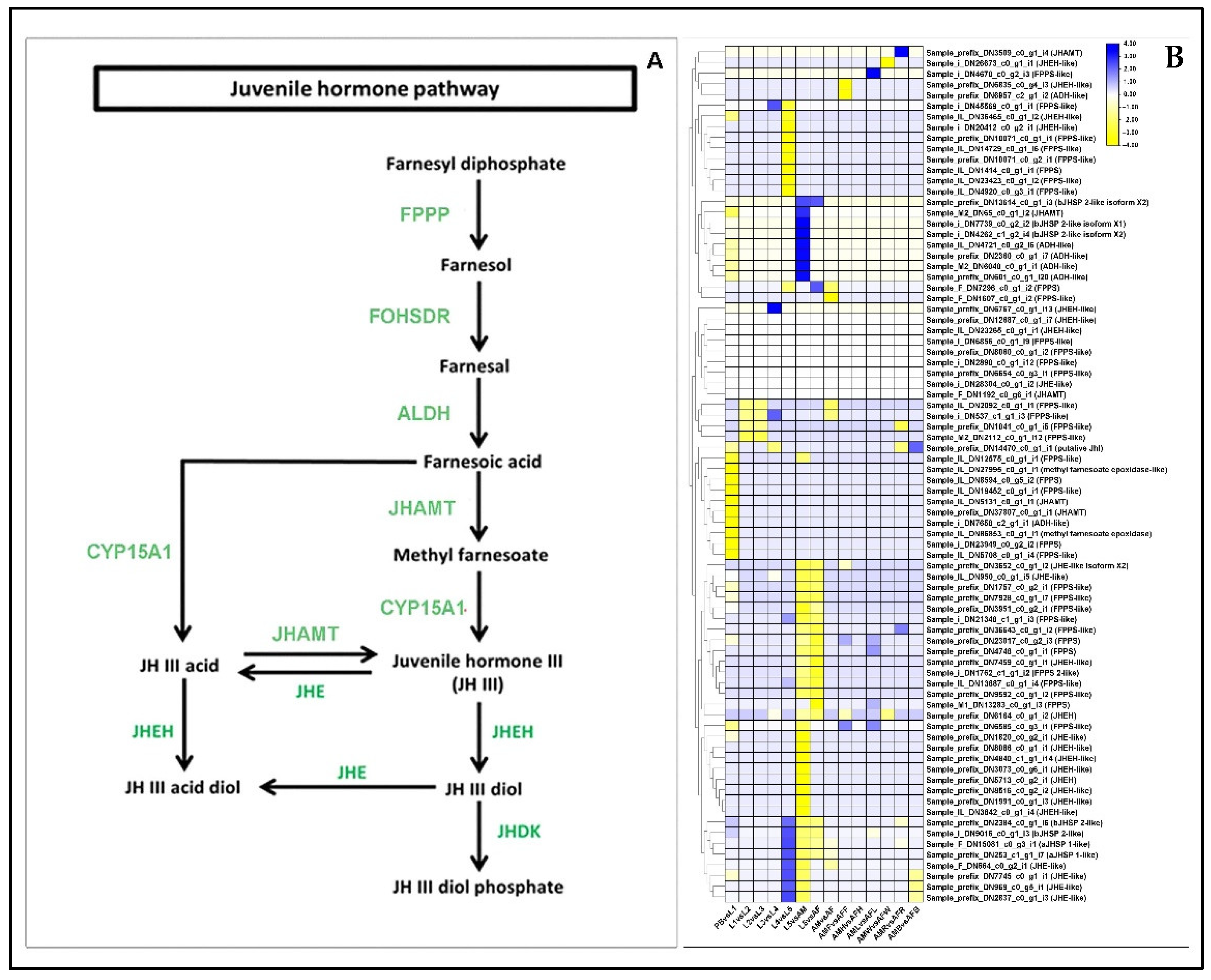
2.10. Differential Regulation of Juvenile-Hormone Signaling Genes
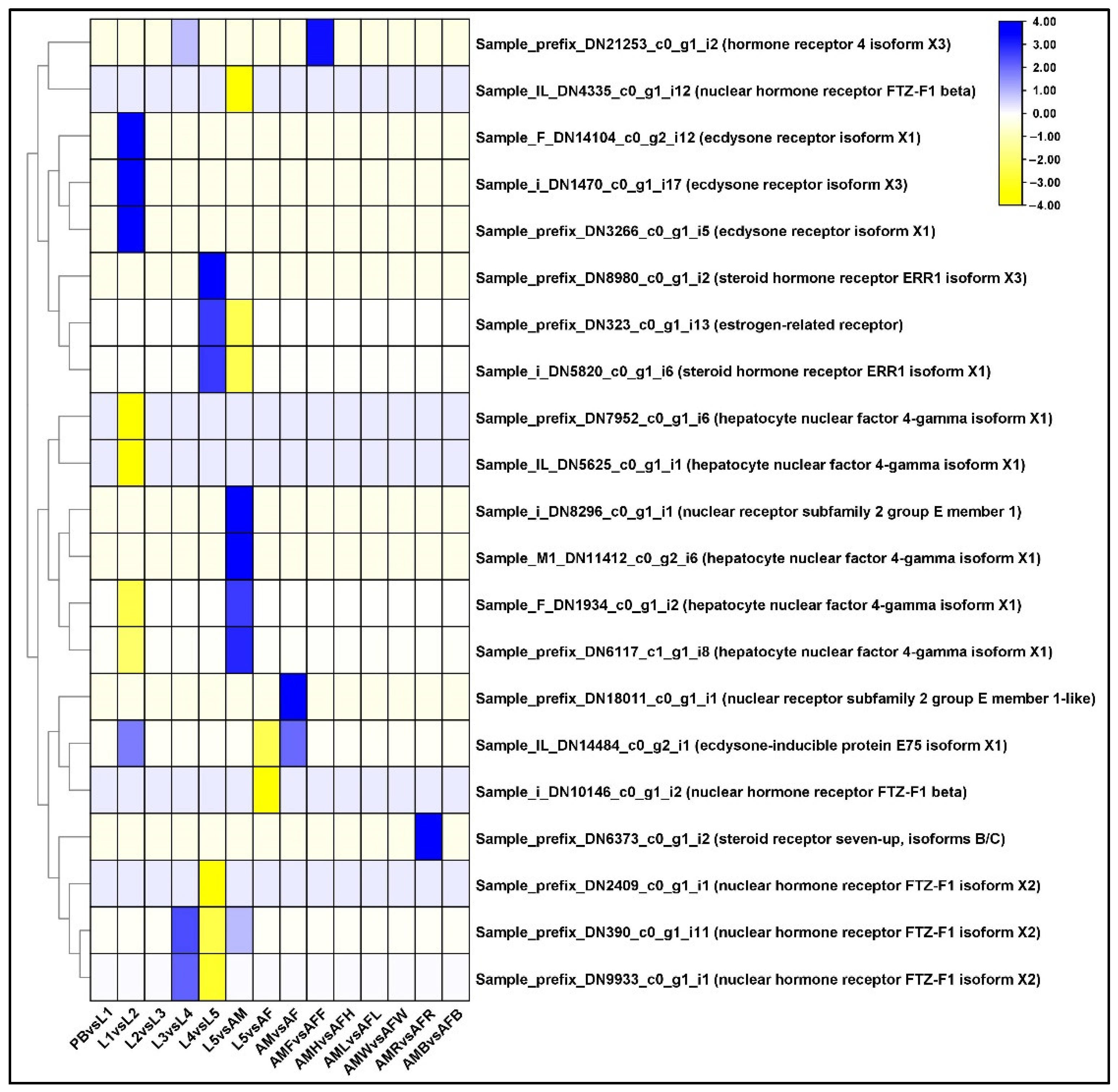
2.11. Differential Regulation of Ecdysone Receptor (EcR) Family Genes
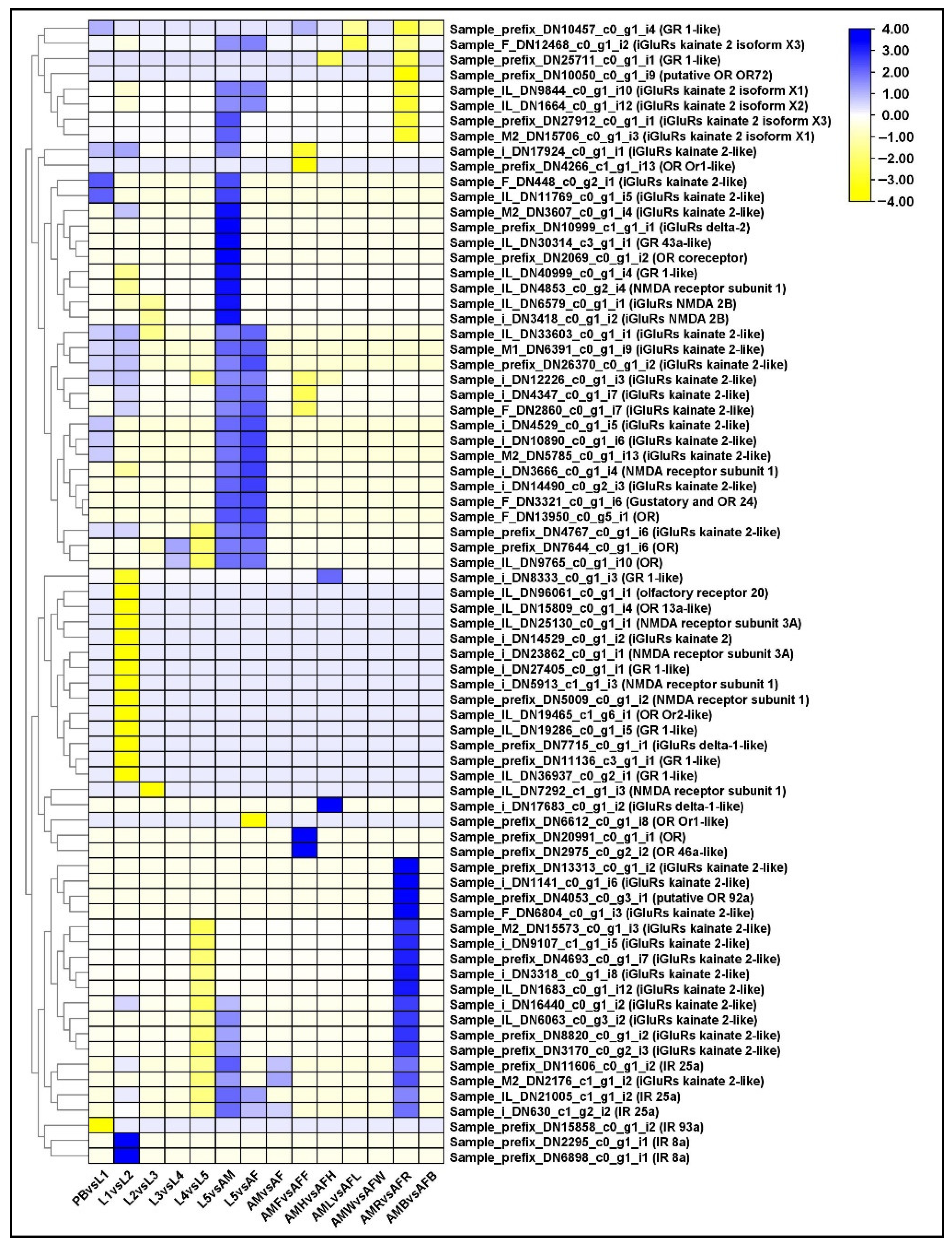
2.12. Differential Regulation of Sensory Receptors
2.13. Differential Regulation of Pathways in Wing and Eye-Spot Development
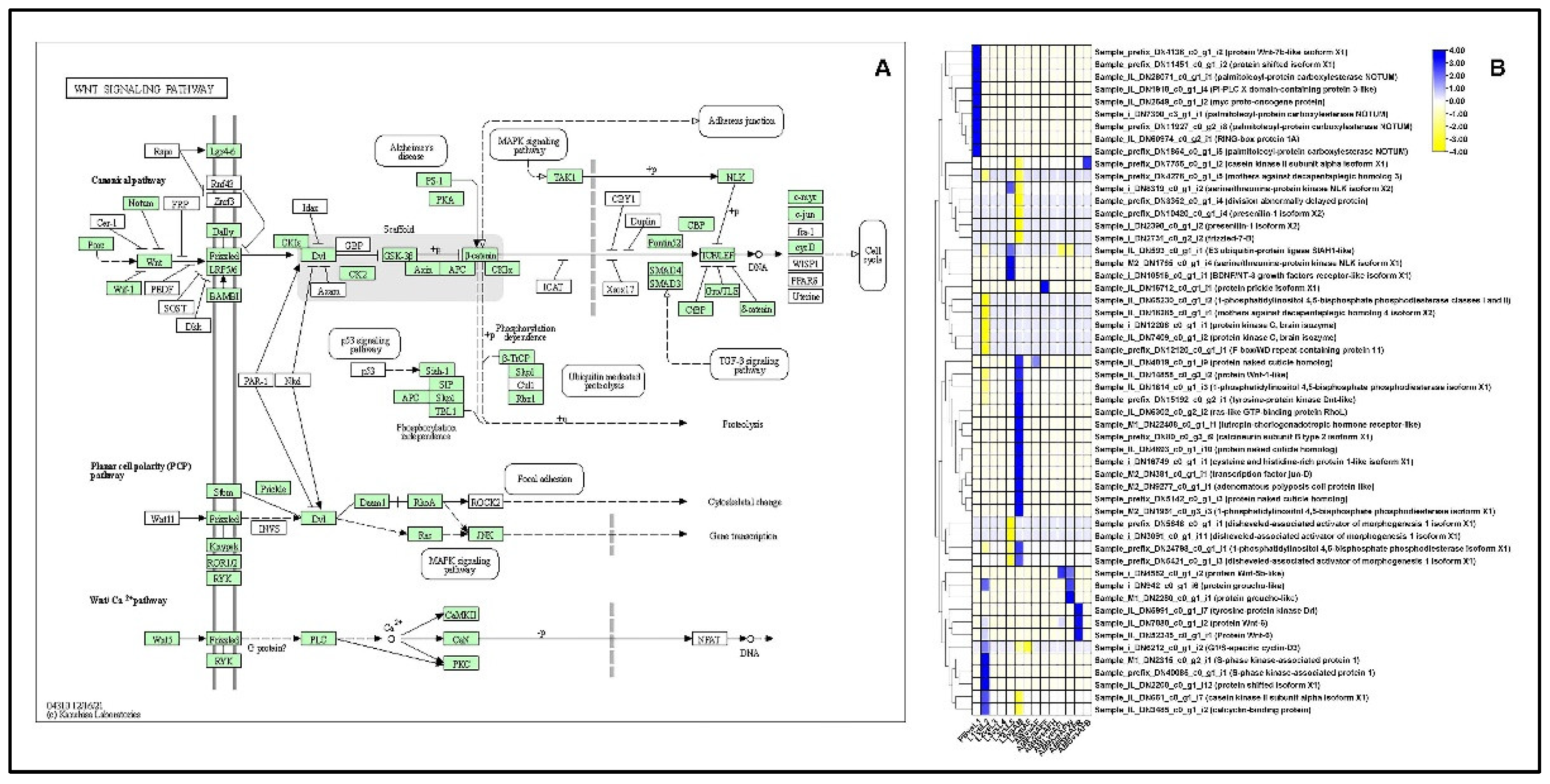
2.13.1. Wnt-Signaling Pathway
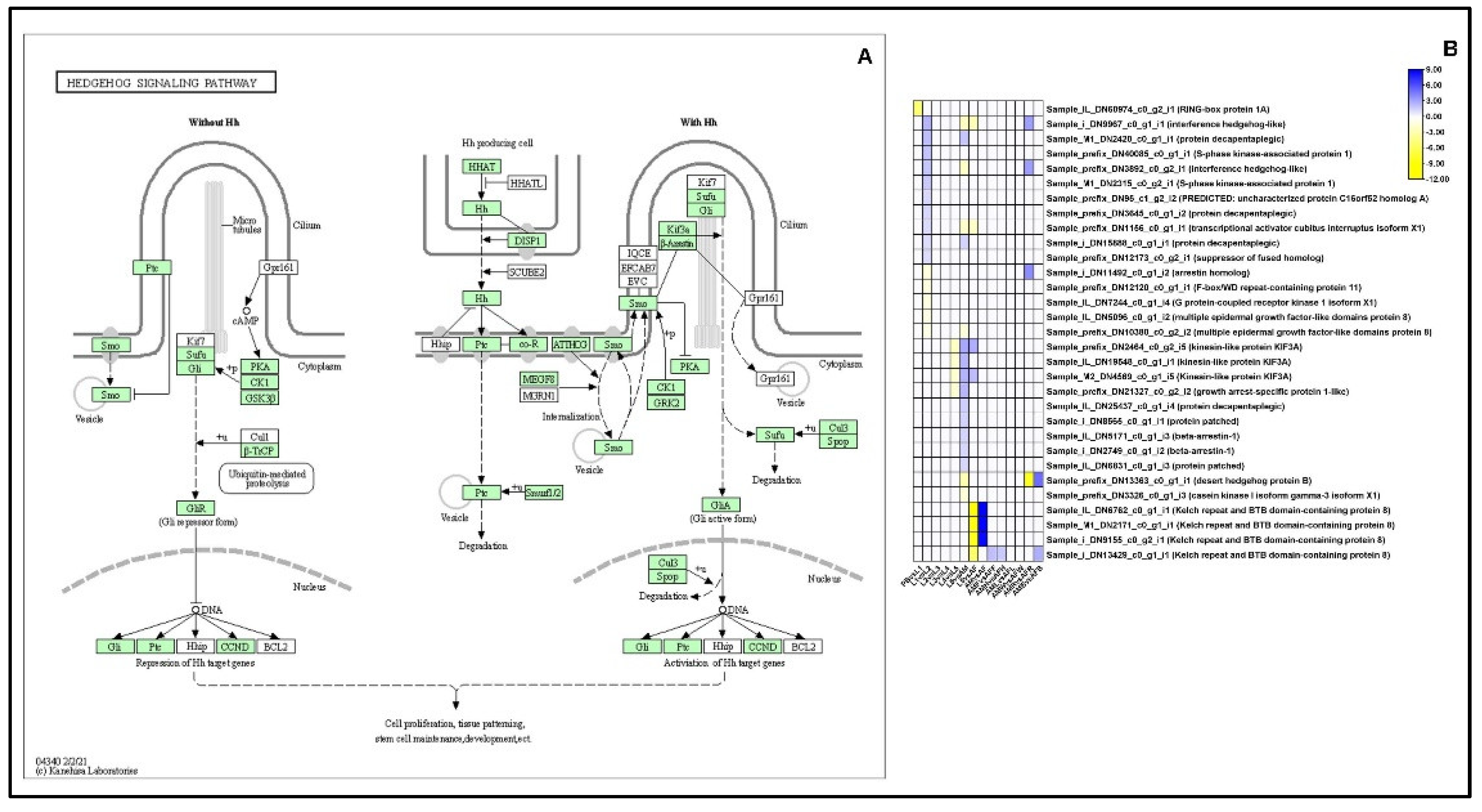
2.13.2. Hedgehog Signaling Pathway
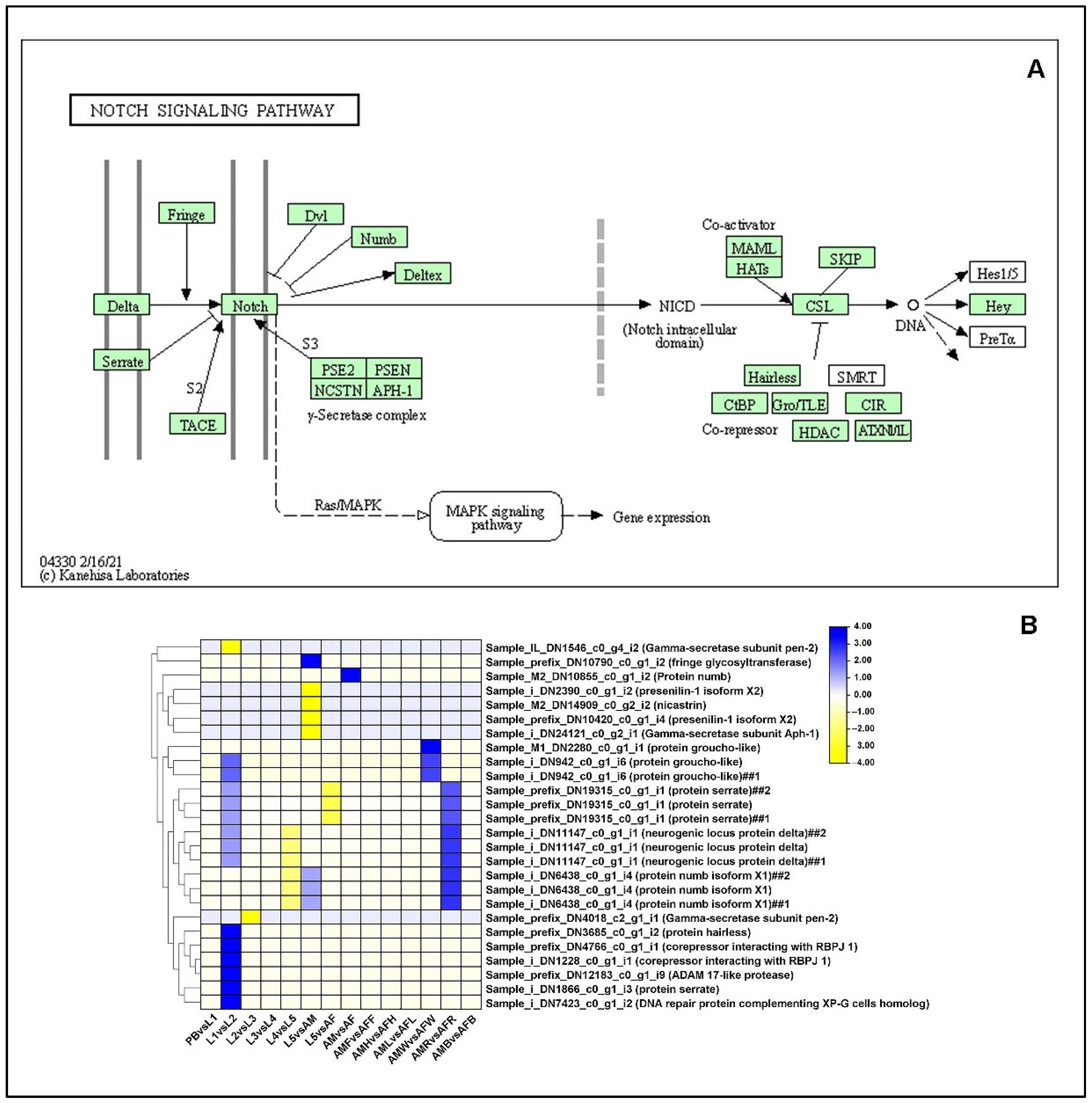
2.13.3. Notch-Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. RNA Extraction and Sequencing
4.3. Assembly Normalization and Quality Assessment
4.4. Functional Annotation and GO Enrichment
4.5. Analysis of Differentially Expressed Genes (DEGs)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 2015, 1, e1500052. [Google Scholar] [CrossRef] [PubMed]
- TODISCO, V.; GRATTON, P.; CESARONI, D.; SBORDONI, V. Phylogeography of Parnassius apollo: Hints on taxonomy and conservation of a vulnerable glacial butterfly invader. Biol. J. Linn. Soc. 2010, 101, 169–183. [Google Scholar] [CrossRef]
- Junker, M.; Konvicka, M.; Zimmermann, K.; Schmitt, T. Gene-flow within a butterfly metapopulation: The marsh fritillary Euphydryas aurinia in western Bohemia (Czech Republic). J. Insect Conserv. 2021, 25, 585–596. [Google Scholar] [CrossRef]
- Kim, M.I.; Baek, J.Y.; Kim, M.J.; Jeong, H.C.; Kim, K.-G.; Bae, C.H.; Han, Y.S.; Jin, B.R.; Kim, I. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol. Cells 2009, 28, 347–363. [Google Scholar] [CrossRef]
- Kim, D.-S.; Park, D.-S.; Kwon, Y.-J.; Suh, S.-J.; Kim, C.-H.; Park, S.-J.; Kim, D.-H.; Kim, J.-S.; Yu, H.-M.; Hwang, J.-S. Metapopulation structure and movement of a threatened butterfly Parnassius bremeri (Lepidoptera: Papilionidae) in Korea. Korean J. Appl. Entomol. 2011, 50, 97–105. [Google Scholar] [CrossRef]
- Park, G.W.; Jung, R.; Woon, K. Sex Pheromones as a tool to overcome Parnassius bremeri Bremer shortfall in conservation biology. (Lepidoptera: Papilionidae). In Proceedings of the Korea Society of Applied Entomology Conference 2016 한국응용곤충학회 정기총회 및 국제 심포지엄; Korean Society of Applied Entomology: Korea, 2016; p. 93. [Google Scholar]
- Park, Y.; Kim, Y.; Park, G.-W.; Lee, J.-O.; Lee, K.-W. Supercooling capacity along with up-regulation of glycerol content in an overwintering butterfly, Parnassius bremeri. J. Asia-Pac. Entomol. 2017, 20, 949–954. [Google Scholar] [CrossRef]
