Three-Way DNA Junction as an End Label for DNA in Atomic Force Microscopy Studies
Abstract
1. Introduction
2. Results
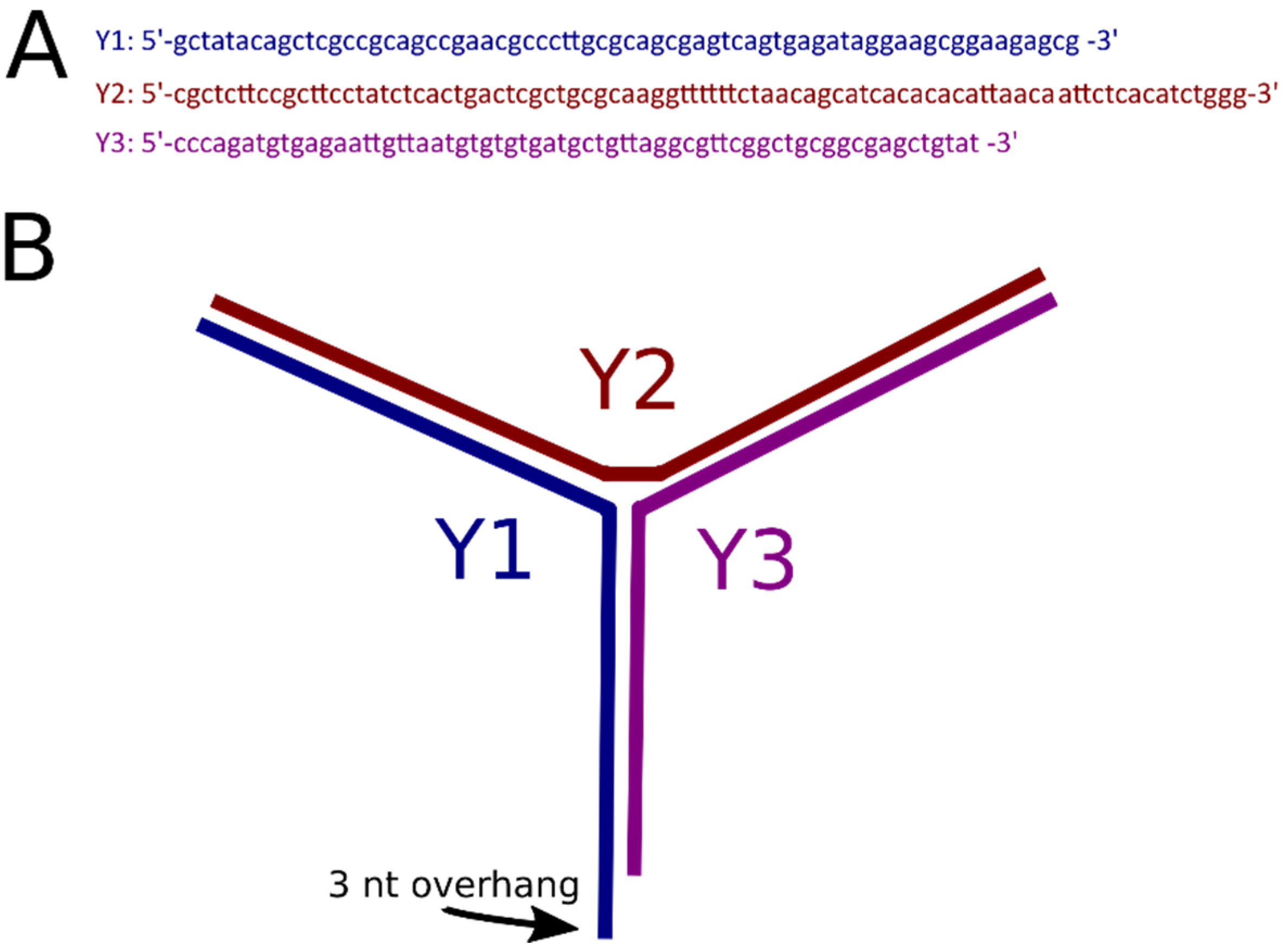
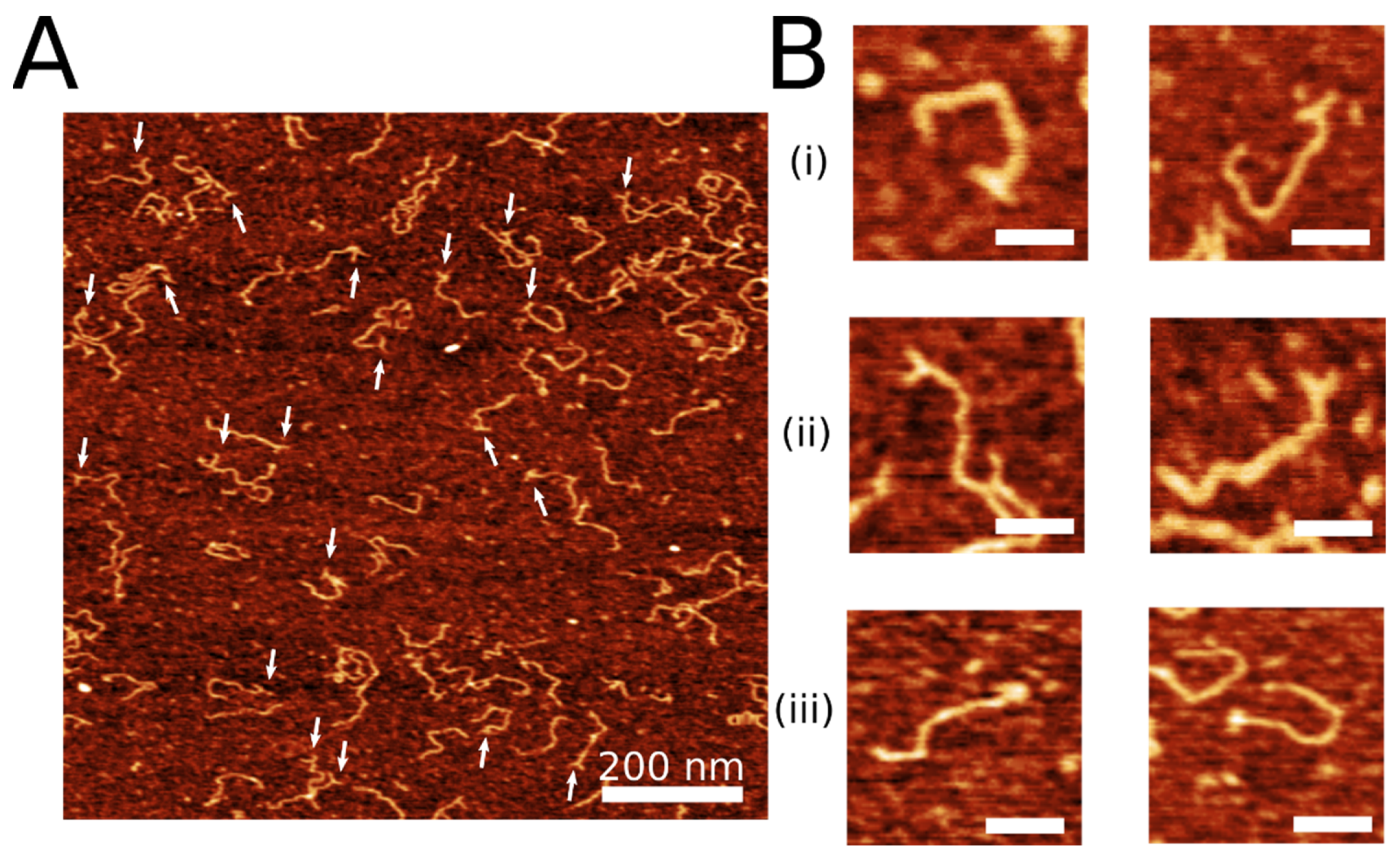
