Genome-Wide Copy Number Variant and High-Throughput Transcriptomics Analyses of Placental Tissues Underscore Persisting Child Susceptibility in At-Risk Pregnancies Cleared in Standard Genetic Testing
Abstract
1. Introduction
2. Results
2.1. Case Mothers Do Not Differ from Controls in Psychological State across Pregnancy following Trisomy Screening Clearance
2.2. Children from Pregnancies Recommended for Follow-Up Prenatal Genetic Testing Carry Higher Likelihood of Negative Developmental and Disease-Related Outcomes
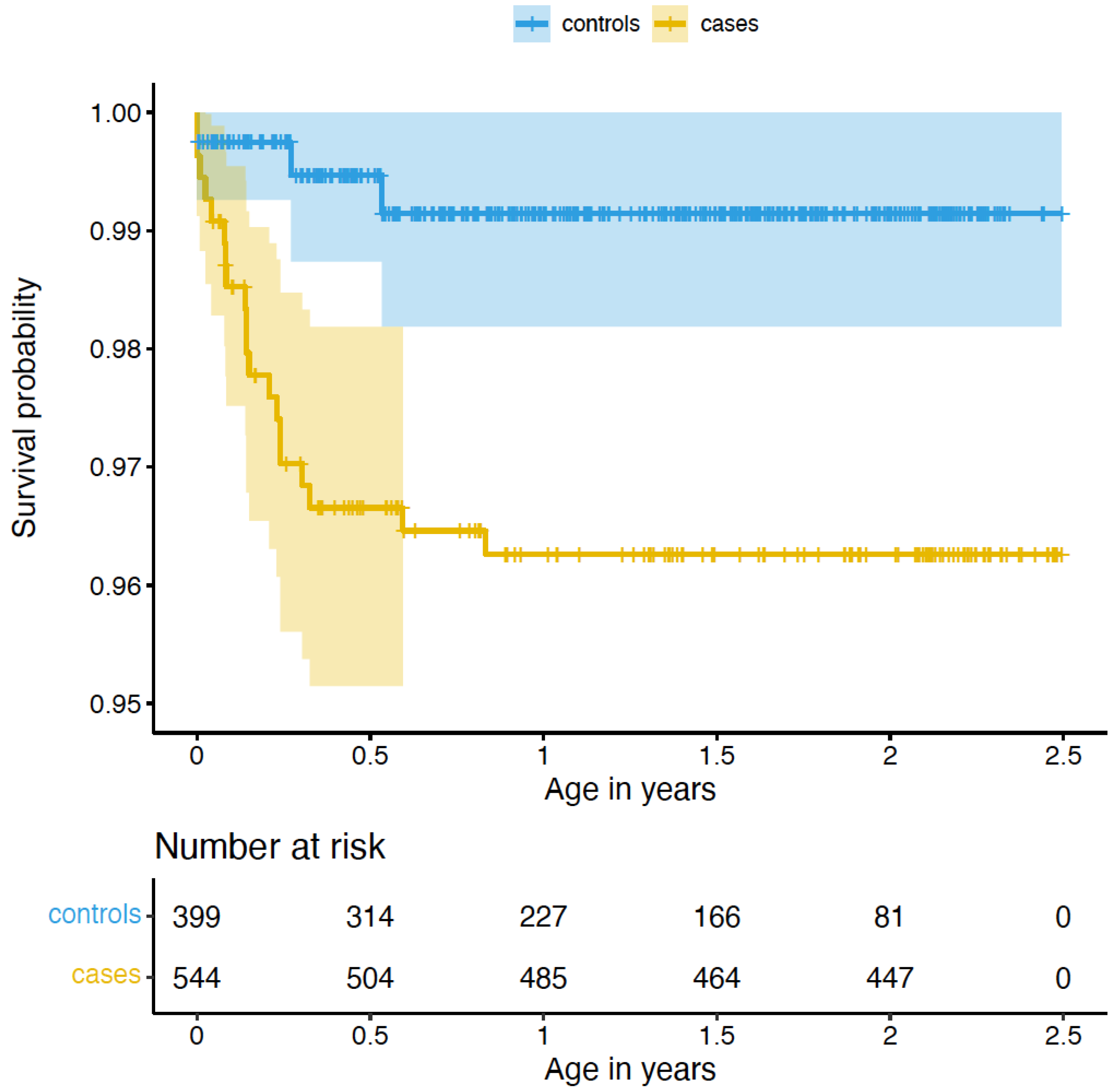
2.2.1. Congenital Malformations
2.2.2. Copy Number Variants
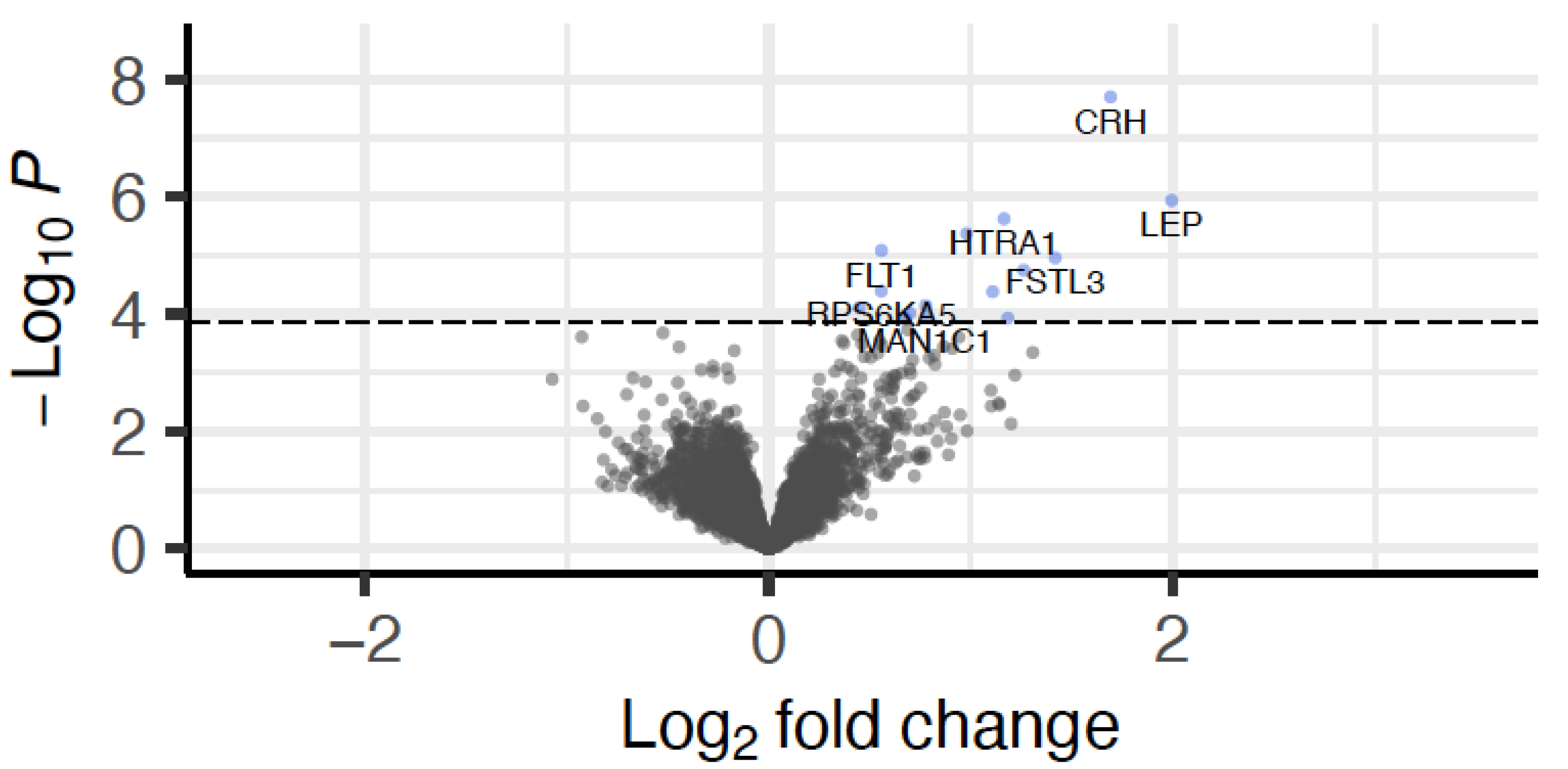
2.3. The Placental Transcriptome in Early Pregnancy Carries Signatures of Case-Control-Associated Negative Developmental Outcomes
3. Discussion
4. Materials and Methods
4.1. Sampling and Phenotypes
4.1.1. Study Samples
4.1.2. Phenotypes
Maternal Characteristics
Child Characteristics
4.2. Biosampling, DNA/RNA Extractions
4.3. Genotyping, Imputation, and MDS Components
4.4. CNV Calling
4.5. RNA Sequencing
4.6. Statistical Analysis
4.6.1. Differences in Congenital Malformations Using Cox Regression
4.6.2. Association of Congenital Malformations with Screening Variables
4.6.3. CNV Associations
4.6.4. Differential Gene Expression
4.6.5. p-Values
4.6.6. Enrichment for Pre-Eclampsia Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carlson, L.M.; Vora, N.L. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet. Gynecol. Clin. N. Am. 2017, 44, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Bardi, F.; Bosschieter, P.; Verheij, J.; Go, A.; Haak, M.; Bekker, M.; Sikkel, E.; Coumans, A.; Pajkrt, E.; Bilardo, C. Is there still a role for nuchal translucency measurement in the changing paradigm of first trimester screening? Prenat Diagn. 2020, 40, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Fruscalzo, A.; Cividino, A.; Rossetti, E.; Maurigh, A.; Londero, A.P.; Driul, L. First trimester PAPP-A serum levels and long-term metabolic outcome of mothers and their offspring. Sci. Rep. 2020, 10, 5131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, T.; Wang, J.; Li, Q.; Wang, H.; Liu, S. Prenatal Diagnostic Value of Chromosomal Microarray in Fetuses with Nuchal Translucency Greater than 2.5 mm. BioMed Res. Int. 2019, 2019, 6504159. [Google Scholar] [CrossRef]
