miRNA: A Promising Therapeutic Target in Cancer
Abstract
1. Introduction
2. miRNA Biogenesis
3. Functions of miRNAs
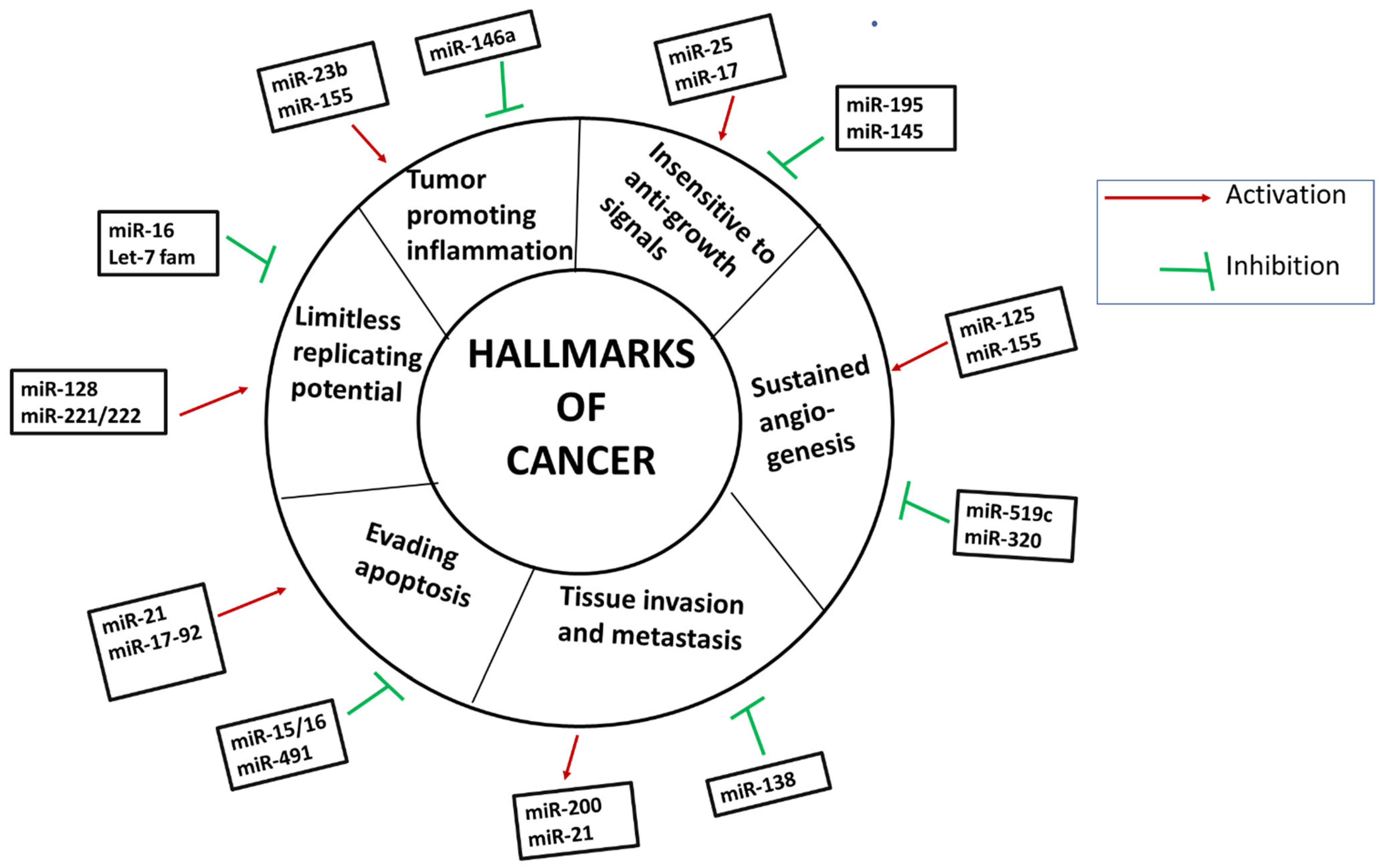
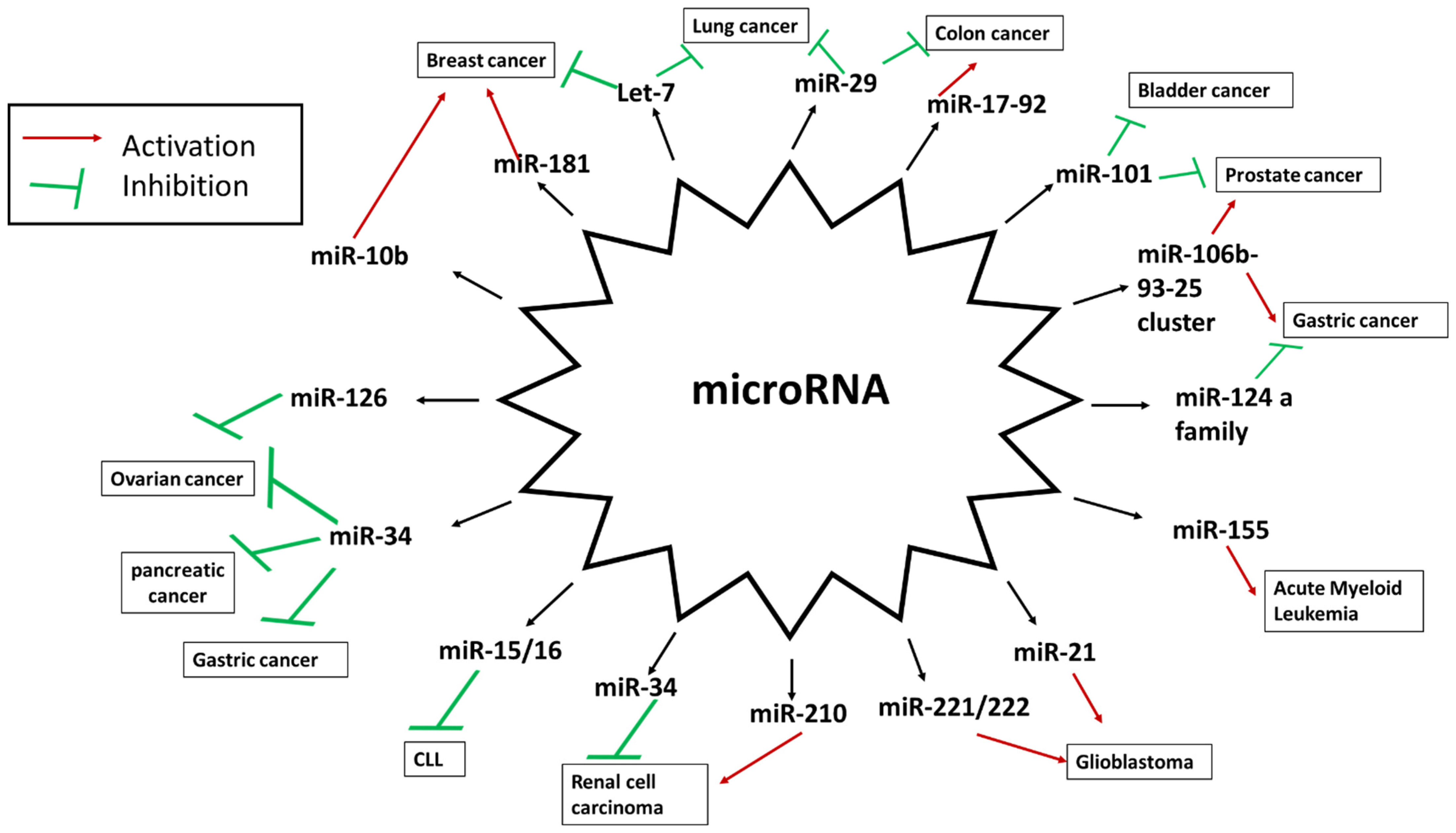
4. miRNA Deregulation in Cancer
4.1. miRNAs as Oncogenes
4.2. miRNAs as Tumor Suppressors
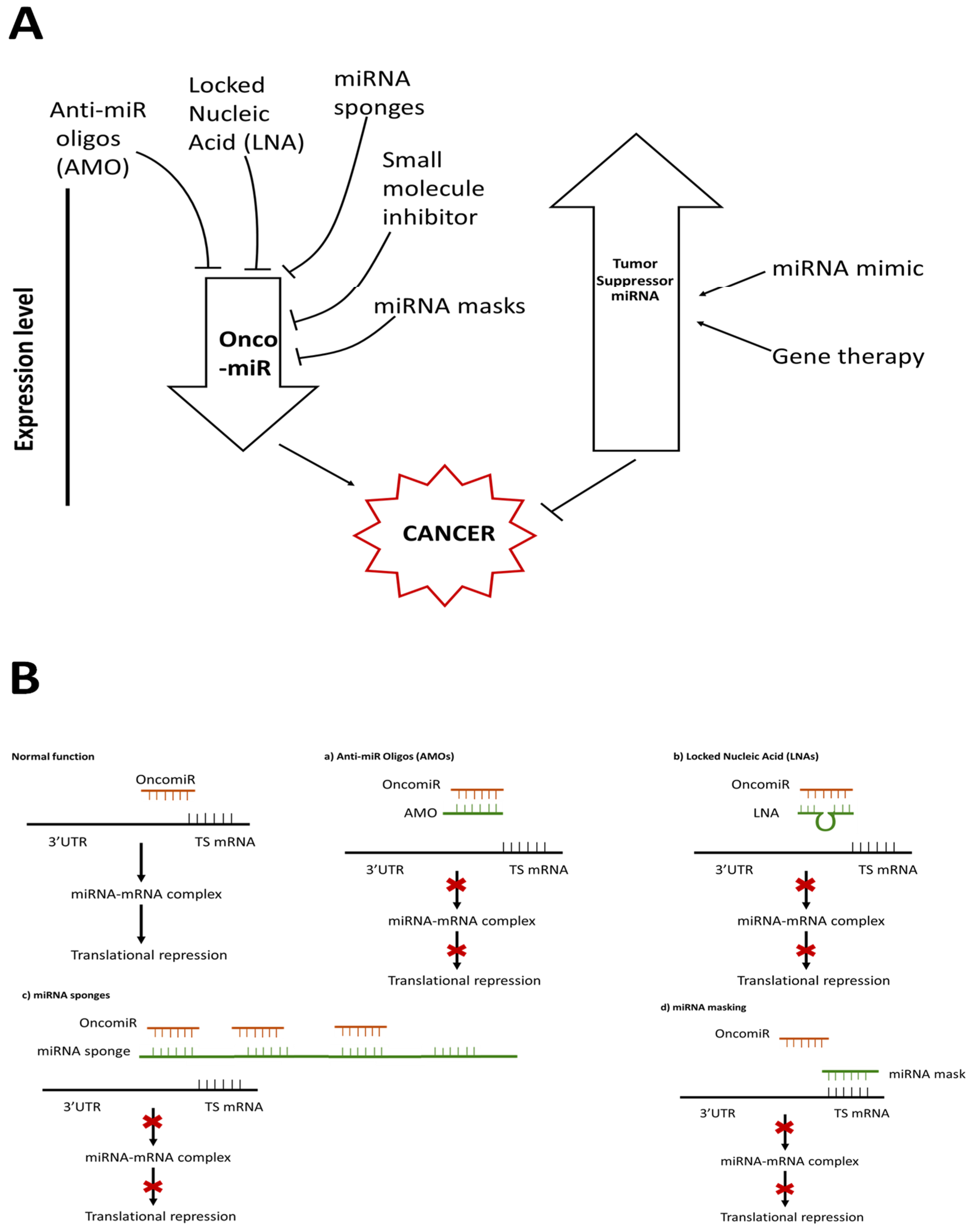
5. miRNA Therapeutics
- The inhibition of oncogenic miRNAs, and hence the restoration of the expression of tumor-suppressing genes that they target; or
- Restoring the expression of tumor-suppressing miRNAs and hence inhibiting the oncogenes that they target
5.1. Therapeutic Approach with Oncogenic miRNAs
5.1.1. Anti-miRNA Oligonucleotides (AMOs)
5.1.2. Modified AMOs
5.1.3. miRNA Sponges
5.1.4. Small Molecule Inhibitor
5.1.5. miRNA Masking
5.2. Therapeutic Approach with Suppressor miRNAs
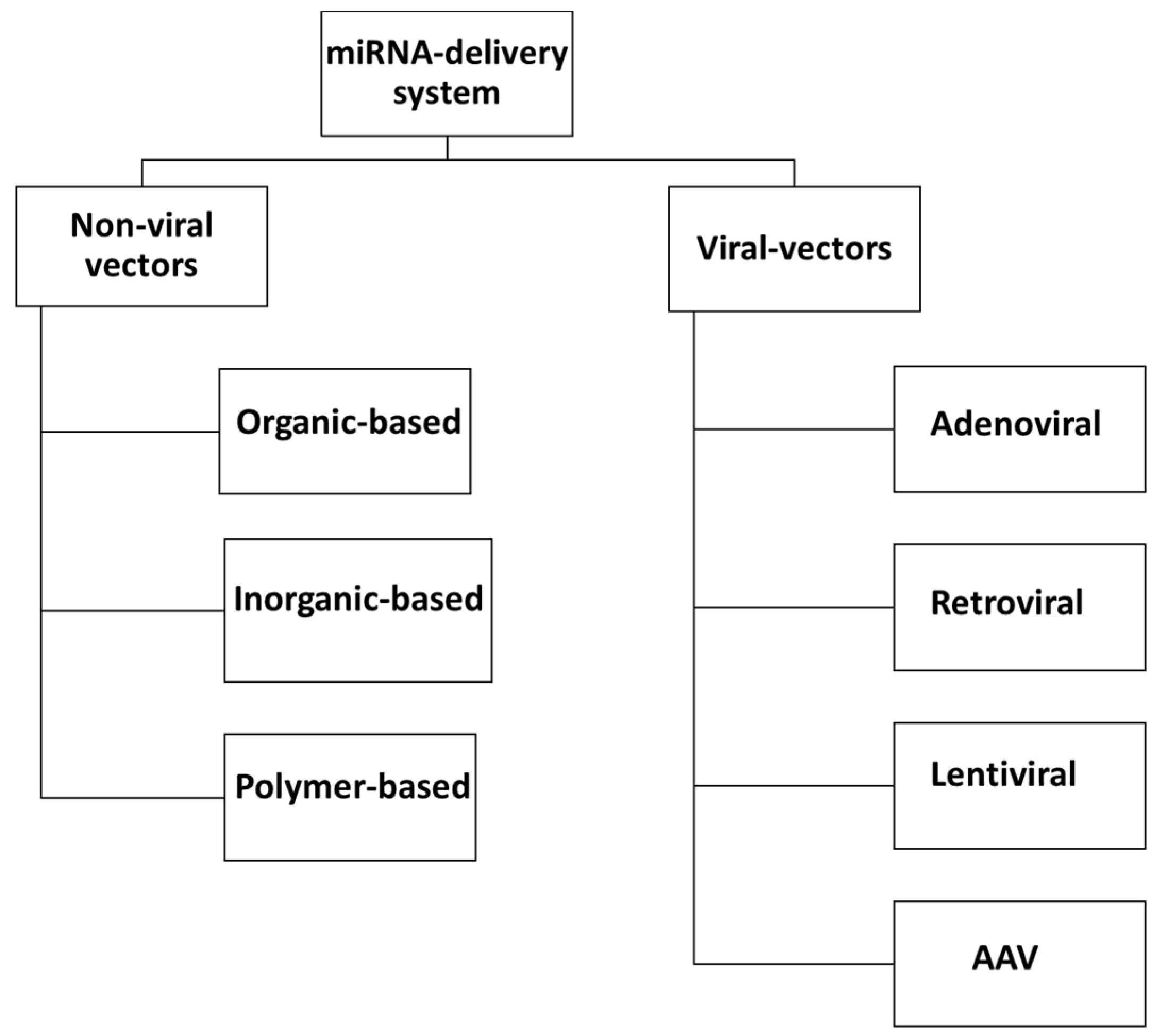
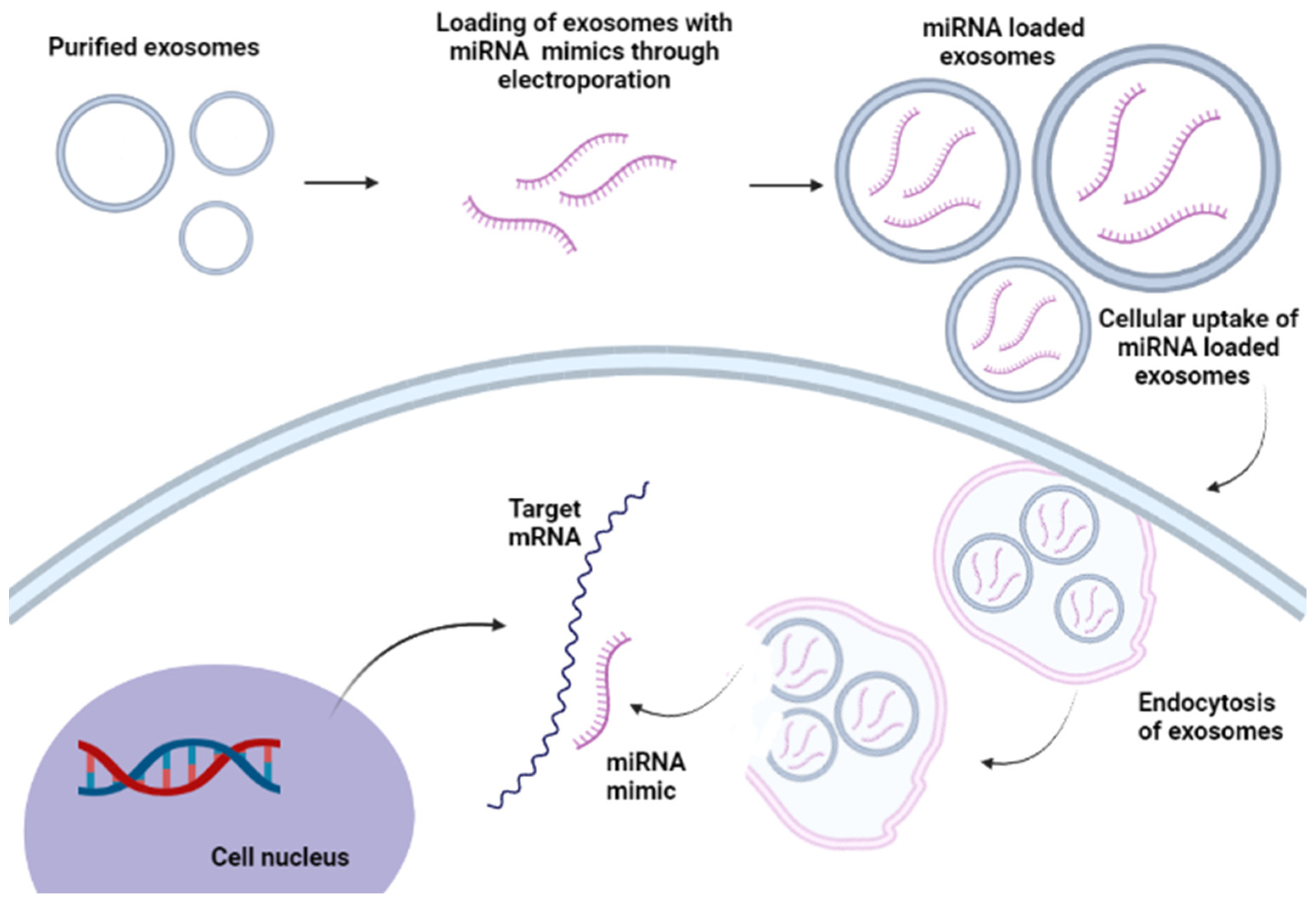
6. miRNA Delivery Systems
6.1. Non-Viral miRNA Delivery System
6.2. Viral-Based miRNA Delivery System
7. Drug Intervention on miRNA Therapy in Cancer
8. Patent Updates for miRNA in Cancer Therapy
9. miRNA-Based Clinical Trials for Cancer Therapy
10. Future Prospects and Challenges
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lindsay, M.A.; Griffiths-Jones, S.; Dalmay, T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013, 54, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Chiu, H.; Domenger, D.; Chuang, C.-F.; Chang, C. The lin-4 microRNA targets the LIN-14 transcription factor to inhibit netrin-mediated axon attraction. Sci. Signal. 2012, 5, ra43. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Arif, M.A.; Seumel, G.I.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Transcriptional control of gene expression by microRNAs. Cell 2010, 140, 111–122. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, C. MicroRNA-directed cleavage of targets: Mechanism and experimental approaches. BMB Rep. 2014, 47, 417. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Mette, M.; Aufsatz, W.; Van der Winden, J.; Matzke, M.; Matzke, A. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000, 19, 5194–5201. [Google Scholar] [CrossRef]
- Sandoval, J.; Méndez González, J.; Nadal, E.; Chen, G.; Carmona, F.J.; Sayols, S.; Moran, S.; Heyn, H.; Vizoso, M.; Gomez, A. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J. Clin. Oncol. 2013, 31, 4140–4147. [Google Scholar] [CrossRef]
- Huang, R.-L.; Gu, F.; Kirma, N.B.; Ruan, J.; Chen, C.-L.; Wang, H.-C.; Liao, Y.-P.; Chang, C.-C.; Yu, M.-H.; Pilrose, J.M. Comprehensive methylome analysis of ovarian tumors reveals hedgehog signaling pathway regulators as prognostic DNA methylation biomarkers. Epigenetics 2013, 8, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.-C.; Munschauer, M. PAR-CliP-a method to identify transcriptome-wide the binding sites of RNA binding proteins. JoVE (J. Vis. Exp.) 2010, 2, e2034. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef]
- Zhou, P.; Xu, W.; Peng, X.; Luo, Z.; Xing, Q.; Chen, X.; Hou, C.; Liang, W.; Zhou, J.; Wu, X. Large-scale screens of miRNA-mRNA interactions unveiled that the 3′ UTR of a gene is targeted by multiple miRNAs. PLoS ONE 2013, 8, e68204. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5′ UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Sassen, S.; Miska, E.A.; Caldas, C. MicroRNA—Implications for cancer. Virchows Arch. 2008, 452, 1–10. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Gabriely, G.; Yi, M.; Narayan, R.S.; Niers, J.M.; Wurdinger, T.; Imitola, J.; Ligon, K.L.; Kesari, S.; Esau, C.; Stephens, R.M. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011, 71, 3563–3572. [Google Scholar] [CrossRef]
- Sasayama, T.; Nishihara, M.; Kondoh, T.; Hosoda, K.; Kohmura, E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer 2009, 125, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Zhou, Q.; Wang, D.; Guan, L.; Yuan, L.; Li, S. The downregulation of microRNA-10b and its role in cervical cancer. Oncol. Res. 2016, 24, 99. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lei, H.; Luo, M.; Wang, Y.; Dong, L.; Ma, Y.; Liu, C.; Song, W.; Wang, F.; Zhang, J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer 2015, 18, 43–54. [Google Scholar] [CrossRef]
- Huang, L.; Lin, J.-X.; Yu, Y.-H.; Zhang, M.-Y.; Wang, H.-Y.; Zheng, M. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS ONE 2012, 7, e33762. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Blaszczyk, J.; Tropea, J.E.; Bubunenko, M.; Routzahn, K.M.; Waugh, D.S.; Court, D.L.; Ji, X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 2001, 9, 1225–1236. [Google Scholar] [CrossRef]
- Romero-Cordoba, S.L.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.-K.; Yeom, K.-H.; Yang, W.-Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, X.; Graves, P.; Zeng, Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA 2012, 18, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Maniataki, E.; Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005, 19, 2979–2990. [Google Scholar] [CrossRef]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Cougot, N.; Babajko, S.; Séraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004, 165, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Sheth, U.; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Valencia-Sanchez, M.A.; Hannon, G.J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar] [CrossRef]