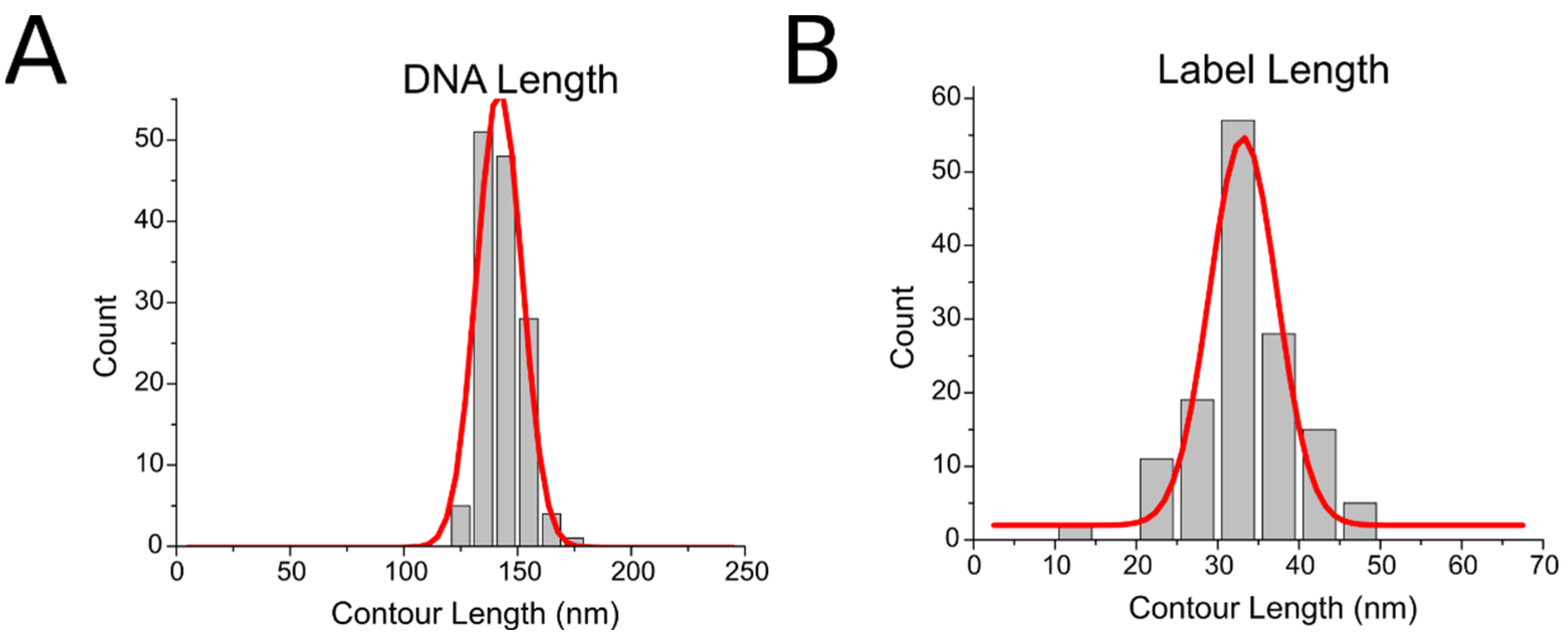
2.1. Coupling of Linear DNA with Three-Way DNA Junction
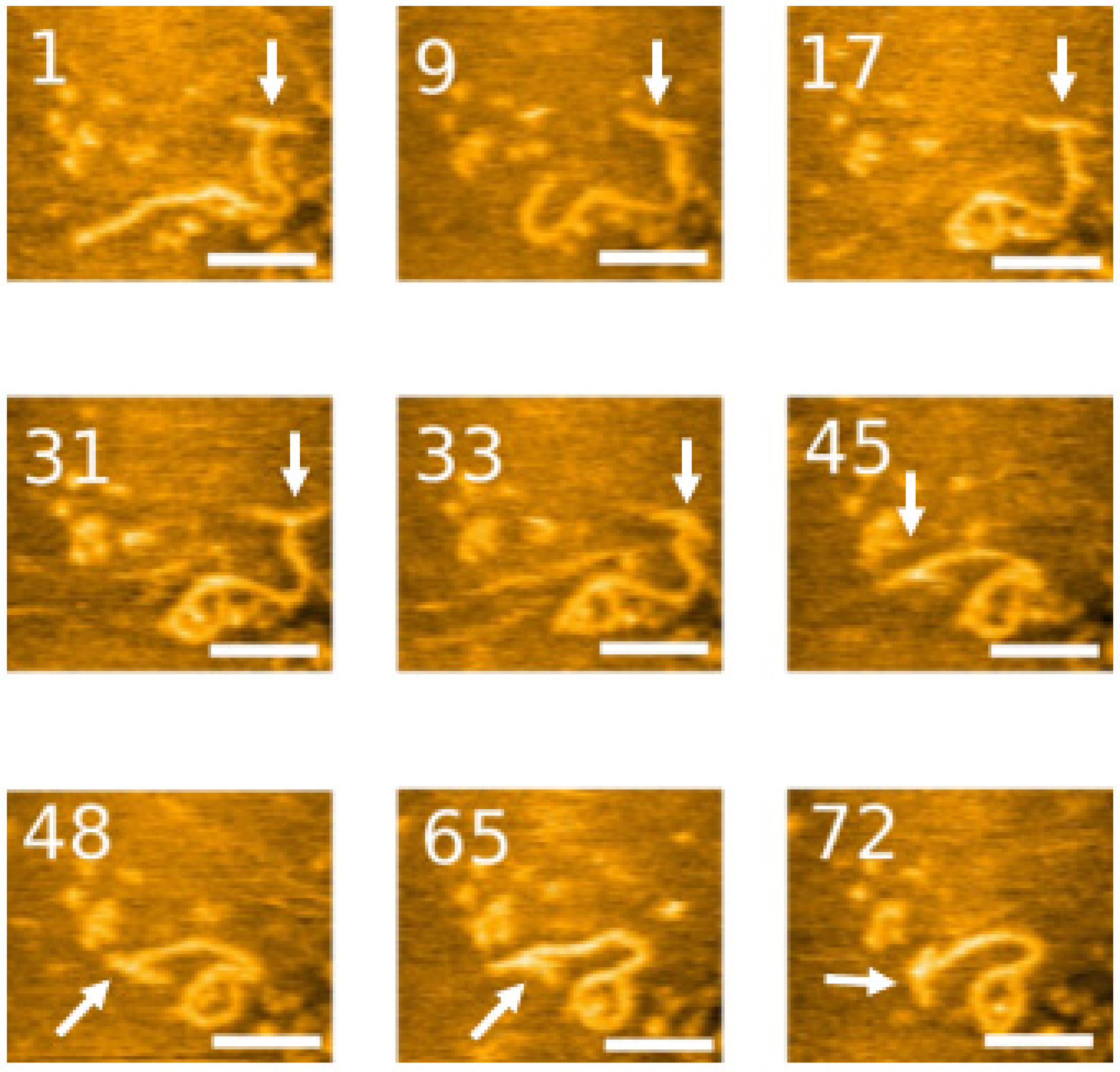
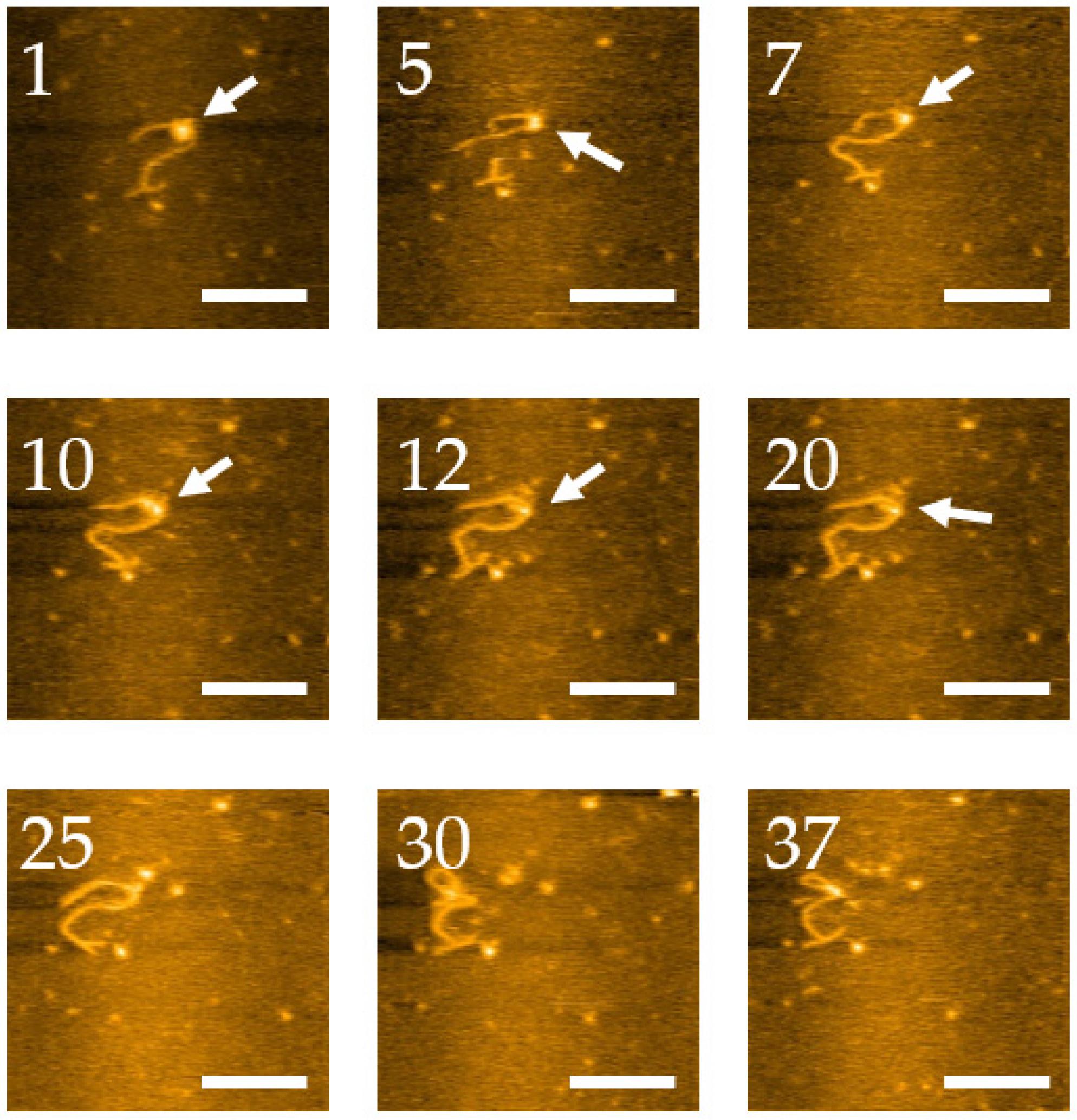
2.2. HS-AFM to Visualize the Dynamics of the 3WJ Directly