- Hui, L.; Hutchinson, B.; Poulton, A.; Halliday, J. Population-based impact of noninvasive prenatal screening on screening and diagnostic testing for fetal aneuploidy. Genet. Med. 2017, 19, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Pös, O.; Budis, J.; Szemes, T. Recent trends in prenatal genetic screening and testing. F1000Research 2019, 8, 764. [Google Scholar] [CrossRef] [PubMed]
- Nadler, H.L.; Gerbie, A.B. Role of Amniocentesis in the Intrauterine Detection of Genetic Disorders. N. Engl. J. Med. 1970, 282, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Kvist, T.; Sammallahti, S.; Lahti-Pulkkinen, M.; Cruceanu, C.; Czamara, D.; Dieckmann, L.; Tontsch, A.; Röh, S.; Rex-Haffner, M.; Wolford, E.; et al. Cohort profile: InTraUterine sampling in early pregnancy (ITU), a prospective pregnancy cohort study in Finland: Study design and baseline characteristics. BMJ Open 2022, 12, e049231. [Google Scholar] [CrossRef] [PubMed]
- Conover, W.J. Pracitcal Nonparametric Statistics, 3rd ed.; Wiley: Hoboken, NJ, USA, 1999. [Google Scholar]
- Southard, A.E.; Edelmann, L.J.; Gelb, B.D. Role of Copy Number Variants in Structural Birth Defects. Pediatrics 2012, 129, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, X.; Wang, D.; Chen, X.; Wang, C.; Zhang, Y.; Xu, M.; Yu, J. Prevalence of chromosomal abnormalities identified by copy number variation sequencing in high-risk pregnancies, spontaneous abortions, and suspected genetic disorders. J. Int. Med. Res. 2019, 47, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.; Heo, J.; Han, S.-S.; Kim, W.J.; Cheong, H.S.; Hong, Y. Difference of copy number variation in blood of patients with lung cancer. Int. J. Biol. Mark. 2020, 36, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Saei, H.; Govahi, A.; Abiri, A.; Eghbali, M.; Abiri, M. Comprehensive transcriptome mining identified the gene expression signature and differentially regulated pathways of the late-onset preeclampsia. Pregnancy Hypertens. 2021, 25, 91–102. [Google Scholar] [CrossRef]
- van Uitert, M.; Moerland, P.D.; Enquobahrie, D.A.; Laivuori, H.; van der Post, J.A.; Ris-Stalpers, C.; Afink, G.B. Meta-Analysis of Placental Transcriptome Data Identifies a Novel Molecular Pathway Related to Preeclampsia. PLoS ONE 2015, 10, e0132468. [Google Scholar] [CrossRef] [PubMed]
- Vennou, K.E.; Kontou, P.I.; Braliou, G.G.; Bagos, P.G. Meta-analysis of gene expression profiles in preeclampsia. Pregnancy Hypertens. Int. J. Women’s Cardiovasc. Health 2019, 19, 52–60. [Google Scholar] [CrossRef]