- Rai, A.; Yamazaki, M.; Takahashi, H.; Nakamura, M.; Kojoma, M.; Suzuki, H.; Saito, K. RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Front. Plant Sci. 2016, 7, 481. [Google Scholar] [CrossRef]
- Powell, J.A. Lepidoptera: Moths, butterflies. In Encyclopedia of Insects; Elsevier: Amsterdam, The Netherlands, 2009; pp. 559–587. [Google Scholar]
- Zhang, L.; Steward, R.A.; Wheat, C.W.; Reed, R.D. High-quality genome assembly and comprehensive transcriptome of the painted lady butterfly Vanessa cardui. Genome Biol. Evol. 2021, 13, evab145. [Google Scholar] [CrossRef]
- Qi, L.; Fang, Q.; Zhao, L.; Xia, H.; Zhou, Y.; Xiao, J.; Li, K.; Ye, G. De novo assembly and developmental transcriptome analysis of the small white butterfly Pieris rapae. PLoS ONE 2016, 11, e0159258. [Google Scholar] [CrossRef]
- Macias-Muñoz, A.; Smith, G.; Monteiro, A.; Briscoe, A.D. Transcriptome-wide differential gene expression in Bicyclus anynana butterflies: Female vision-related genes are more plastic. Mol. Biol. Evol. 2016, 33, 79–92. [Google Scholar] [CrossRef]
- Podsiadlowski, L.; Tunström, K.; Espeland, M.; Wheat, C.W. The genome assembly and annotation of the Apollo butterfly Parnassius apollo, a flagship species for conservation biology. Genome Biol. Evol. 2021, 13, evab122. [Google Scholar] [CrossRef]
- He, J.-W.; Dong, Z.-W.; Hu, P.; Liu, W.; Zhang, R.; Liu, G.-C.; Zhao, R.-P.; Wan, W.-T.; Wang, W.; Li, X.-Y. Integrated analysis of transcriptome and proteome to reveal pupal color switch in Papilio xuthus butterflies. Front. Genet. 2021, 12, 795115. [Google Scholar] [CrossRef]
- Wan, W.-T.; Dong, Z.-W.; Ren, Y.-D.; Yang, J.; Pan, X.-Y.; He, J.-W.; Chang, Z.; Liu, W.; Liu, G.-C.; Zhao, R.-P. Chromatin accessibility profiling provides insights into larval cuticle color and adult longevity in butterflies. Zool. Res. 2021, 42, 614. [Google Scholar] [CrossRef]
- Lee, K.-W.; Kim, J.-G.; Veerappan, K.; Chung, H.; Natarajan, S.; Kim, K.-Y.; Park, J. Utilizing Red Spotted Apollo Butterfly Transcriptome to Identify Antimicrobial Peptide Candidates against Porphyromonas gingivalis. Insects 2021, 12, 466. [Google Scholar] [CrossRef]
- Gurevich, E.V.; Gurevich, V.V. Arrestins: Ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006, 7, 1–10. [Google Scholar] [CrossRef]
- Choi, J.; Roche, H.; Caquet, T. Characterization of superoxide dismutase activity in Chironomus riparius Mg.(Diptera, Chironomidae) larvae—A potential biomarker. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1999, 124, 73–81. [Google Scholar] [CrossRef]
