Evodiamine and Rutaecarpine as Potential Anticancer Compounds: A Combined Computational Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Antitumor Activities of EVO and RUT
2.2. DFT Calculation Studies
2.2.1. The Natural Population Analysis (NPA) Charge
2.2.2. Molecular Electrostatic Potential (MEP)
2.2.3. Frontier Molecular Orbitals (FMOs)
2.2.4. Reactivity Descriptors
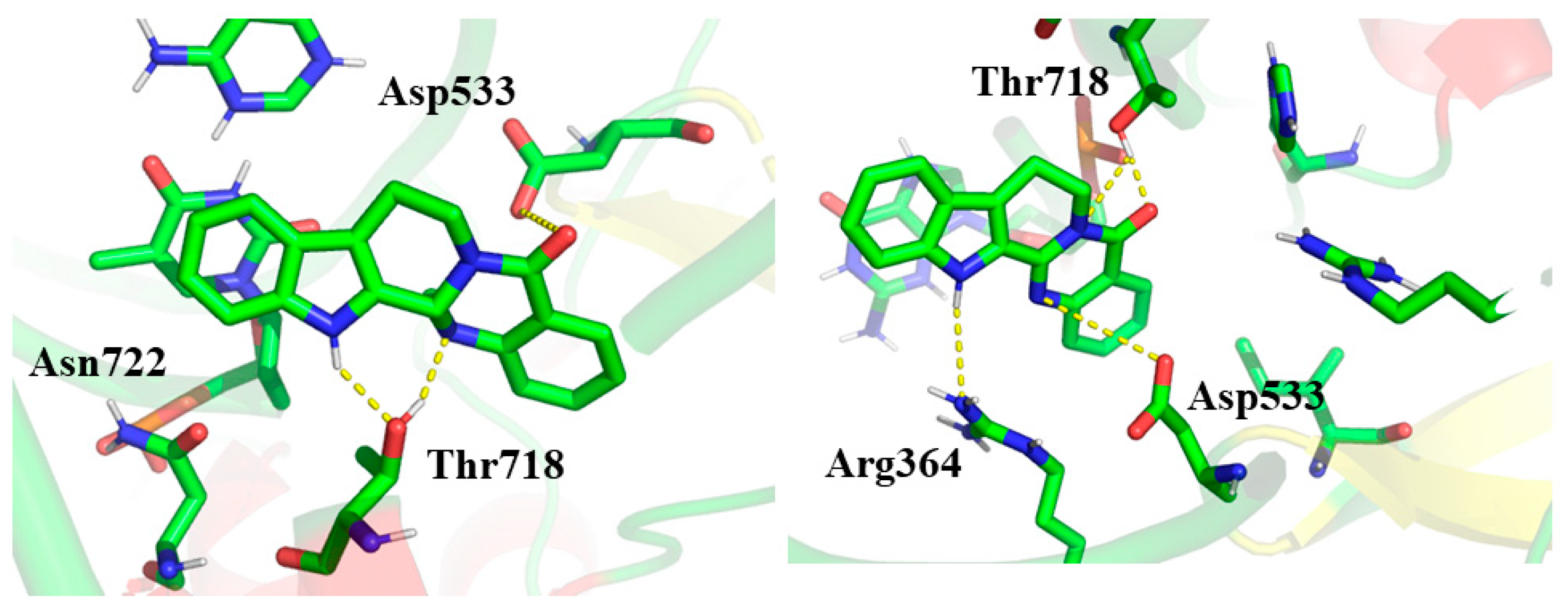
2.3. Molecular Docking Studies
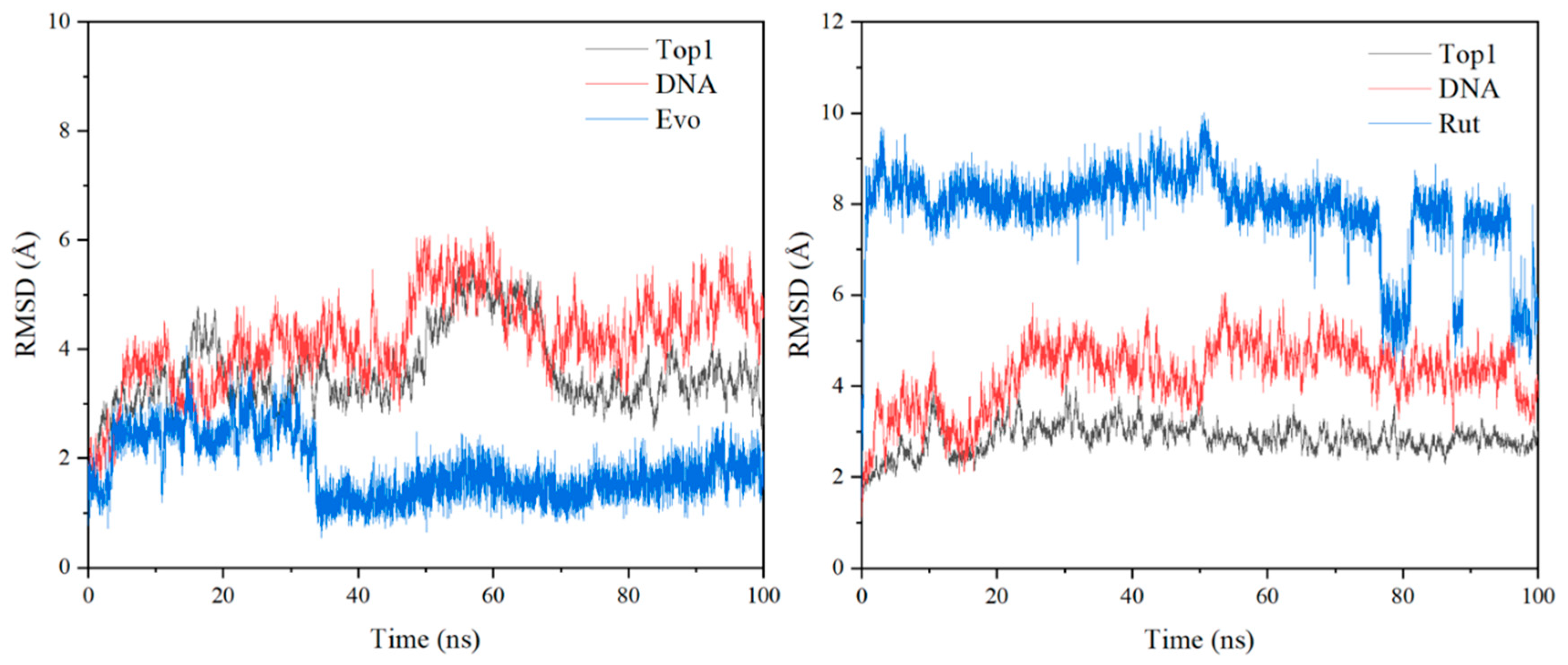
2.4. MD Simulation Studies
2.5. Binding Free Energies
2.6. Free Energy Decomposition
3. Materials and Methods
3.1. Cell Culture and MTT Assay
3.2. Density Functional Theory (DFT) Calculations
3.3. Molecular Docking
3.4. Molecular Dynamics (MD) Simulations
3.5. Binding Free Energy Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, Z.; Morris-Natschke, S.L.; Lee, K.H. Strategies for the optimization of natural leads to anticancer drugs or drug candidates. Med. Res. Rev. 2016, 36, 32–91. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Tan, M.; Gong, J.; Tan, W.; Bian, B.; Chen, M.; Wang, Y. Fighting fire with fire: Poisonous Chinese herbal medicine for cancer therapy. J. Ethnopharmacol. 2012, 140, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Liu, J.; Xu, S.; Zhu, Z.; Xu, J. The structural modification of natural products for novel drug discovery. Expert Opin. Drug Discovery. 2017, 12, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, W.; Yao, H.; Xu, J. Insights into drug discovery from natural products through structural modification. Fitoterapia. 2015, 103, 231–241. [Google Scholar] [CrossRef]
- Guo, Z. The modification of natural products for medical use. Acta Pharm. Sin. B. 2017, 7, 119–136. [Google Scholar] [CrossRef]