- Paulson, K.R.; Kamath, A.M.; Alam, T.; Bienhoff, K.; Abady, G.G.; Abbas, J.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abd-Elsalam, S.M.; et al. Global, regional, and national progress towards Sustainable Development Goal 3.2 for neonatal and child health: All-cause and cause-specific mortality findings from the Global Burden of Disease Study 2019. Lancet 2021, 398, 870–905. [Google Scholar] [CrossRef]
- de Knegt, V.E.; Hedley, P.L.; Kanters, J.K.; Thagaard, I.N.; Krebs, L.; Christiansen, M.; Lausten-Thomsen, U. The Role of Leptin in Fetal Growth during Pre-Eclampsia. Int. J. Mol. Sci. 2021, 22, 4569. [Google Scholar] [CrossRef]
- Sitras, V.; Paulssen, R.; Grønaas, H.; Leirvik, J.; Hanssen, T.; Vårtun, A.; Acharya, G. Differential Placental Gene Expression in Severe Preeclampsia. Placenta 2009, 30, 424–433. [Google Scholar] [CrossRef]
- Kassotaki, I.; Valsamakis, G.; Mastorakos, G.; Grammatopoulos, D.K. Placental CRH as a Signal of Pregnancy Adversity and Impact on Fetal Neurodevelopment. Front. Endocrinol. 2021, 12, 714214. [Google Scholar] [CrossRef]
- Leviton, A.; Allred, E.N.; Kuban, K.C.; O’Shea, T.M.; Paneth, N.; Majzoub, J. ELGAN study investigators Brain disorders associated with corticotropin-releasing hormone expression in the placenta among children born before the 28th week of gestation. Acta Paediatr. 2015, 105, e7–e11. [Google Scholar] [CrossRef]
- Karteris, E.; Vatish, M.; Hillhouse, E.W.; Grammatopoulos, D.K. Preeclampsia Is Associated with Impaired Regulation of the Placental Nitric Oxide-Cyclic Guanosine Monophosphate Pathway by Corticotropin-Releasing Hormone (CRH) and CRH-Related Peptides. J. Clin. Endocrinol. Metab. 2005, 90, 3680–3687. [Google Scholar] [CrossRef]
- Founds, S.A.; Ren, D.; Roberts, J.M.; Jeyabalan, A.; Powers, R.W. Follistatin-Like 3 Across Gestation in Preeclampsia and Uncomplicated Pregnancies Among Lean and Obese Women. Reprod. Sci. 2014, 22, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Macintire, K.; Tuohey, L.; Ye, L.; Palmer, K.; Gantier, M.; Tong, S.; Kaitu’U-Lino, T.J. PAPPA2 is increased in severe early onset pre-eclampsia and upregulated with hypoxia. Reprod. Fertil. Dev. 2014, 26, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Gurusinghe, S.; Wallace, E.M.; Lim, R. The relationship between Activin A and anti-angiogenic factors in the development of pre-eclampsia. Pregnancy Hypertens. Int. J. Women’s Cardiovasc. Health 2014, 4, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xing, F.; He, Y.; Zong, S.; Luo, C.; Li, C.; Duan, T.; Wang, K.; Zhou, Q. Elevated HTRA1 and HTRA4 in severe preeclampsia and their roles in trophoblast functions. Mol. Med. Rep. 2018, 18, 2937–2944. [Google Scholar] [CrossRef]