- Ji, Q.; Hao, X.; Zhang, M.; Tang, W.; Yang, M.; Li, L.; Xiang, D.; DeSano, J.T.; Bommer, G.T.; Fan, D. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE 2009, 4, e6816. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Lee, S.K.-W.; Teng, Y.; Wong, H.-K.; Ng, T.-K.; Huang, L.; Lei, P.; Choy, K.-W.; Liu, Y.; Zhang, M.; Lam, D.S.-C. MicroRNA-145 regulates human corneal epithelial differentiation. PLoS ONE 2011, 6, e21249. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Klein, M.E.; Impey, S.; Goodman, R.H. Role reversal: The regulation of neuronal gene expression by microRNAs. Curr. Opin. Neurobiol. 2005, 15, 507–513. [Google Scholar] [CrossRef]
- Pinter, R.; Hindges, R. Perturbations of microRNA function in mouse dicer mutants produce retinal defects and lead to aberrant axon pathfinding at the optic chiasm. PLoS ONE 2010, 5, e10021. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.N.; Bellon, A.; Baudet, M.-L. microRNAs in axon guidance. Front. Cell. Neurosci. 2014, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Nakao, H.; Kiyonari, H.; Abe, T.; Aizawa, S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011, 31, 3407–3422. [Google Scholar] [CrossRef] [PubMed]
- Giraldez, A.J.; Cinalli, R.M.; Glasner, M.E.; Enright, A.J.; Thomson, J.M.; Baskerville, S.; Hammond, S.M.; Bartel, D.P.; Schier, A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science 2005, 308, 833–838. [Google Scholar] [CrossRef]
- Baudet, M.-L.; Zivraj, K.H.; Abreu-Goodger, C.; Muldal, A.; Armisen, J.; Blenkiron, C.; Goldstein, L.D.; Miska, E.A.; Holt, C.E. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat. Neurosci. 2012, 15, 29–38. [Google Scholar] [CrossRef]
- Bellon, A.; Iyer, A.; Bridi, S.; Lee, F.C.; Ovando-Vazquez, C.; Corradi, E.; Longhi, S.; Roccuzzo, M.; Strohbuecker, S.; Naik, S. miR-182 regulates Slit2-mediated axon guidance by modulating the local translation of a specific mRNA. Cell Rep. 2017, 18, 1171–1186. [Google Scholar] [CrossRef]
- Nagaoka, K.; Zhang, H.; Watanabe, G.; Taya, K. Epithelial cell differentiation regulated by MicroRNA-200a in mammary glands. PLoS ONE 2013, 8, e65127. [Google Scholar] [CrossRef]
- Liang, B.; Chen, Y.; Yuan, W.; Qin, F.; Zhang, Q.; Deng, N.; Liu, X.; Ma, X.; Zhang, X.; Zhang, B. Down-regulation of miRNA-451a and miRNA-486-5p involved in benzene-induced inhibition on erythroid cell differentiation in vitro and in vivo. Arch. Toxicol. 2018, 92, 259–272. [Google Scholar] [CrossRef]
- Cai, B.; Li, J.; Wang, J.; Luo, X.; Ai, J.; Liu, Y.; Wang, N.; Liang, H.; Zhang, M.; Chen, N. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem Cells 2012, 30, 1746–1755. [Google Scholar] [CrossRef]
- Zehir, A.; Hua, L.L.; Maska, E.L.; Morikawa, Y.; Cserjesi, P. Dicer is required for survival of differentiating neural crest cells. Dev. Biol. 2010, 340, 459–467. [Google Scholar] [CrossRef]
- Ni, Y.; Zhang, K.; Liu, X.; Yang, T.; Wang, B.; Fu, L.; Lan, A.; Zhou, Y. miR-21 promotes the differentiation of hair follicle-derived neural crest stem cells into Schwann cells. Neural Regen. Res. 2014, 9, 828. [Google Scholar] [PubMed]
- Schneider, M.; Andersen, D.C.; Silahtaroglu, A.; Lyngbæk, S.; Kauppinen, S.; Hansen, J.L.; Sheikh, S.P. Cell-specific detection of microRNA expression during cardiomyogenesis by combined in situ hybridization and immunohistochemistry. J. Mol. Histol. 2011, 42, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, S.; Shcherbata, H.; Fischer, K.; Nakahara, K.; Carthew, R.; Ruohola-Baker, H. Stem cell division is regulated by the microRNA pathway. Nature 2005, 435, 974–978. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Guo, J.; Li, L.; Liu, H.; Lu, C.; Jiang, Y.; Cui, S. MicroRNA-7a2 is required for the development of pituitary stem cells. Stem Cells Dev. 2022, 31, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Lodish, H.F.; Zhou, B.; Liu, G.; Chen, C.-Z. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 2008, 8, 120–130. [Google Scholar] [CrossRef]
- Witten, L.W.; Cheng, C.J.; Slack, F.J. miR-155 drives oncogenesis by promoting and cooperating with mutations in the c-Kit oncogene. Oncogene 2019, 38, 2151–2161. [Google Scholar] [CrossRef]
- Mattiske, S.; Suetani, R.J.; Neilsen, P.M.; Callen, D.F. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1236–1243. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Boldin, M.P.; Taganov, K.D.; Nicoll, J.; Paquette, R.L.; Baltimore, D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008, 205, 585–594. [Google Scholar] [CrossRef]
- Toki, T.; Katsuoka, F.; Kanezaki, R.; Xu, G.; Kurotaki, H.; Sun, J.; Kamio, T.; Watanabe, S.; Tandai, S.; Terui, K. Transgenic expression of BACH1 transcription factor results in megakaryocytic impairment. Blood 2005, 105, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Dragone, L.L.; Myers, M.D.; White, C.; Sosinowski, T.; Weiss, A. SRC-like adaptor protein regulates B cell development and function. J. Immunol. 2006, 176, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Tenen, D. C/EBPbeta is required for’emergency’granulopoiesis. Nat. Immunol. 2006, 7, 732–739. [Google Scholar]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; Van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Thai, T.-H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.-H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef]