- Wall, M.E.; Wani, M.C.; Cook, C.E.; Palmer, K.H.; McPhail, A.T.; Sim, G.A. The Isolation and Structure of Camptothecin, a Novel Alkaloidal Leukemia and Tumor Inhibitor from Camptotheca acuminata. J. Am. Chem. Soc. 1996, 88, 3888–3890. [Google Scholar] [CrossRef]
- Hsiang, Y.H.; Hertzberg, R.; Hecht, S.; Liu, L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985, 260, 14873–14878. [Google Scholar] [CrossRef]
- Garcia-Carbonero, R.; Supko, J.G. Current perspectives on the clinical experience, pharmacology, and continued development of the camptothecins. Clin. Cancer Res. 2002, 8, 641–661. [Google Scholar]
- Boudjedir, A.; Kraim, K.; Saihi, Y.; Attoui-Yahia, O.; Ferkous, F.; Nacereddine, A.K. A computational molecular docking study of camptothecin similars as inhibitors for topoisomerase Ⅰ. Struct. Chem. 2021, 32, 689–697. [Google Scholar] [CrossRef]
- Capranico, G.; Marinello, J.; Chillemi, G. Type I DNA Topoisomerases. J. Med. Chem. 2017, 60, 2169–2192. [Google Scholar] [CrossRef] [PubMed]
- Redinbo, M.R.; Stewart, L.; Kuhn, P.; Champoux, J.J.; Hol, W.G.J. Crystal Structures of Human Topoisomerase I in Covalent and Noncovalent Complexes with DNA. Science 1998, 279, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.; Redinbo, M.R.; Qiu, X.Y.; Hol, W.G.J.; Champoux, J.J. Model for the Mechanism of Human Topoisomerase I. Science 1998, 279, 1534–1541. [Google Scholar] [CrossRef]
- Martín-Encinas, E.; Selas, A.; Palacios, F.; Alonso, C. The design and discovery of topoisomerase I inhibitors as anticancer therapies. Expert Opin. Drug Dis. 2022, 17, 581–601. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.H.; Liao, Z.Y.; Pommier, Y. Non-camptothecin DNA topoisomerase I inhibitors in cancer therapy. Curr. Top. Med. Chem. 2003, 3, 305–320. [Google Scholar]