- Dauwalder, B.; Tsujimoto, S.; Moss, J.; Mattox, W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002, 16, 2879–2892. [Google Scholar] [CrossRef]
- Chanchay, P.; Vongsangnak, W.; Thancharoen, A.; Sriboonlert, A. Reconstruction of insect hormone pathways in an aquatic firefly, Sclerotia aquatilis (Coleoptera: Lampyridae), using RNA-seq. PeerJ 2019, 7, e7428. [Google Scholar] [CrossRef]
- Barish, S.; Li, Q.; Pan, J.W.; Soeder, C.; Jones, C.; Volkan, P.C. Transcriptional profiling of olfactory system development identifies distal antenna as a regulator of subset of neuronal fates. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Kim, M.J.; Jeong, S.Y.; Kim, S.S.; Kim, I. Development and validation of microsatellite markers for the endangered red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae), in South Korea. Int. J. Ind. Entomol. 2017, 35, 30–38. [Google Scholar]
- Matter, S.; Wick, A.; Gaydos, M.; Frantz, M. Egg viability and larval contribution to fecundity of Parnassius smintheus Doubleday (Papilionidae). J.-Lepid. Soc. 2006, 60, 228. [Google Scholar]
- Wang, X.; Fan, Y.; Ge, Q.; Xu, J.; Taha, R.H.; Yuan, Y.; Chen, K. Time-Course Transcriptome Analysis Reveals Global Gene Expression Profiling and Dynamic Developmental Signatures across Complete Life Cycle of Bombyx mori. Processes 2021, 9, 1730. [Google Scholar] [CrossRef]
- McCorkle, D.V.; Hammond, P. Observations on the biology of Parnassius clodius (Papilionidae) in the Pacific Northwest. J. Lepid. Soc. 1985, 39, 156–162. [Google Scholar]
- Scott, J.A. The Butterflies of North America: A Natural History and Field Guide; Stanford University Press: Stanford, CA, USA, 1992. [Google Scholar]
- Maumus, F.; Fiston-Lavier, A.-S.; Quesneville, H. Impact of transposable elements on insect genomes and biology. Curr. Opin. Insect Sci. 2015, 7, 30–36. [Google Scholar] [CrossRef]
- Ding, D.; Lipshitz, H.D. Spatially regulated expression of retrovirus-like transposons during Drosophila melanogaster embryogenesis. Genet. Res. 1994, 64, 167–181. [Google Scholar] [CrossRef]
- Jiang, F.; Yang, M.; Guo, W.; Wang, X.; Kang, L. Large-scale transcriptome analysis of retroelements in the migratory locust, Locusta migratoria. PLoS ONE 2012, 7, e40532. [Google Scholar] [CrossRef]