- Founds, S.; Conley, Y.; Lyons-Weiler, J.; Jeyabalan, A.; Hogge, W.A.; Conrad, K. Altered Global Gene Expression in First Trimester Placentas of Women Destined to Develop Preeclampsia. Placenta 2009, 30, 15–24. [Google Scholar] [CrossRef]
- Hayeems, R.Z.; Campitelli, M.; Ma, X.; Huang, T.; Walker, M.; Guttmann, A. Rates of prenatal screening across health care regions in Ontario, Canada: A retrospective cohort study. CMAJ Open 2015, 3, E236–E243. [Google Scholar] [CrossRef][Green Version]
- Zhu, H.; Jin, X.; Xu, Y.; Zhang, W.; Liu, X.; Jin, J.; Qian, Y.; Dong, M. Efficiency of non-invasive prenatal screening in pregnant women at advanced maternal age. BMC Pregnancy Childbirth 2021, 21, 86. [Google Scholar] [CrossRef]
- Kim, Y.-N.; Choi, D.-W.; Kim, D.S.; Park, E.-C.; Kwon, J.-Y. Maternal age and risk of early neonatal mortality: A national cohort study. Sci. Rep. 2021, 11, 814. [Google Scholar] [CrossRef]
- Radloff, L.S. The CES-D Scale: A Self-Report Depression Scale for Research in the General Population. Appl. Psychol Meas. 1977, 1, 385–401. [Google Scholar] [CrossRef]
- Cohen, S.; Kamarck, T.; Mermelstein, R. A global measure of perceived stress. J. Health Soc. Behav. 1983, 24, 385–396. [Google Scholar] [CrossRef]
- Spielberger, C.D. State-Trait Anxiety Inventory: Bibliography, 2nd ed; Consulting Psychologists Press: Washington, DC, USA, 1989. [Google Scholar]
- Natamba, B.K.; Achan, J.; Arbach, A.; Oyok, T.O.; Ghosh, S.; Mehta, S.; Stoltzfus, R.J.; Griffiths, J.K.; Young, S.L. Reliability and validity of the center for epidemiologic studies-depression scale in screening for depression among HIV-infected and -uninfected pregnant women attending antenatal services in northern Uganda: A cross-sectional study. BMC Psychiatry 2014, 14, 303. [Google Scholar] [CrossRef] [PubMed]
- Sund, R. Quality of the Finnish Hospital Discharge Register: A systematic review. Scand. J. Public Health 2012, 40, 505–515. [Google Scholar] [CrossRef]
- Dieckmann, L.; Lahti-Pulkkinen, M.; Kvist, T.; Lahti, J.; DeWitt, P.E.; Cruceanu, C.; Laivuori, H.; Sammallahti, S.; Villa, P.M.; Suomalainen-König, S.; et al. Characteristics of epigenetic aging across gestational and perinatal tissues. Clin. Epigenet. 2021, 13, 97. [Google Scholar] [CrossRef]
- Delaneau, O.; Marchini, J.; Zagury, J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods 2011, 9, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Marchini, J.; Howie, B.; Myers, S.; McVean, G.; Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007, 39, 906–913. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.; Hakonarson, H.; Bucan, M. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef]