- Liao, J.F.; Chiou, W.F.; Shen, Y.C.; Wang, G.J.; Chen, C.F. Anti-inflammatory and anti-infectious effects of Evodia rutaecarpa (Wuzhuyu) and its major bioactive components. Chin. Med. 2011, 6, 6–13. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Z.; Zhou, X.; Cai, Z.; Gong, X.; Zhou, C. Quality evaluation of Evodia rutaecarpa (Juss.) Benth by high performance liquid chromatography with photodiode-array detection. J. Pharmaceut. Biomed. 2008, 48, 1230–1236. [Google Scholar] [CrossRef]
- Hu, C.; Li, Y. Research progress in pharmacological actions of evodiamine and rutaecarpine. Chin. Pharmacol. Bull. 2003, 19, 1084–1087. [Google Scholar]
- Lee, S.H.; Son, J.K.; Jeong, B.S.; Jeong, T.C.; Chang, H.W.; Lee, E.S.; Jahng, Y. Progress in the studies on rutaecarpine. Molecules 2008, 13, 272–300. [Google Scholar] [CrossRef]
- Jong-Keun, S.; Hyeun, W.C.; Yurngdong, J. Progress in Studies on Rutaecarpine II—Synthesis and Structure-Biological Activity Relationships. Molecules 2015, 20, 10800–10821. [Google Scholar]
- Fujii, I.; Kobayashi, Y.; Hirayama, N. Molecular structure of two indole alkaloids, evodiamine and rutecarpine, from evodia fruit. Z. Kristallogr. 2000, 215, 762–765. [Google Scholar]
- Jiang, J.; Hu, C. Evodiamine: A novel anti-cancer alkaloid from Evodia rutaecarpa. Molecules 2009, 14, 1852–1859. [Google Scholar] [CrossRef]
- Lu, J.J.; Bao, J.L.; Chen, X.P.; Huang, M.; Wang, Y.T. Alkaloids isolated from natural herbs as the anticancer agents. Evid. Based Complement. Altern. Med. 2012, 2012, 485042. [Google Scholar] [CrossRef]
- Guo, H.; Yang, Y.Y.; Yu, D.L.; Wang, C.L.; Zhang, S. Synthesis, Structure Characterization of Rutaecarpine and Its Antitumor Activity in vivo. Chem. Bioeng. 2010, 27, 37–40. [Google Scholar]
- Yang, Y.Y.; Guo, H.; Wang, C.L.; Zhang, S. Synthesis as well as structural characterization and in vivo antitumor activity evaluation of evodiamine. Chem. Res. 2011, 22, 22–25. [Google Scholar]