- Kankare, M.; Parker, D.J.; Merisalo, M.; Salminen, T.S.; Hoikkala, A. Transcriptional differences between diapausing and non-diapausing D. montana females reared under the same photoperiod and temperature. PLoS ONE 2016, 11, e0161852. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Simon, S.; Sagasser, S.; Saccenti, E.; Brugler, M.R.; Schranz, M.E.; Hadrys, H.; Amato, G.; DeSalle, R. Comparative transcriptomics reveal developmental turning points during embryogenesis of a hemimetabolous insect, the damselfly Ischnura elegans. Sci. Rep. 2017, 7, 13547. [Google Scholar] [CrossRef]
- Charles, J.-P. The regulation of expression of insect cuticle protein genes. Insect Biochem. Mol. Biol. 2010, 40, 205–213. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, J.; Sun, G.; Raikhel, A. The early gene Broad is involved in the ecdysteroid hierarchy governing vitellogenesis of the mosquito Aedes aegypti. J. Mol. Endocrinol. 2004, 33, 743–761. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ou, J.; Liu, Y.; Wu, Q.; Wen, L.; Zheng, S.; Li, S.; Feng, Q.; Liu, L. Transcriptomic analysis of the testicular fusion in Spodoptera litura. BMC Genom. 2020, 21, 171. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liu, W.; Zhao, R.; He, J.; Dong, Z.; Chen, L.; Wan, W.; Chang, Z.; Wang, W.; Li, X. Genome-wide identification and gene-editing of pigment transporter genes in the swallowtail butterfly Papilio xuthus. BMC Genom. 2021, 22, 120. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Fujiwara, H. Molecular Mechanisms of Larval Color Pattern Switch in the Swallowtail Butterfly. In Diversity and Evolution of Butterfly Wing Patterns; Springer: Berlin/Heidelberg, Germany, 2017; pp. 271–286. [Google Scholar]
- Yamamoto, K.; Ozakiya, Y.; Uno, T. Localization of an aldo-keto reductase (AKR2E4) in the silkworm Bombyx mori (Lepidoptera: Bombycidae). J. Insect Sci. 2017, 17, 5. [Google Scholar] [CrossRef]
- Shelby, K.S.; Webb, B.A. Polydnavirus infection inhibits synthesis of an insect plasma protein, arylphorin. J. Gen. Virol. 1994, 75, 2285–2292. [Google Scholar] [CrossRef]
- Han, Q.; Li, G.; Li, J. Purification and characterization of chorion peroxidase from Aedes aegypti eggs. Arch. Biochem. Biophys. 2000, 378, 107–115. [Google Scholar] [CrossRef][Green Version]
- Zhou, X.; Riddiford, L.M. Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development 2002, 129, 2259–2269. [Google Scholar] [CrossRef]
- Swevers, L.; Iatrou, K. Ecdysteroids and ecdysteroid signaling pathways during insect oogenesis. In Ecdysone: Structures and Functions; Springer: Berlin/Heidelberg, Germany, 2009; pp. 127–164. [Google Scholar]
- González-Santoyo, I.; Córdoba-Aguilar, A. Phenoloxidase: A key component of the insect immune system. Entomol. Exp. Appl. 2012, 142, 1–16. [Google Scholar] [CrossRef]