- Li, Q.-J.; Chau, J.; Ebert, P.J.; Sylvester, G.; Min, H.; Liu, G.; Braich, R.; Manoharan, M.; Soutschek, J.; Skare, P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007, 129, 147–161. [Google Scholar] [CrossRef]
- Wolfe, F.; Clauw, D.J.; Fitzcharles, M.A.; Goldenberg, D.L.; Katz, R.S.; Mease, P.; Russell, A.S.; Russell, I.J.; Winfield, J.B.; Yunus, M.B. The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res. 2010, 62, 600–610. [Google Scholar] [CrossRef]
- Braun, A.; Evdokimov, D.; Frank, J.; Sommer, C.; Üçeyler, N. MiR103a-3p and miR107 are related to adaptive coping in a cluster of fibromyalgia patients. PLoS ONE 2020, 15, e0239286. [Google Scholar] [CrossRef]
- Cheng, C.-Y.; Chen, S.-P.; Liao, Y.-C.; Fuh, J.-L.; Wang, Y.-F.; Wang, S.-J. Elevated circulating endothelial-specific microRNAs in migraine patients: A pilot study. Cephalalgia 2018, 38, 1585–1591. [Google Scholar] [CrossRef] [PubMed]
- Uzuner, E.; Ulu, G.T.; Gürler, S.B.; Baran, Y. The role of MiRNA in cancer: Pathogenesis, diagnosis, and treatment. In miRNomics; Springer: Berlin/Heidelberg, Germany, 2022; pp. 375–422. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Li, C.; Feng, Y.; Coukos, G.; Zhang, L. Therapeutic microRNA strategies in human cancer. AAPS J. 2009, 11, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Rakheja, D.; Chen, K.S.; Liu, Y.; Shukla, A.A.; Schmid, V.; Chang, T.-C.; Khokhar, S.; Wickiser, J.E.; Karandikar, N.J.; Malter, J.S. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun. 2014, 5, 4802. [Google Scholar] [CrossRef]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sánchez-Céspedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, M.; Gu, J.; Lin, J.; Ye, Y.; Tan, W.; Tamboli, P.; Wood, C.; Wu, X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 2010, 29, 5724–5728. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.-W.; Liao, Y.-L.; Wu, C.-W.; Hu, L.-Y.; Li, S.-C.; Chan, W.-C.; Ho, M.-R.; Lai, C.-H.; Kao, H.-W.; Fang, W.-L. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics 2011, 6, 1189–1197. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, Z.; Ren, X.; Chen, K.; Xin, S. MicroRNA-124 (MiR-124) Inhibits Cell Proliferation, Metastasis and Invasion in Colorectal Cancer by Downregulating Rho-Associated Protein Kinase 1(ROCK1). Cell Physiol. Biochem. 2016, 38, 1785–1795. [Google Scholar] [CrossRef]
- Lujambio, A.; Ropero, S.; Ballestar, E.; Fraga, M.F.; Cerrato, C.; Setién, F.; Casado, S.; Suarez-Gauthier, A.; Sanchez-Cespedes, M.; Gitt, A. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007, 67, 1424–1429. [Google Scholar] [CrossRef]
- Bandres, E.; Agirre, X.; Bitarte, N.; Ramirez, N.; Zarate, R.; Roman-Gomez, J.; Prosper, F.; Garcia-Foncillas, J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer 2009, 125, 2737–2743. [Google Scholar] [CrossRef]
- Sethupathy, P.; Collins, F.S. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008, 24, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.J.; Mishra, P.J.; Banerjee, D.; Bertino, J.R. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle 2008, 7, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Luciano, D.J.; Mirsky, H.; Vendetti, N.J.; Maas, S. RNA editing of a miRNA precursor. RNA 2004, 10, 1174–1177. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17–92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef]
- Zhu, H.; Han, C.; Wu, T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis 2015, 36, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef]
- Yaman Agaoglu, F.; Kovancilar, M.; Dizdar, Y.; Darendeliler, E.; Holdenrieder, S.; Dalay, N.; Gezer, U. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumor Biol. 2011, 32, 583–588. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Z.; Gao, C.; Chen, P.; Chen, J.; Liu, W.; Xiao, S.; Lu, H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Investig. 2008, 88, 1358–1366. [Google Scholar] [CrossRef]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef]
- Simonian, M.; Mosallayi, M.; Mirzaei, H. Circulating miR-21 as novel biomarker in gastric cancer: Diagnostic and prognostic biomarker. J. Cancer Res. Ther. 2018, 14, 475. [Google Scholar] [PubMed]
- Si, M.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef] [PubMed]
- Sicard, F.; Gayral, M.; Lulka, H.; Buscail, L.; Cordelier, P. Targeting miR-21 for the therapy of pancreatic cancer. Mol. Ther. 2013, 21, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Blansit, K.; Kiet, T.; Sherman, A.; Wong, G.; Earle, C.; Bourguignon, L.Y. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol. Oncol. 2014, 132, 739–744. [Google Scholar] [CrossRef]
- Li, L.; Gong, Y.; Tang, J.; Yan, C.; Li, L.; Peng, W.; Cheng, Z.; Yu, R.; Xiang, Q.; Deng, C. ZBTB28 inhibits breast cancer by activating IFNAR and dual blocking CD24 and CD47 to enhance macrophages phagocytosis. Cell. Mol. Life Sci. 2022, 79, 83. [Google Scholar] [CrossRef]
- Neilsen, P.M.; Noll, J.E.; Mattiske, S.; Bracken, C.P.; Gregory, P.A.; Schulz, R.B.; Lim, S.P.; Kumar, R.; Suetani, R.J.; Goodall, G.J. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 2013, 32, 2992–3000. [Google Scholar] [CrossRef]
- Lei, T.; Xiao, B.; He, Y.; Sun, Z.; Li, L. High expression of ZNF652 promotes carcinogenesis and progression of breast cancer. Nan Fang Yi Ke Da Xue Xue Bao = J. South. Med. Univ. 2020, 40, 1732–1739. [Google Scholar]