- Guo, H.; Zhang, S.; Zhang, K.Y.; Zhu, H.Y.; Liu, J.L.; Liu, D.M.; Wang, C.L.; Su, Z. Antitumor Activity and Theoretical Calculation of Evodiamine and Rutaecarpine. Chin. J. Struct. Chem. 2016, 35, 1174–1180. [Google Scholar]
- Chen, M.C.; Yu, C.H.; Wang, S.W.; Pu, H.F.; Kan, S.F.; Lin, L.C.; Chi, C.W.; Ho, L.L.; Lee, C.H.; Wang, P.S. Anti-proliferative effects of evodiamine on human thyroid cancer cell line ARO. J. Cell. Biochem. 2010, 110, 1495–1503. [Google Scholar] [CrossRef]
- Yang, L.; Liu, X.; Wu, D.; Zhang, M.; Ran, G.; Bi, Y.; Huang, H. Growth inhibition and induction of apoptosis in SGC-7901 human gastric cancer cells by evodiamine. Mol. Med. Rep. 2014, 9, 1147–1152. [Google Scholar] [CrossRef]
- Shyu, K.G.; Lin, S.; Lee, C.C.; Chen, E.; Lin, L.C.; Wang, B.W.; Tsai, S.C. Evodiamine inhibits in vitro angiogenesis: Implication for antitumorgenicity. Life Sci. 2006, 78, 2234–2243. [Google Scholar] [CrossRef]
- Chan, L.F.A.; Chang, W.S.; Chen, L.M.; Lee, C.M.; Chen, C.E.; Lin, C.M.; Hwang, J.L. Evodiamine Stabilizes Topoisomerase I-DNA Cleavable Complex to Inhibit Topoisomerase I Activity. Molecules 2009, 14, 1342–1352. [Google Scholar] [CrossRef]
- Dong, G.; Sheng, C.; Wang, S.; Miao, Z.; Yao, J.; Zhang, W. Selection of evodiamine as a novel topoisomerase I inhibitor by structure-based virtual screening and hit optimization of evodiamine derivatives as antitumor agents. J. Med. Chem. 2010, 53, 7521–7531. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Wang, S.; Miao, Z.; Yao, J.; Zhang, Y.; Guo, Z.; Zhang, W.; Sheng, C. New tricks for an old natural product: Discovery of highly potent evodiamine derivatives as novel antitumor agents by systemic structure-activity relationship analysis and biological evaluations. J. Med. Chem. 2012, 55, 7593–7613. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.L.; Li, G.; Moon, D.C.; Lee, C.S.; Woo, M.H.; Lee, E.S.; Jahng, Y.; Chang, H.W.; Lee, S.H.; Son, J.K. Cytotoxicity and DNA topoisomerase inhibitory activity of constituents isolated from the fruits of Evodia officinalis. Arch. Pharm. Res. 2006, 29, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Seung, H.L.; Eung-Seok, L.; Chong-Soon, L.; Yurngdong, J. New Topoisomerases Inhibitors: Synthesis of Rutaecarpine Derivatives and Their Inhibitory Activity against Topoisomerases. Arch. Pharm. Res. 2012, 35, 785–789. [Google Scholar]
- Patrykei, S.; Korobko, Y.; Ogorodniichuk, O.; Garazd, M.; Polishchuk, P.; Gurská, S.; Hajdúch, M.; Lesyk, R. Synthesis and evaluation of the anticancer activity of some semisynthetic derivatives of rutaecarpine and evodiamine. Synthetic Commun. 2021, 51, 3237–3245. [Google Scholar] [CrossRef]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, E.M.; Govender, T.; Naicker, T.; Kruger, H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021, 224, 113705. [Google Scholar] [CrossRef]
- Jakhar, R.; Dangi, M.; Khichi, A.; Chhillar, A.K. Relevance of Molecular Docking Studies in Drug Designing. Curr. Bioinf. 2020, 15, 270–278. [Google Scholar] [CrossRef]
- Zou, Y.; Ewalt, J.; Ng, H.L. Recent Insights from Molecular Dynamics Simulations for G Protein-Coupled Receptor Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4237. [Google Scholar] [CrossRef]
- Nehra, B.; Mathew, B.; Chawla, P.A. A Medicinal Chemist’s Perspective towards Structure Activity Relationship of Heterocycle based Anticancer Agents. Curr. Top. Med. Chem. 2022, 22, 493–528. [Google Scholar] [CrossRef]
- Reed, A.E.; Weinstock, R.B.; Weinhold, F. Natural population analysis. J. Chem. Phys. 1985, 83, 735–746. [Google Scholar] [CrossRef]
- Al-Otaibi, J.S.; Almuqrin, A.H.; Mary, Y.S.; Mary, Y.S.; Alsenoy, C.V. DFT and molecular docking studies of self-assembly of sulfone analogues and graphene. J. Mol. Mod. 2020, 26, 273. [Google Scholar] [CrossRef]