- Diskin, S.J.; Li, M.; Hou, C.; Yang, S.; Glessner, J.; Hakonarson, H.; Bucan, M.; Maris, J.M.; Wang, K. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008, 36, e126. [Google Scholar] [CrossRef]
- Lin, C.-F.; Naj, A.C.; Wang, L.-S. Analyzing Copy Number Variation Using SNP Array Data: Protocols for Calling CNV and Association Tests. Curr. Protoc. Hum. Genet. 2013, 79, 1–27. [Google Scholar] [CrossRef]
- Li, Y.R.; Glessner, J.T.; Coe, B.P.; Li, J.; Mohebnasab, M.; Chang, X.; Connolly, J.; Kao, C.; Wei, Z.; Bradfield, J.; et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat. Commun. 2020, 11, 255. [Google Scholar] [CrossRef]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257, Correction in Genet. Med. 2021, 23, 2230. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Gong, S.; Gaccioli, F.; Dopierala, J.; Sovio, U.; Cook, E.; Volders, P.J.; Martens, L.; Kirk, P.D.W.; Richardson, S.; Smith, G.C.S.; et al. The RNA landscape of the human placenta in health and disease. Nat. Commun. 2021, 12, 2639. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]

| Maternal Characteristics | Cases (n = 544) | Controls (n = 399) | p-Value 5 |
|---|---|---|---|
| Age at delivery, years, mean (SD), CI | 35.54 (5.5) [35.52–35.56] | 33.71 (4.2) [33.69–33.73] | 7.16 × 10−9 |
| Antenatal corticosteroid treatment, n (%) | 23 (4.2%) | 9 (2.3%) | 1.41 × 10−1 |
| Cesarean section, n (%) | 120 (22.1%) | 79 (19.8%) | 4.48 × 10−1 |
| Diabetes Disorders in pregnancy, n (%) | 120 (22.06%) | 77 (19.3%) | 3.43 × 10−1 |
| Early pregnancy BMI, median (IQR), CI | 23.43 (5.23) [23.05–23.74] | 22.65 (4.24) [22.27–23.03] | 1.42 × 10−3 |
| Hypertensive Disorders in pregnancy, n (%) | 46 (8.5%) | 20 (5.0%) | 5.50 × 10−2 |
| Primiparous, n (%) | 221 (40.6%) | 243 (60.9%) | 1.15 × 10−9 |
| Smoking during pregnancy 1, n (%) | 36 (6.6%) | 4 (1.0%) | 1.83 × 10−5 |
| Thyroid disorders 2, n (%) | 9 (1.7%) | 8 (2.0%) | 8.79 × 10−1 |
| Child characteristics | |||
| Birth weight, g, median (IQR), CI | 3533 (675) [3470–3585] | 3562 (610) [3508–3620] | 3.83 × 10−1 |
| Gestational age at birth, weeks, median (IQR), CI | 40.00 (2.0) [40.00–40.14] | 40.14 (1.7) [40.00–40.29] | 5.17 × 10−1 |
| Preterm birth 3 (<37 weeks), n (%) | 33 (6.1%) | 13 (3.3%) | 6.80 × 10−2 |
| Sex, girl, (%) | 256 (47.1%) | 204 (51.1%) | 2.42 × 10−1 |