- Kan, H.; Kim, C.-H.; Kwon, H.-M.; Park, J.-W.; Roh, K.-B.; Lee, H.; Park, B.-J.; Zhang, R.; Zhang, J.; Söderhäll, K. Molecular Control of Phenoloxidase-induced Melanin Synthesis in an Insect. J. Biol. Chem. 2008, 283, 25316–25323. [Google Scholar] [CrossRef]
- Cerenius, L.; Söderhäll, K. The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 2004, 198, 116–126. [Google Scholar] [CrossRef]
- Yang, G.; Zhu, W.; Kim, K.; Byun, S.Y.; Choi, G.; Wang, K.; Cha, J.S.; Cho, H.-S.; Oldfield, E.; No, J.H. In vitro and in vivo investigation of the inhibition of Trypanosoma brucei cell growth by lipophilic bisphosphonates. Antimicrob. Agents Chemother. 2015, 59, 7530–7539. [Google Scholar] [CrossRef]
- Futahashi, R.; Fujiwara, H. Juvenile hormone regulates butterfly larval pattern switches. Science 2008, 319, 1061. [Google Scholar] [CrossRef]
- Tusun, A.; Li, M.; Liang, X.; Yang, T.; Yang, B.; Wang, G. Juvenile hormone epoxide hydrolase: A promising target for hemipteran pest management. Sci. Rep. 2017, 7, 789. [Google Scholar] [CrossRef]
- Abdel-Latief, M.; Garbe, L.A.; Koch, M.; Ruther, J. An epoxide hydrolase involved in the biosynthesis of an insect sex attractant and its use to localize the production site. Proc. Natl. Acad. Sci. USA 2008, 105, 8914–8919. [Google Scholar] [CrossRef]
- Mello, T.R.; Aleixo, A.C.; Pinheiro, D.G.; Nunes, F.M.; Bitondi, M.M.; Hartfelder, K.; Barchuk, A.R.; Simões, Z.L. Developmental regulation of ecdysone receptor (EcR) and EcR-controlled gene expression during pharate-adult development of honeybees (Apis mellifera). Front. Genet. 2014, 5, 445. [Google Scholar] [CrossRef]
- Iga, M.; Kataoka, H. Recent studies on insect hormone metabolic pathways mediated by cytochrome P450 enzymes. Biol. Pharm. Bull. 2012, 35, 838–843. [Google Scholar] [CrossRef]
- Ono, H. Ecdysone differentially regulates metamorphic timing relative to 20-hydroxyecdysone by antagonizing juvenile hormone in Drosophila melanogaster. Dev. Biol. 2014, 391, 32–42. [Google Scholar] [CrossRef]
- Truman, J.W.; Talbot, W.S.; Fahrbach, S.E.; Hogness, D.S. Ecdysone receptor expression in the CNS correlates with stage-specific responses to ecdysteroids during Drosophila and Manduca development. Development 1994, 120, 219–234. [Google Scholar] [CrossRef]
- Miura, T. Proximate mechanisms and evolution of caste polyphenism in social insects: From sociality to genes. Ecol. Res. 2004, 19, 141–148. [Google Scholar] [CrossRef]