- Shafieyoon, P.; Mehdipour, E.; Mary, Y.S. Synthesis, characterization and biological investigation of glycine based sulfonamide derivative and its complex: Vibration assignment, HOMO-LUMO analysis, MEP and molecular docking. J. Mol. Struct. 2019, 1181, 244–252. [Google Scholar] [CrossRef]
- Mary, Y.S.; Miniyar, P.B.; Mary, Y.S.; Resmi, K.S.; Panicker, C.Y.; Armakovic, S.; Armakovic, S.J.; Thomas, R.; Sureshkumar, B. Synthesis and spectroscopic study of three new oxadiazole derivatives with detailed computational evaluation of their reactivity and pharmaceutical potential. J. Mol. Struct. 2018, 1173, 469–480. [Google Scholar] [CrossRef]
- Mohamed, H.; Hoda, A.A.; Ghadah, A.; Omaima, A.A. Investigation of Some Antiviral N-Heterocycles as COVID 19 Drug: Molecular Docking and DFT Calculations. Int. J. Mol. Sci. 2020, 21, 3922. [Google Scholar]
- Geerlings, P.; Proft, F.D.; Langenaeker, W. Conceptual density functional theory. Chem. Rev. 2003, 103, 1793–1873. [Google Scholar] [CrossRef]
- Kumar, C.P.; Santanab, G.; Soma, D. Update 2 of: Electrophilicity index. Chem. Rev. 2011, 111, PR43–PR75. [Google Scholar]
- Parr, R.G.; Szentpály, V.L.; Liu, S. Electrophilicity index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Jorgensen, W.L. The many roles of computation in drug discovery. Science 2004, 303, 1813–1818. [Google Scholar] [CrossRef]
- Meng, X.Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comp. Aid. Drug. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Staker, B.L.; Feese, M.D.; Cushman, M.; Pommier, Y.; Zembower, D.; Stewart, L.; Burgin, A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005, 48, 2336–2345. [Google Scholar] [CrossRef]