- Yang, C.C.; Hung, P.S.; Wang, P.W.; Liu, C.J.; Chu, T.H.; Cheng, H.W.; Lin, S.C. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 397–404. [Google Scholar] [CrossRef]
- Strotbek, M.; Schmid, S.; Sánchez-González, I.; Boerries, M.; Busch, H.; Olayioye, M.A. miR-181 elevates Akt signaling by co-targeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int. J. Cancer 2017, 140, 2310–2320. [Google Scholar] [CrossRef]
- Tian, Y.; Fu, X.; Li, Q.; Wang, Y.; Fan, D.; Zhou, Q.; Kuang, W.; Shen, L. MicroRNA-181 serves an oncogenic role in breast cancer via the inhibition of SPRY4. Mol. Med. Rep. 2018, 18, 5603–5613. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.J.; Liu, J.; Wang, X.; Qu, L.X. microRNA-181 promotes prostate cancer cell proliferation by regulating DAX-1 expression. Exp. Ther. Med. 2014, 8, 1296–1300. [Google Scholar] [CrossRef]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-X.; Hu, Q.; Qiu, M.-T.; Zhong, S.-L.; Xu, J.-J.; Tang, J.-H.; Zhao, J.-H. miR-221/222: Promising biomarkers for breast cancer. Tumor Biol. 2013, 34, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Yang, J.; Guo, Z.; Hu, Y.; Sheng, H.; Gao, H.; Yu, H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am. J. Transl. Res. 2014, 6, 391. [Google Scholar] [PubMed]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer 2008, 123, 372–379. [Google Scholar] [CrossRef]
- Shen, J.; Wan, R.; Hu, G.; Yang, L.; Xiong, J.; Wang, F.; Shen, J.; He, S.; Guo, X.; Ni, J. miR-15b and miR-16 induce the apoptosis of rat activated pancreatic stellate cells by targeting Bcl-2 in vitro. Pancreatology 2012, 12, 91–99. [Google Scholar] [CrossRef]
- Musumeci, M.; Coppola, V.; Addario, A.; Patrizii, M.; Maugeri-Sacca, M.; Memeo, L.; Colarossi, C.; Francescangeli, F.; Biffoni, M.; Collura, D. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 2011, 30, 4231–4242. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef]
- Buechner, J.; Tømte, E.; Haug, B.; Henriksen, J.; Løkke, C.; Flaegstad, T.; Einvik, C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer 2011, 105, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Qin, S.; Fan, C.; Xu, C.; Du, N.; Ren, H. Let-7: A regulator of the ERα signaling pathway in human breast tumors and breast cancer stem cells. Oncol. Rep. 2013, 29, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
- Slusarz, A.; Pulakat, L. The two faces of miR-29. J. Cardiovasc. Med. (Hagerstown Md.) 2015, 16, 480. [Google Scholar] [CrossRef]
- Yu, P.N.; Yan, M.D.; Lai, H.C.; Huang, R.L.; Chou, Y.C.; Lin, W.C.; Yeh, L.T.; Lin, Y.W. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int. J. Cancer 2014, 134, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wu, Z.; Wang, Y.; Chen, F.; Liu, Y. Expression of miR-29 and STAT3 in osteosarcoma and its effect on proliferation regulation of osteosarcoma cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7275–7282. [Google Scholar] [PubMed]
- Zhang, W.; Qian, J.-X.; Yi, H.-L.; Yang, Z.-D.; Wang, C.-F.; Chen, J.-Y.; Wei, X.-Z.; Fu, Q.; Ma, H. The microRNA-29 plays a central role in osteosarcoma pathogenesis and progression. Mol. Biol. 2012, 46, 557–562. [Google Scholar] [CrossRef]
- Corney, D.C.; Hwang, C.-I.; Matoso, A.; Vogt, M.; Flesken-Nikitin, A.; Godwin, A.K.; Kamat, A.A.; Sood, A.K.; Ellenson, L.H.; Hermeking, H. Frequent Downregulation of miR-34 Family in Human Ovarian CancersmiR-34 in Ovarian Cancer. Clin. Cancer Res. 2010, 16, 1119–1128. [Google Scholar] [CrossRef]
- Ji, Q.; Hao, X.; Meng, Y.; Zhang, M.; DeSano, J.; Fan, D.; Xu, L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 2008, 8, 266. [Google Scholar] [CrossRef]
- Xu, Y.; An, Y.; Wang, Y.; Zhang, C.; Zhang, H.; Huang, C.; Jiang, H.; Wang, X.; Li, X. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 2013, 29, 2019–2024. [Google Scholar] [CrossRef]
- Nassirpour, R.; Mehta, P.P.; Yin, M.-J. miR-122 regulates tumorigenesis in hepatocellular carcinoma by targeting AKT3. PLoS ONE 2013, 8, e79655. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 2012, 413, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Garofalo, M.; Martelli, M.P.; Briesewitz, R.; Wang, L.; Fernandez-Cymering, C.; Volinia, S.; Liu, C.-G.; Schnittger, S.; Haferlach, T. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA 2008, 105, 3945–3950. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-G.; Gu, J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012, 36, e61–e67. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer—An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
- Hutvágner, G.; Simard, M.J.; Mello, C.C.; Zamore, P.D.; Joyce, G. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004, 2, e98. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef]
- Vester, B.; Wengel, J. LNA (locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Corsten, M.F.; Miranda, R.; Kasmieh, R.; Krichevsky, A.M.; Weissleder, R.; Shah, K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell–delivered S-TRAIL in human gliomas. Cancer Res. 2007, 67, 8994–9000. [Google Scholar] [CrossRef]
- Griveau, A.; Bejaud, J.; Anthiya, S.; Avril, S.; Autret, D.; Garcion, E. Silencing of miR-21 by locked nucleic acid–lipid nanocapsule complexes sensitize human glioblastoma cells to radiation-induced cell death. Int. J. Pharm. 2013, 454, 765–774. [Google Scholar] [CrossRef]