- Hartfelder, K.; Engels, W. 2 Social insect polymorphism: Hormonal regulation of plasticity in development and reproduction in the honeybee. Curr. Top. Dev. Biol. 1998, 40, 45–77. [Google Scholar]
- Mane-Padros, D.; Cruz, J.; Cheng, A.; Raikhel, A.S. A critical role of the nuclear receptor HR3 in regulation of gonadotrophic cycles of the mosquito Aedes aegypti. PLoS ONE 2012, 7, e45019b. [Google Scholar] [CrossRef]
- Guo, A.-Y.; Zhu, Q.-H.; Chen, X.; Luo, J.-C. GSDS: A gene structure display server. Yi Chuan = Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Gouti, M.; Metzis, V.; Briscoe, J. The route to spinal cord cell types: A tale of signals and switches. Trends Genet. 2015, 31, 282–289. [Google Scholar] [CrossRef]
- Xu, H.; Turlings, T.C. Plant volatiles as mate-finding cues for insects. Trends Plant Sci. 2018, 23, 100–111. [Google Scholar] [CrossRef]
- Fleischer, J.; Pregitzer, P.; Breer, H.; Krieger, J. Access to the odor world: Olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 2018, 75, 485–508. [Google Scholar] [CrossRef]
- Croset, V.; Rytz, R.; Cummins, S.F.; Budd, A.; Brawand, D.; Kaessmann, H.; Gibson, T.J.; Benton, R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010, 6, e1001064. [Google Scholar] [CrossRef]
- Koh, T.-W.; He, Z.; Gorur-Shandilya, S.; Menuz, K.; Larter, N.K.; Stewart, S.; Carlson, J.R. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 2014, 83, 850–865. [Google Scholar] [CrossRef]
- Zhang, Y.V.; Ni, J.; Montell, C. The molecular basis for attractive salt-taste coding in Drosophila. Science 2013, 340, 1334–1338. [Google Scholar] [CrossRef]
- Xu, Y.-L.; He, P.; Zhang, L.; Fang, S.-Q.; Dong, S.-L.; Zhang, Y.-J.; Li, F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genom. 2009, 10, 632. [Google Scholar] [CrossRef]
- Senthilan, P.R.; Piepenbrock, D.; Ovezmyradov, G.; Nadrowski, B.; Bechstedt, S.; Pauls, S.; Winkler, M.; Möbius, W.; Howard, J.; Göpfert, M.C. Drosophila auditory organ genes and genetic hearing defects. Cell 2012, 150, 1042–1054. [Google Scholar] [CrossRef]
- Weston, M.C.; Schuck, P.; Ghosal, A.; Rosenmund, C.; Mayer, M.L. Conformational restriction blocks glutamate receptor desensitization. Nat. Struct. Mol. Biol. 2006, 13, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-Y.; Xu, W.; Dong, S.-L.; Zhu, J.-Y.; Xu, Y.-X.; Anderson, A. Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2018, 99, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Fenner, J.; Benson, C.; Rodriguez-Caro, L.; Ren, A.; Papa, R.; Martin, A.; Hoffmann, F.; Range, R.; Counterman, B.A. Wnt genes in wing pattern development of Coliadinae butterflies. Front. Ecol. Evol. 2020, 8, 197. [Google Scholar] [CrossRef]