- Lauria, A.; Ippolito, M.; Almerico, A.M. Molecular docking approach on the Topoisomerase I inhibitors series included in the NCI anti-cancer agents mechanism database. J. Mol. Model. 2007, 13, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Redinbo, M.R.; Stewart, L.; Champoux, J.J.; Hol, W.G.J. Structural flexibility in human topoisomerase Ⅰ revealed in multiple non-isomorphous crystal structures. J. Mol. Biol. 1999, 292, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, J.; Persico, M. Molecular interactions in solution: An overview of methods based on continuous distributions of the solvent. Chem. Rev. 1994, 94, 2027–2094. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision D.02; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Morris, G.; Huey, R.; Lindstrom, W.; Sanner, M.; Belew, R.; Goodsell, D.; Olson, A. AutoDock4 and AutoDockTools4: Automated docking with selective receptor 828 flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. FF14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from FF99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.; Cheatham, T.E., III; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. Amber 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Roe, D.R.; Cheatham, T.E. Parallelization of CPPTRAJ Enables Large Scale Analysis of Molecular Dynamics Trajectory Data. J. Comput. Chem. 2018, 39, 2110–2117. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ AND CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Srinivasan, J.; Cheatham, T.E.; Cieplak, P.; Kollman, P.A.; Case, D.A. Continuum Solvent Studies of the Stability of DNA, RNA, and Phosphoramidate—DNA Helices. J. Am. Chem. Soc. 1998, 120, 9401–9409. [Google Scholar] [CrossRef]
- Lee, M.S.; Salsbury, F.R.; Olson, M.A. An Efficient Hybrid Explicit/Implicit Solvent Method for Biomolecular Simulations. J. Comput. Chem. 2004, 25, 1967–1978. [Google Scholar] [CrossRef]