- Heo, Y.-A. Golodirsen: First Approval. Drugs 2020, 80, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.R.; Maruyama, R.; Yokota, T. Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 2017, 11, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.; Das, U.; Bose, C.; Bhadra, J.; Sinha, S. Self-transfecting GMO-PMO and PMO-GMO chimeras enable gene silencing in vitro and in vivo zebrafish model and NANOG Inhibition Induce the Apoptosis in Breast and Prostate Cancer Cells. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fabbri, E.; Brognara, E.; Borgatti, M.; Lampronti, I.; Finotti, A.; Bianchi, N.; Sforza, S.; Tedeschi, T.; Manicardi, A.; Marchelli, R. miRNA therapeutics: Delivery and biological activity of peptide nucleic acids targeting miRNAs. Epigenomics 2011, 3, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, S.; Na, H.K.; Min, D.H. MicroRNA-responsive drug release system for selective fluorescence imaging and photodynamic therapy in vivo. Adv. Healthc. Mater. 2016, 5, 2386–2395. [Google Scholar] [CrossRef] [PubMed]
- Dhuri, K.; Pradeep, S.P.; Shi, J.; Anastasiadou, E.; Slack, F.J.; Gupta, A.; Zhong, X.-B.; Bahal, R. Simultaneous Targeting of Multiple oncomiRs with Phosphorothioate or PNA-Based Anti-miRs in Lymphoma Cell Lines. Pharm. Res. 2022. [Google Scholar] [CrossRef]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Yu, R.Z.; Grundy, J.S.; Geary, R.S. Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin. Drug Metab. Toxicol. 2013, 9, 169–182. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nature Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef]
- Gumireddy, K.; Young, D.D.; Xiong, X.; Hogenesch, J.B.; Huang, Q.; Deiters, A. Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. 2008, 120, 7592–7594. [Google Scholar] [CrossRef]
- Melo, S.; Villanueva, A.; Moutinho, C.; Davalos, V.; Spizzo, R.; Ivan, C.; Rossi, S.; Setien, F.; Casanovas, O.; Simo-Riudalbas, L. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. USA 2011, 108, 4394–4399. [Google Scholar] [CrossRef] [PubMed]
- Watashi, K.; Yeung, M.L.; Starost, M.F.; Hosmane, R.S.; Jeang, K.-T. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J. Biol. Chem. 2010, 285, 24707–24716. [Google Scholar] [CrossRef] [PubMed]
- Hei, Y.-Y.; Wang, S.; Xi, X.-X.; Wang, H.-P.; Guo, Y.; Xin, M.; Jiang, C.; Lu, S.; Zhang, S.-Q. Design, synthesis, and evaluation of fluoroquinolone derivatives as MicroRNA-21 small-molecule inhibitors. J. Pharm. Anal. 2022, 12, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Yang, B.; Lin, H.; Lu, Y.; Luo, X.; Wang, Z. Retracted: Novel approaches for gene-specific interference via manipulating actions of microRNAs: Examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol. 2007, 212, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Trang, P.; Wiggins, J.F.; Patrawala, L.; Cheng, A.; Ford, L.; Weidhaas, J.B.; Brown, D.; Bader, A.G.; Slack, F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008, 7, 759–764. [Google Scholar] [CrossRef]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, X.Y.; Lu, A.; Wang, X.Y.; Jiang, L.X.; Wang, J.C. Non-viral vectors for RNA delivery. J. Control Release 2022, 342, 241–279. [Google Scholar] [CrossRef]
- Yang, N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015, 5, 179–181. [Google Scholar] [CrossRef]
- Wu, Y.; Crawford, M.; Mao, Y.; Lee, R.J.; Davis, I.C.; Elton, T.S.; Lee, L.J.; Nana-Sinkam, S.P. Therapeutic delivery of microRNA-29b by cationic lipoplexes for lung cancer. Mol. Ther. Nucleic Acids 2013, 2, e84. [Google Scholar] [CrossRef]
- Huang, X.; Schwind, S.; Yu, B.; Santhanam, R.; Wang, H.; Hoellerbauer, P.; Mims, A.; Klisovic, R.; Walker, A.R.; Chan, K.K. Targeted Delivery of microRNA-29b by Transferrin-Conjugated Anionic Lipopolyplex Nanoparticles: A Novel Therapeutic Strategy in Acute Myeloid LeukemiaLipopolyplex Nanoparticles for miR-29b Delivery in AML. Clin. Cancer Res. 2013, 19, 2355–2367. [Google Scholar] [CrossRef] [PubMed]
- Sukumar, U.K.; Bose, R.J.; Malhotra, M.; Babikir, H.A.; Afjei, R.; Robinson, E.; Zeng, Y.; Chang, E.; Habte, F.; Sinclair, R. Intranasal delivery of targeted polyfunctional gold–iron oxide nanoparticles loaded with therapeutic microRNAs for combined theranostic multimodality imaging and presensitization of glioblastoma to temozolomide. Biomaterials 2019, 218, 119342. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.S.; Lee, J.Y.; Choi, Y.S.; Chong, P.C.; Park, Y.J. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials 2013, 34, 4347–4359. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.F.; Zhu, Y.L.; Sun, B.; Hu, F.H.; Tian, T.; Li, S.C.; Xiao, Z.D. PLGA-based gene delivering nanoparticle enhance suppression effect of miRNA in HePG2 cells. Nanoscale Res. Lett. 2011, 6, 447. [Google Scholar] [CrossRef]
- Kasar, S.; Salerno, E.; Yuan, Y.; Underbayev, C.; Vollenweider, D.; Laurindo, M.F.; Fernandes, H.; Bonci, D.; Addario, A.; Mazzella, F.; et al. Systemic in vivo lentiviral delivery of miR-15a/16 reduces malignancy in the NZB de novo mouse model of chronic lymphocytic leukemia. Genes Immun. 2012, 13, 109–119. [Google Scholar] [CrossRef]
- Shin, K.-J.; Wall, E.A.; Zavzavadjian, J.R.; Santat, L.A.; Liu, J.; Hwang, J.-I.; Rebres, R.; Roach, T.; Seaman, W.; Simon, M.I.; et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 13759–13764. [Google Scholar] [CrossRef]