- Bänziger, C.; Soldini, D.; Schütt, C.; Zipperlen, P.; Hausmann, G.; Basler, K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 2006, 125, 509–522. [Google Scholar] [CrossRef]
- Bolognesi, R.; Beermann, A.; Farzana, L.; Wittkopp, N.; Lutz, R.; Balavoine, G.; Brown, S.J.; Schröder, R. Tribolium Wnts: Evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev. Genes Evol. 2008, 218, 193–202. [Google Scholar] [CrossRef]
- Holzem, M.; Braak, N.; Brattström, O.; McGregor, A.P.; Breuker, C.J. Wnt gene expression during early embryogenesis in the nymphalid butterfly Bicyclus anynana. Front. Ecol. Evol. 2019, 7, 468. [Google Scholar] [CrossRef]
- Han, X.; Wang, M.; Liu, C.; Trush, O.; Takayama, R.; Akiyama, T.; Naito, T.; Tomomizu, T.; Imamura, K.; Sato, M. DWnt4 and DWnt10 regulate morphogenesis and arrangement of columnar units via Fz2/PCP signaling in the Drosophila brain. Cell Rep. 2020, 33, 108305. [Google Scholar] [CrossRef]
- Cho, I.K.; Chang, C.L.; Li, Q.X. Diet-induced over-expression of flightless-I protein and its relation to flightlessness in Mediterranean fruit fly, Ceratitis capitata. PLoS ONE 2013, 8, e81099. [Google Scholar] [CrossRef]
- Tong, X.; Lindemann, A.; Monteiro, A. Differential involvement of Hedgehog signaling in butterfly wing and eyespot development. PLoS ONE 2012, 7, e51087. [Google Scholar] [CrossRef]
- Yao, S.; Lum, L.; Beachy, P. The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 2006, 125, 343–357. [Google Scholar] [CrossRef]
- Mito, T.; Inoue, Y.; Kimura, S.; Miyawaki, K.; Niwa, N.; Shinmyo, Y.; Ohuchi, H.; Noji, S. Involvement of hedgehog, wingless, and dpp in the initiation of proximodistal axis formation during the regeneration of insect legs, a verification of the modified boundary model. Mech. Dev. 2002, 114, 27–35. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Crystal, A.S.; Morais, V.A.; Pierson, T.C.; Pijak, D.S.; Carlin, D.; Lee, V.M.-Y.; Doms, R.W. Membrane topology of γ-secretase component PEN-2. J. Biol. Chem. 2003, 278, 20117–20123. [Google Scholar] [CrossRef]