| Screening variables | |||
| β-hCG level (MoM, ug/L), median (IQR), CI | 1.31 (1.17) [1.30–1.73] | 0.98 (0.74) [0.93–1.08] | 1.20 × 10−11 |
| NT, MoM mm, median (IQR), CI | 1.80 (1.15) [1.58–1.88] | 0.84 (0.32) [0.83–0.88] | 1.58 × 10−36 |
| PAPP-A level, MoM, mU/L, median (IQR), CI | 0.75 (0.80) [0.75–1.08] | 1.12 (0.73) [1.09–1.20] | 3.45 × 10−18 |
| Risk for Down syndrome 4, median (IQR), CI | 0.65 (1.01) [0.63–0.89] | 0.01 (0.2) [0.008–0.11] | 3.06 × 10−101 |
| Risk for trisomy 18 4, median (IQR), CI | 0.003 (0.01) [0.003–0.014] | 0.001 (0.00) [0.001–0.001] | 1.15 × 10−64 |
| Phenotype | Cases (n = 260) | Controls (n = 353) | p-Value 1 |
|---|---|---|---|
| CESD 19 gestational weeks, median (IQR), CI | 9 (9) [9–14] | 9 (8) [9–11] | 0.32 |
| CESD 26 gestational weeks, median (IQR), CI | 8 (10) [9–12] | 9 (8) [9–11] | 0.35 |
| CESD 38 gestational weeks, median (IQR), CI | 8 (9) [8–11] | 9 (9) [9–10] | 0.43 |
| Cohen’s perceived stress scale 19 gestational weeks, median (IQR), CI | 5 (4.06) [6–8] | 6 (4) [6–7] | 0.48 |
| Cohen’s 26 gestational weeks, median (IQR), CI | 5 (5) [5–7] | 5.5 (3) [6–6] | 0.30 |
| Cohen’s perceived stress scale 38 gestational weeks, median (IQR), CI | 5 (4) [5–6] | 5 (4) [4.5–6] | 0.87 |
| STAI 19 gestational weeks, median (IQR). CI | 37 (11) [39–42] | 38 (9) [38.95–41] | 0.74 |
| STAI 26 gestational weeks, median (IQR), CI | 36 (9) [37–40] | 37 (10) [37–39] | 0.38 |
| STAI 38 gestational weeks, median (IQR), CI | 36 (10.42) [36–39] | 37 (10) [36–38] | 0.70 |
| Gene 1 | Position (hg19) 2 | Log2 (FC) 3 | p-Value 4 | Adjusted p-Value 5 | Correlation CVS and Placenta 6 |
|---|---|---|---|---|---|
| LEP | chr7: 127,881,331–127,897,682 | 1.99 | 1.16 × 10−6 | 5.28 × 10−3 | r = 0.27; p = 0.01 |
| CRH | chr8: 67,088,612–67,090,846 | 1.69 | 1.99 × 10−8 | 1.81 × 10−4 | r = 0.14; p = 0.17 |
| FSTL3 | chr19: 676,389–683,392 | 1.42 | 1.11 × 10−5 | 1.68 × 10−2 | r = 0.26; p = 0.01 |
| PAPPA2 | chr1: 176,432,307–176,811,970 | 1.26 | 1.79 × 10−5 | 2.32 × 10−2 | r = 0.30; p < 0.01 |
| INHBA | chr7: 41,733,514–41,818,976 | 1.18 | 1.18 × 10−4 | 7.36 × 10−2 | r = 0.23; p = 0.03 |
| HTRA1 | chr10: 124,221,041–124,274,424 | 1.16 | 2.38 × 10−6 | 7.22 × 10−3 | r = 0.30; p < 0.01 |
| DHRS2 | chr14: 24,105,573–24,114,848 | 1.11 | 4.16 × 10−5 | 4.20 × 10−2 | r < 0.01; p = 0.97 |
| HS3ST3B1 | chr17: 14,204,506–14,249,492 | 0.98 | 4.27 × 10−6 | 9.71 × 10−3 | r = 0.13; p = 0.23 |
| ANXA4 | chr2: 69,969,127–70,053,596 | 0.78 | 7.15 × 10−5 | 6.47 × 10−2 | r < 0.01; p = 0.97 |