- Craig, V.; Tzankov, A.; Flori, M.; Schmid, C.; Bader, A.; Müller, A. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia 2012, 26, 2421–2424. [Google Scholar] [CrossRef]
- Ye, D.; Wang, G.; Liu, Y.; Huang, W.; Wu, M.; Zhu, S.; Jia, W.; Deng, A.M.; Liu, H.; Kang, J. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells 2012, 30, 1645–1654. [Google Scholar] [CrossRef]
- Schultz, B.R.; Chamberlain, J.S. Recombinant adeno-associated virus transduction and integration. Mol. Ther. 2008, 16, 1189–1199. [Google Scholar] [CrossRef]
- Kim, M.W.; Kwon, S.H.; Choi, J.H.; Lee, A. A Promising Biocompatible Platform: Lipid-Based and Bio-Inspired Smart Drug Delivery Systems for Cancer Therapy. Int. J. Mol. Sci. 2018, 19, 3859. [Google Scholar] [CrossRef]
- Dehghankelishadi, P.; Maritz, M.F.; Badiee, P.; Thierry, B. High density lipoprotein nanoparticle as delivery system for radio-sensitising miRNA: An investigation in 2D/3D head and neck cancer models. Int. J. Pharm. 2022, 617, 121585. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.F.; Weirauch, U.; Thomas, M.; Grünweller, A.; Hartmann, R.K.; Aigner, A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011, 71, 5214–5224. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhou, X.; Mei, M.; Yuan, X.-B.; Han, L.; Wang, G.-X.; Jia, Z.-F.; Xu, P.; Pu, P.-Y.; Kang, C.-S. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer 2010, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Yeom, J.-H.; Ko, J.-J.; Han, M.S.; Lee, K.; Na, S.-Y.; Bae, J. Effective delivery of anti-miRNA DNA oligonucleotides by functionalized gold nanoparticles. J. Biotechnol. 2011, 155, 287–292. [Google Scholar] [CrossRef]
- Crew, E.; Rahman, S.; Razzak-Jaffar, A.; Mott, D.; Kamundi, M.; Yu, G.; Tchah, N.; Lee, J.; Bellavia, M.; Zhong, C.-J. MicroRNA conjugated gold nanoparticles and cell transfection. Anal. Chem. 2012, 84, 26–29. [Google Scholar] [CrossRef]
- Cui, C.; Guo, T.; Zhang, S.; Yang, M.; Cheng, J.; Wang, J.; Kang, J.; Ma, W.; Nian, Y.; Sun, Z.; et al. Bacteria-derived outer membrane vesicles engineered with over-expressed pre-miRNA as delivery nanocarriers for cancer therapy. Nanomed. Nanotechnol. Biol. Med. 2022, 45, 102585. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, X.; Huang, H.; Wu, J. Exosome based miRNA delivery strategy for disease treatment. Chin. Chem. Lett. 2022, 33, 1693–1704. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent advances in miRNA delivery systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Herrera-Carrillo, E.; Liu, Y.P.; Berkhout, B. Improving miRNA delivery by optimizing miRNA expression cassettes in viral vectors. Hum. Gene Ther. Methods 2017, 28, 177–190. [Google Scholar] [CrossRef]
- Bikram, M.; Lee, M.; Chang, C.W.; Janát-Amsbury, M.M.; Kern, S.E.; Kim, S.W. Long-circulating DNA-complexed biodegradable multiblock copolymers for gene delivery: Degradation profiles and evidence of dysopsonization. J. Control Release 2005, 103, 221–233. [Google Scholar] [CrossRef]
- Qian, X.; Long, L.; Shi, Z.; Liu, C.; Qiu, M.; Sheng, J.; Pu, P.; Yuan, X.; Ren, Y.; Kang, C. Star-branched amphiphilic PLA-b-PDMAEMA copolymers for co-delivery of miR-21 inhibitor and doxorubicin to treat glioma. Biomaterials 2014, 35, 2322–2335. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Ju, S.; Wang, Y.; Wang, H.; Zhong, R. Myeloma cell adhesion to bone marrow stromal cells confers drug resistance by microRNA-21 up-regulation. Leuk Lymphoma 2011, 52, 1991–1998. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Han, L.; Deng, L.; Zhang, Y.; Shen, H.; Gong, T.; Zhang, Z.; Sun, X. Dual drugs (microRNA-34a and paclitaxel)-loaded functional solid lipid nanoparticles for synergistic cancer cell suppression. J. Control Release 2014, 194, 228–237. [Google Scholar] [CrossRef]
- Gandham, S.K.; Rao, M.; Shah, A.; Trivedi, M.S.; Amiji, M.M. Combination microRNA-based cellular reprogramming with paclitaxel enhances therapeutic efficacy in a relapsed and multidrug-resistant model of epithelial ovarian cancer. Mol. Ther. Oncolytics 2022, 25, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Normann, L.S.; Aure, M.R.; Leivonen, S.-K.; Haugen, M.H.; Hongisto, V.; Kristensen, V.N.; Mælandsmo, G.M.; Sahlberg, K.K. MicroRNA in combination with HER2-targeting drugs reduces breast cancer cell viability in vitro. Sci. Rep. 2021, 11, 10893. [Google Scholar] [CrossRef] [PubMed]
- Nieuweboer, A.J.; de Morrée, E.S.; de Graan, A.J.; Sparreboom, A.; de Wit, R.; Mathijssen, R.H. Inter-patient variability in docetaxel pharmacokinetics: A review. Cancer Treat. Rev. 2015, 41, 605–613. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, N. How nanotechnology can enhance docetaxel therapy. Int. J. Nanomed. 2013, 8, 2927–2941. [Google Scholar] [CrossRef]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, X.; Lv, Y.; Xin, X.; Qin, C.; Han, X.; Yang, L.; He, W.; Yin, L. Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Sci. Rep. 2017, 7, 46186. [Google Scholar] [CrossRef]
- Shah, M.Y.; Calin, G.A. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip. Rev. RNA 2014, 5, 537–548. [Google Scholar] [CrossRef]