- Gaillard, T.; Simonson, T. Pairwise Decomposition of an MMGBSA Energy Function for Computational Protein Design. J. Comput. Chem. 2014, 35, 1371–1387. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
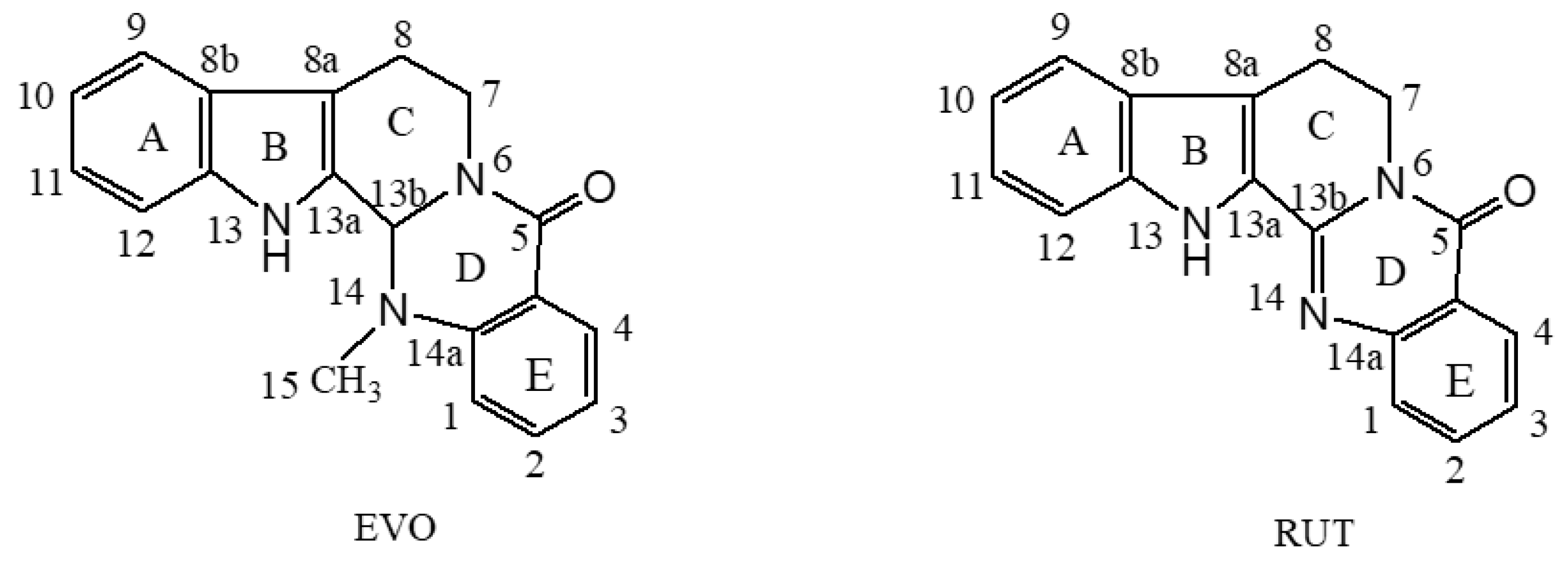
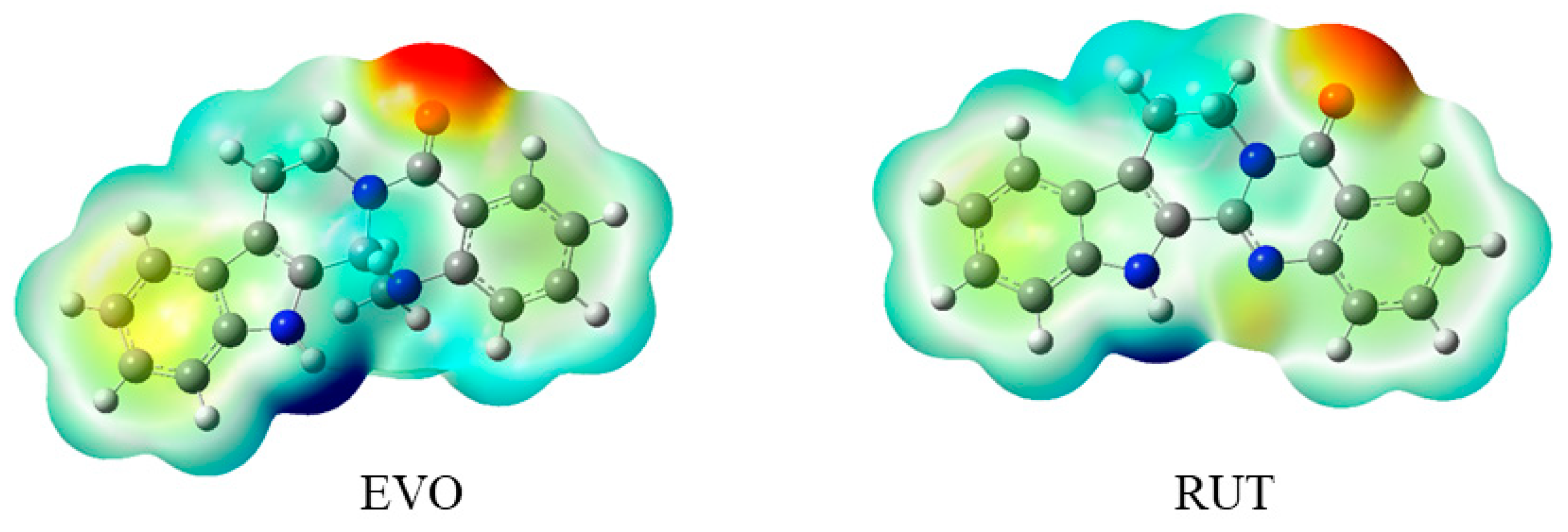
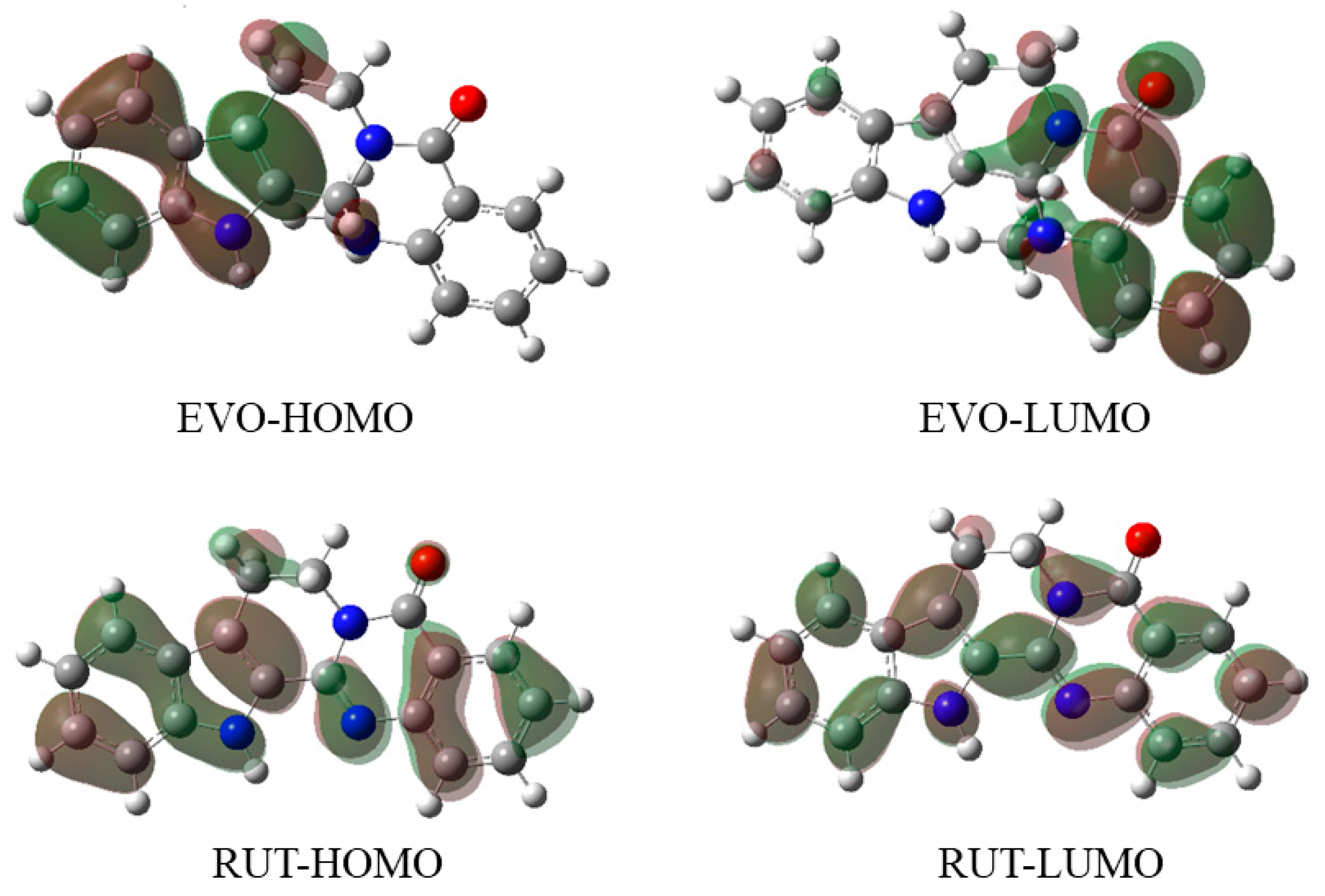
| Atom | EVO | RUT | Atom | EVO | RUT |
|---|---|---|---|---|---|
| C1 | −0.302 | −0.190 | C8 | −0.737 | −0.407 |
| C2 | −0.182 | −0.184 | C9 | −0.745 | −0.183 |
| C3 | −0.268 | −0.209 | C10 | −0.592 | −0.223 |
| C4 | −0.616 | −0.148 | C11 | −0.226 | −0.194 |
| C5 | 0.334 | 0.676 | C12 | −0.284 | −0.233 |
| N6 | −0.675 | −0.474 | N13 | −0.602 | −0.524 |
| C7 | −0.299 | −0.153 | N14 | −0.645 | −0.525 |
| O5 | −0.713 | −0.622 | C15 | −0.768 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Guo, H.; Zhou, J.; Wang, Y.; Yan, H.; Jin, R.; Tang, Y. Evodiamine and Rutaecarpine as Potential Anticancer Compounds: A Combined Computational Study. Int. J. Mol. Sci. 2022, 23, 11513. https://doi.org/10.3390/ijms231911513
Liu J, Guo H, Zhou J, Wang Y, Yan H, Jin R, Tang Y. Evodiamine and Rutaecarpine as Potential Anticancer Compounds: A Combined Computational Study. International Journal of Molecular Sciences. 2022; 23(19):11513. https://doi.org/10.3390/ijms231911513
Chicago/Turabian StyleLiu, Jingli, Hui Guo, Jing Zhou, Yuwei Wang, Hao Yan, Ruyi Jin, and Yuping Tang. 2022. "Evodiamine and Rutaecarpine as Potential Anticancer Compounds: A Combined Computational Study" International Journal of Molecular Sciences 23, no. 19: 11513. https://doi.org/10.3390/ijms231911513
APA StyleLiu, J., Guo, H., Zhou, J., Wang, Y., Yan, H., Jin, R., & Tang, Y. (2022). Evodiamine and Rutaecarpine as Potential Anticancer Compounds: A Combined Computational Study. International Journal of Molecular Sciences, 23(19), 11513. https://doi.org/10.3390/ijms231911513