- Siebel, C.; Lendahl, U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Grabherr, M.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from rNA-seq data without a reference genome. Nat. Biotech. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Nishimura, O.; Hara, Y.; Kuraku, S. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 2017, 33, 3635–3637. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Deng, Y.; Li, F.; Rieske, L.K.; Sun, L.-L.; Sun, S.-H. Transcriptome sequencing for identification of diapause-associated genes in fall webworm, Hyphantria cunea Drury. Gene 2018, 668, 229–236. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35 (Suppl. S2), W182–W185. [Google Scholar] [CrossRef]


| Measure | Value |
|---|---|
| Number of genes | 72,161 |
| Number of transcripts | 124,158 |
| Average contig length (bp) | 1043.2 |
| Median contig length (bp) | 627 |
| Total assembled bases | 75,278,310 |
| GC content of unigene (%) | 45.43 |
| Minimum sequence length (bp) | 297 |
| Maximum sequence length (bp) | 48,967 |
| Number of genes > 1 kb | 44,218 |
| Number of genes > 5 kb | 472 |
| N50 (bp) | 1560 |
| Features | Results |
| Complete BUSCOs (C) | 954 (94.17%) |
| Complete + Partial | 965 (95.26%) |
| Complete and single-copy BUSCOs (S) | 365 (36%) |
| Complete and duplicated BUSCOs (D) | 589 (58.1%) |
| Fragmented BUSCOs (F) | 11 (1.1%) |
| Missing BUSCOs (M) | 48 (4.8%) |
| Total BUSCO groups searched | 1013 |
| BUSCO version | BUSCO_v5.3.1 |
| Selected ortholog set | Arthropoda |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.-W.; Denison, M.I.J.; Veerappan, K.; Srinivasan, S.; Park, B.; Natarajan, S.; Chung, H.; Park, J. Developmental Transcriptome Analysis of Red-Spotted Apollo Butterfly, Parnassius bremeri. Int. J. Mol. Sci. 2022, 23, 11533. https://doi.org/10.3390/ijms231911533
Lee K-W, Denison MIJ, Veerappan K, Srinivasan S, Park B, Natarajan S, Chung H, Park J. Developmental Transcriptome Analysis of Red-Spotted Apollo Butterfly, Parnassius bremeri. International Journal of Molecular Sciences. 2022; 23(19):11533. https://doi.org/10.3390/ijms231911533
Chicago/Turabian StyleLee, Kang-Woon, Michael Immanuel Jesse Denison, Karpagam Veerappan, Sridhar Srinivasan, Bohyeon Park, Sathishkumar Natarajan, Hoyong Chung, and Junhyung Park. 2022. "Developmental Transcriptome Analysis of Red-Spotted Apollo Butterfly, Parnassius bremeri" International Journal of Molecular Sciences 23, no. 19: 11533. https://doi.org/10.3390/ijms231911533
APA StyleLee, K.-W., Denison, M. I. J., Veerappan, K., Srinivasan, S., Park, B., Natarajan, S., Chung, H., & Park, J. (2022). Developmental Transcriptome Analysis of Red-Spotted Apollo Butterfly, Parnassius bremeri. International Journal of Molecular Sciences, 23(19), 11533. https://doi.org/10.3390/ijms231911533