- Lindow, M.; Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, H.; Wang, J.; Gumireddy, K.; Li, A.; Yao, W.; Tang, K.; Xiao, W.; Hu, J.; Xiao, H.; et al. miRNA-34a suppresses cell proliferation and metastasis by targeting CD44 in human renal carcinoma cells. J. Urol. 2014, 192, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The Risks of miRNA Therapeutics: In a Drug Target Perspective. Drug Des. Devel. Ther. 2021, 15, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef]
- Reid, G.; Pel, M.E.; Kirschner, M.; Cheng, Y.; Mugridge, N.; Weiss, J.; Williams, M.; Wright, C.; Edelman, J.J.; Vallely, M.; et al. Restoring expression of miR-16: A novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO 2013, 24, 3128–3135. [Google Scholar] [CrossRef] [PubMed]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Poltronieri, P.; D’Urso, P.I.; Mezzolla, V.; D’Urso, O.F. Potential of anti-cancer therapy based on anti-miR-155 oligonucleotides in glioma and brain tumours. Chem. Biol. Drug Des. 2013, 81, 79–84. [Google Scholar] [CrossRef]
- Querfeld, C.; Pacheco, T.; Foss, F.M.; Halwani, A.S.; Porcu, P.; Seto, A.G.; Ruckman, J.; Landry, M.L.; Jackson, A.L.; Pestano, L.A.; et al. Preliminary Results of a Phase 1 Trial Evaluating MRG-106, a Synthetic microRNA Antagonist (LNA antimiR) of microRNA-155, in Patients with CTCL. Blood 2016, 128, 1829. [Google Scholar] [CrossRef]
- Foss, F.M.; Querfeld, C.; Porcu, P.; Kim, Y.H.; Pacheco, T.; Halwani, A.S.; DeSimone, J.; William, B.M.; Seto, A.G.; Ruckman, J.; et al. Phase 1 trial evaluating MRG-106, a synthetic inhibitor of microRNA-155, in patients with cutaneous t-cell lymphoma (CTCL). J. Clin. Oncol. 2017, 35, 7564. [Google Scholar] [CrossRef]
- Sheedy, P.; Medarova, Z. The fundamental role of miR-10b in metastatic cancer. Am. J. Cancer Res. 2018, 8, 1674–1688. [Google Scholar] [PubMed]
- Ma, L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Yigit, M.V.; Ghosh, S.K.; Kumar, M.; Petkova, V.; Kavishwar, A.; Moore, A.; Medarova, Z. Context-dependent differences in miR-10b breast oncogenesis can be targeted for the prevention and arrest of lymph node metastasis. Oncogene 2013, 32, 1530–1538. [Google Scholar] [CrossRef]
- Guessous, F.; Alvarado-Velez, M.; Marcinkiewicz, L.; Zhang, Y.; Kim, J.; Heister, S.; Kefas, B.; Godlewski, J.; Schiff, D.; Purow, B.; et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J. Neuro-Oncol. 2013, 112, 153–163. [Google Scholar] [CrossRef]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar] [CrossRef]
- Ghosh, D.; Nandi, S.; Bhattacharjee, S. Combination therapy to checkmate Glioblastoma: Clinical challenges and advances. Clin. Transl. Med. 2018, 7, 33. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, D.-Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef]
- Seinen, E.; Burgerhof, J.G.; Jansen, R.C.; Sibon, O.C. RNAi experiments in D. melanogaster: Solutions to the overlooked problem of off-targets shared by independent dsRNAs. PLoS ONE 2010, 5, e13119. [Google Scholar] [CrossRef]
| Patent No. | Country of Origin | Patent Statement | Inventors | Applicant | Publication Date |
|---|---|---|---|---|---|
| WO2022169922A1 | USA | Compositions and methods for treating disease associated with dux4 overexpression. miR-675 inhibits DUX4 expression | Saad Nizar, Harper Scott | Res Inst Nationwide Childrens Hospital, USA | 11 August 2022 |
| WO2022170133A1 | USA | MicroRNA liver cancer markers and uses thereof | Rotroff Daniel, Aucejo Federico | Cleveland Clinic, USA | 11 August 2022 |
| WO2022164028A1 | Korea | Pharmaceutical composition, for preventing or treating liver fibrosis, containing mir-486-5p as an active ingredient | Jung Youngmi et al. | Pusan National University | 4 August 2022 |
| WO2022153846A1 | Japan | Composition for treating cancer or suppressing angiogenesis, including a microRNA agonist selected from the group consisting of miR-139-3p agonists, miR-214-3p agonists or a combination thereof. | Iwamoto Hideki et al. | Kurume University | 21 July 2022 |
| WO2022136226A1 | France | miRNA composition comprising 11 specific miRNAs and its use in the treatment of cancer | Jauliac Sébastien | Institut National de la Santé et de la Recherche Médicale | 30 June 2022 |
| WO2022103026A1 | Korea | Biomarker for diagnosing metastasis of cervical cancer and uses thereof (hsa-miR-1228-5p, hsa-miR-3200-3p, hsa-miR-146a-3p, hsa-miR-33a-5p, hsa-miR-6815-5p) | Cho O Yeon | Ajou University | 19 May 2022 |
| CN114432331A | China | Application of miR-138mimic in preparation of ovarian cancer stem cell medicine for inhibiting YAP and WWTR1 high expression | Zhou Qi et al. | Chongqing University Cancer Hospital | 6 May 2022 |
| WO2022072336A1 | USA | Drug-like molecules and methods for the therapeutic targeting of microrna-21 | Shortridge Matthew | University of Washington | 7 April 2022 |
| CN114177293A | China | Application of targeting miR-493/HIF-1alpha/PDK1 in preparation of drug-resistant drugs | Liu Wenjing et al. | Henan Cancer Hospital | 15 March 2022 |
| CN114099684A | China | Application of miR-32-5p in preparation of medicine for improving the sensitivity of tumor cells to dihydroartemisinin | Li Yujie et al. | Institute of Materia Medica of CAMS | 1 March 2022 |
| CN114053294A | China | Application of miR-150 and exosomes loaded with miR-150 simulants for the preparation of medicine for treating colorectal cancer | Zhou Jian et al. | Zhongshan Hospital Fudan University | 18 February 2022 |