| MAN1C1 | chr1: 25,943,959–26,111,258 | 0.78 | 1.15 × 10−4 | 7.36 × 10−2 | r = 0.05; p = 0.60 |
| MROH1 | chr8:145,202,919–145,316,843 | 0.70 | 9.64 × 10−5 | 7.30 × 10−2 | r = −0.14; p = 0.17 |
| SEMA7A | chr15: 74,701,630-74,726,299 | 0.68 | 1.21 × 10−4 | 7.36 × 10−2 | r = 0.07; p = 0.53 |
| FLT1 | chr13: 28,874,483–29,069,265 | 0.56 | 8.27 × 10−6 | 1.50 × 10−2 | r = 0.32; p < 0.01 |
| RPS6KA5 | chr14: 91,337,167–91,526,993 | 0.56 | 4.06 × 10−5 | 4.20 × 10−2 | r = 0.30; p < 0.01 |
| HEXIM1 | chr17: 43,224,684–43,229,468 | 0.45 | 7.83 × 10−5 | 6.47 × 10−2 | r < 0.01; p = 0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czamara, D.; Cruceanu, C.; Lahti-Pulkkinen, M.; Dieckmann, L.; Ködel, M.; Sauer, S.; Rex-Haffner, M.; Sammallahti, S.; Kajantie, E.; Laivuori, H.; et al. Genome-Wide Copy Number Variant and High-Throughput Transcriptomics Analyses of Placental Tissues Underscore Persisting Child Susceptibility in At-Risk Pregnancies Cleared in Standard Genetic Testing. Int. J. Mol. Sci. 2022, 23, 11448. https://doi.org/10.3390/ijms231911448
Czamara D, Cruceanu C, Lahti-Pulkkinen M, Dieckmann L, Ködel M, Sauer S, Rex-Haffner M, Sammallahti S, Kajantie E, Laivuori H, et al. Genome-Wide Copy Number Variant and High-Throughput Transcriptomics Analyses of Placental Tissues Underscore Persisting Child Susceptibility in At-Risk Pregnancies Cleared in Standard Genetic Testing. International Journal of Molecular Sciences. 2022; 23(19):11448. https://doi.org/10.3390/ijms231911448
Chicago/Turabian StyleCzamara, Darina, Cristiana Cruceanu, Marius Lahti-Pulkkinen, Linda Dieckmann, Maik Ködel, Susann Sauer, Monika Rex-Haffner, Sara Sammallahti, Eero Kajantie, Hannele Laivuori, and et al. 2022. "Genome-Wide Copy Number Variant and High-Throughput Transcriptomics Analyses of Placental Tissues Underscore Persisting Child Susceptibility in At-Risk Pregnancies Cleared in Standard Genetic Testing" International Journal of Molecular Sciences 23, no. 19: 11448. https://doi.org/10.3390/ijms231911448
APA StyleCzamara, D., Cruceanu, C., Lahti-Pulkkinen, M., Dieckmann, L., Ködel, M., Sauer, S., Rex-Haffner, M., Sammallahti, S., Kajantie, E., Laivuori, H., Lahti, J., Räikkönen, K., & Binder, E. B. (2022). Genome-Wide Copy Number Variant and High-Throughput Transcriptomics Analyses of Placental Tissues Underscore Persisting Child Susceptibility in At-Risk Pregnancies Cleared in Standard Genetic Testing. International Journal of Molecular Sciences, 23(19), 11448. https://doi.org/10.3390/ijms231911448