| CN113755601A | China | Melanoma molecular marker and application thereof in the early diagnosis and treatment of melanoma (miR-196a, miR-27a-5p and miR-27b-5p) | Kong Yun et al. | Beijing Baiaosike Biomedical Technology Co., Ltd. Kangtai Medical Laboratory Service Hebei Co., Ltd. | 7 December 2021 |
| CN113713104A | China | Application of miR-345-3p in preparation of breast cancer treatment drug | Chen Tingmei, Zeng Qian | International Institute of In Vitro Diagnostics Chongqing Medical University | 30 November 2021 |
| CN113637762A | China | Application of miRNAs related to melanoma in the diagnosis and treatment of melanoma (miR-1321 and miR-139-5p) | Cui Minglu et al. | Mld Biotech Co., Ltd. | 12 November 2021 |
| CN113621707A | China | Application of hsa-miR-190b for the preparation of products for diagnosing and/or treating tumors | Yin Mengxiong, Shang Chuangeng, Ma Jiahui, Song Shuliang; Zhang E | University Shandong | 9 November 2021 |
| N113637762A | China | Application of miRNAs related to melanoma in the diagnosis and treatment of melanoma | Cui Minglu et al. | Beijing Baiaosike Biomedical Technology Co., Ltd. | 15 October 2021 |
| CN113476618A | China | Application of miR-199a-3p in the preparation of medicine for treating nasopharyngeal carcinoma | Luo Haiqing et al. | Guangdong Medical University | 8 October 2021 |
| CN112941183A | China | Application of non-coding gene miR-187-5p in primary liver cancer diagnosis and treatment | Zhou Jun et al. | Wuhan University of Science and Technology | 11 June 2021 |
| CN112791187A | China | Application of miR-142-5p in the preparation of medicine for treating chronic myelogenous leukemia | Wang Shuzhen et al. | China Pharmaceutical University | 14 May 2021 |
| CN111961727A | China | Application of miR-588 and the target gene thereof in gastric cancer | Xu Songxiao et al. | Zhejiang Cancer Hospital | 20 November 2020 |
| CN111961722A | China | Application of miR-887 in breast cancer diagnosis and treatment | He Jingsong | Peking University Shenzhen Hospital China | 20 November 2020 |
| WO2018181877 A1 | Japan | Cancer stem cell growth inhibitor using miRNA | Xin Wu et al. | Cancer Stem Tech Inc, Japan | 4th October 2018 |
| WO2018157026A1 | USA | Treatment of tumors with miRNA targeting CDK4/CDK6 | Amriti R Lulla and Wafik S EL-Deiry | Institute for Cancer Research and The Research Institute of Fox Chase Cancer Center, USA | 30th August 2018 |
| CN108452307 A | China | Application of human miRNA-493-3p inhibitor in preparing medicine for treating renal fibrosis | Rui Du et al. | The Fourth Military Medical University, China | 28th August 2018 |
| US7642348 B2 | USA | miRNA for the diagnosis, prognosis and treatment of prostate cancer; linear amplification and labeling for hybridization techniques such as Luminex and microarray analysis | Itzhak Bentwich et al. | Rosetta Genomics | 5 January 2010 |
| 7825229 B2 | USA | miRNAs; diagnosis, prognosis, and treatments; drug screening; linear amplification and labeling for hybridization techniques such as Luminex and microarray analysis; gene expression inhibition | Itzhak Bentwich et al. | Rosetta Genomics | 2 November 2010 |
| Target miRNA | Drug Name | Company | Disease | Study | Clinical Trial Number, Phase Status |
|---|---|---|---|---|---|
| miR-122 | Miravirsen | SantarisPharma | Hepatitis C virus (HCV) infection | Multiple Ascending Dose Study of Miravirsen in Treatment-Naïve Chronic Hepatitis C Subjects | NCT01200420; Phase II (Completed) |
| miR-34 | MRX34 | miRNA Therapeutics | Liver cancer, Lymphoma, melanoma | A multicentre phase I study of mrx34, microrna miR-rx34 liposomal injection. Five serious immune-related adverse events Pharmacodynamics study of MRX34, microRNA liposomal injection in melanoma patients with biopsy-accessible lesions (MRX34-102) | NCT01829971; Phase 1 (Terminated) NCT02862145; Phase 1 (Withdrawn) |
| miR-16 | MesomiR-1 | EnGeneIC | Mesothelioma, lung cancer | MesomiR 1: A Phase I study of TargomiRs as second- or third-line treatment for patients with recurrent MPM and NSCLC | NCT02369198; Phase 1 (Completed) |
| miR-155 | Cobomarsen (MRG-106) | miRagen therapeutics | T-cell lymphoma/ mycosis fungoides | Safety, tolerability and pharmacokinetics of MRG-106 in patients with mycosis fungoides (MF), CLL, DLBCL or ATLL Efficacy and safety of Cobomarsen (MRG-106) vs. active comparator in subjects with mycosis fungoides (SOLAR) | NCT02580552; Phase 1 (Completed) NCT03713320; Phase 2 (Terminated) |
| miR-10b | Regulus Therapeutics | Glioma | Evaluating the expression levels of microRNA-10b in patients with gliomas | NCT01849952; (recruiting) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. https://doi.org/10.3390/ijms231911502
Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: A Promising Therapeutic Target in Cancer. International Journal of Molecular Sciences. 2022; 23(19):11502. https://doi.org/10.3390/ijms231911502
Chicago/Turabian StyleMenon, Amrutha, Noraini Abd-Aziz, Kanwal Khalid, Chit Laa Poh, and Rakesh Naidu. 2022. "miRNA: A Promising Therapeutic Target in Cancer" International Journal of Molecular Sciences 23, no. 19: 11502. https://doi.org/10.3390/ijms231911502
APA StyleMenon, A., Abd-Aziz, N., Khalid, K., Poh, C. L., & Naidu, R. (2022). miRNA: A Promising Therapeutic Target in Cancer. International Journal of Molecular Sciences, 23(19), 11502. https://doi.org/10.3390/ijms231911502