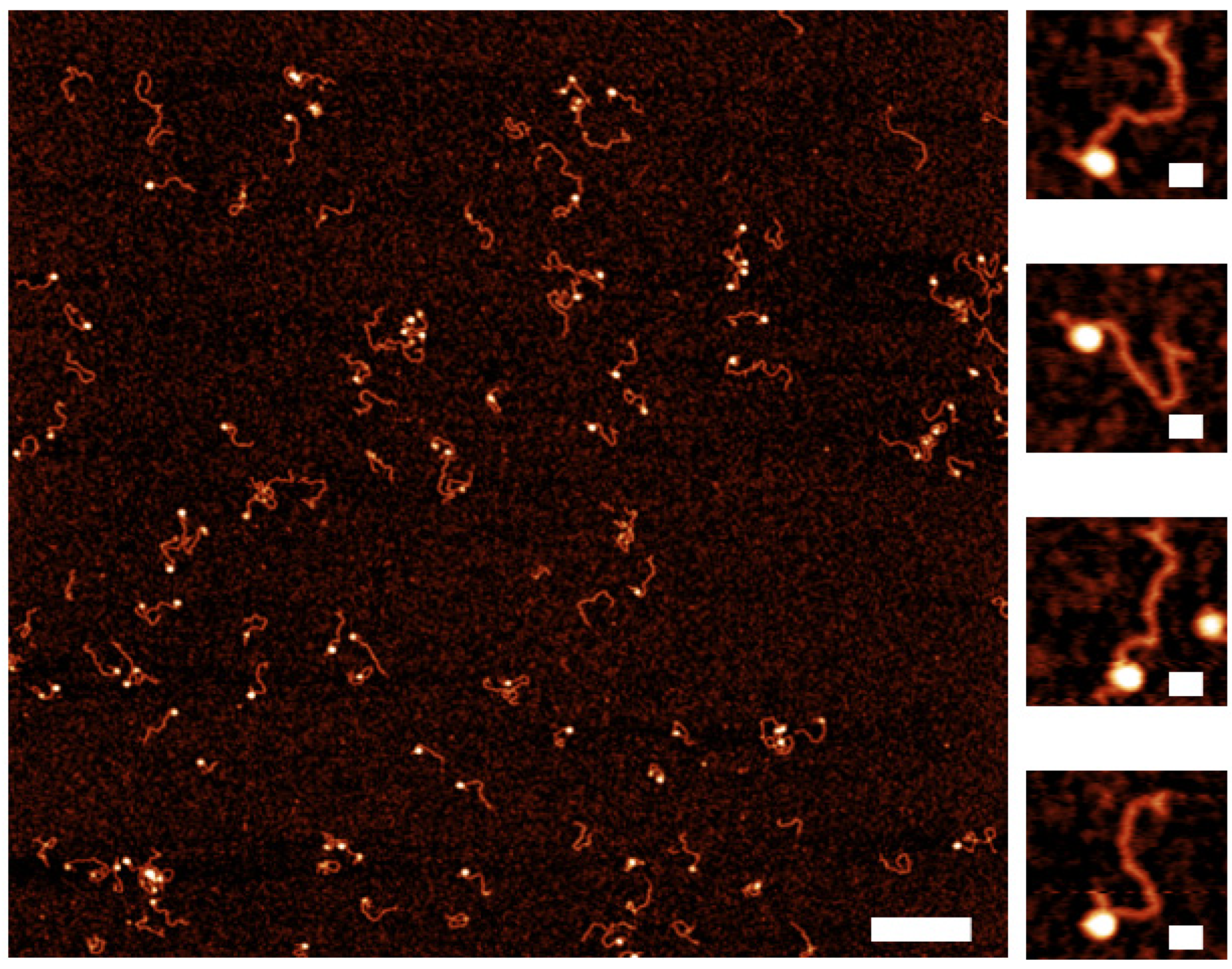
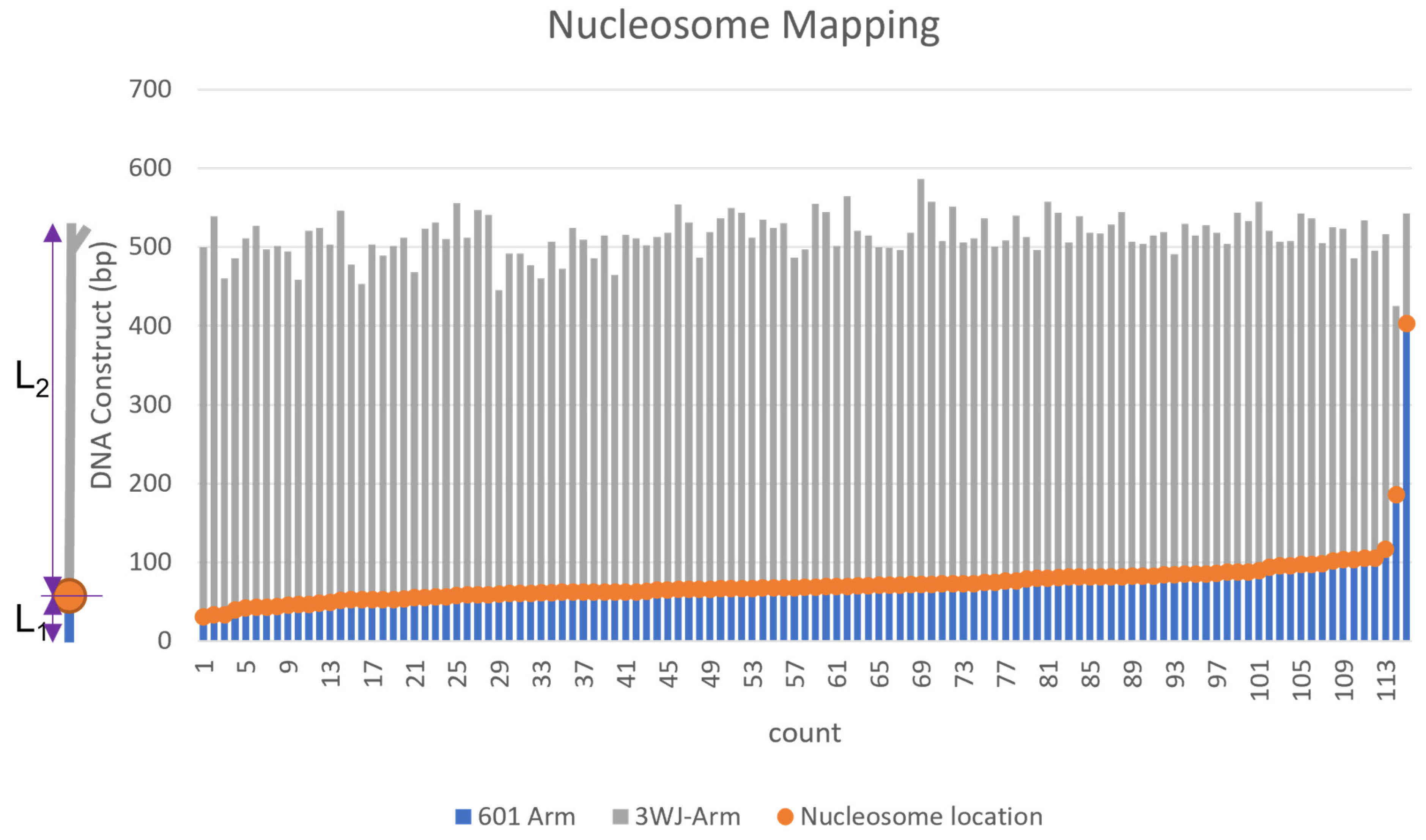
2.3. Assembly of Nucleosomes on Y-DNA Substrate
2.4. Dynamics of the Nucleosome Arrays
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides for 3WJ Assembly
4.2. Preparation of 3WJ End Label
4.3. Preparation of Full DNA Construct
4.4. Assembly of Nucleosome Complexes
4.5. Atomic Force Microscopy Imaging of Dry Samples in Air
4.6. High-Speed Atomic Force Microscopy Imaging in Aqueous Buffer
4.7. Image Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lyubchenko, Y.L. AFM Methods for DNA Analysis. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2000; pp. 1–24. [Google Scholar]
- Neish, C.S.; Martin, I.L.; Henderson, R.M.; Edwardson, J.M. Direct Visualization of Ligand-Protein Interactions Using Atomic Force Microscopy. Br. J. Pharm. 2002, 135, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M.; Adler, M.; Pignataro, B.; Lenhert, S.; Gao, S.; Chi, L.; Fuchs, H.; Blohm, D. Self-Assembly of DNA-Streptavidin Nanostructures and Their Use as Reagents in Immuno-PCR. Nucleic Acids Res. 1999, 27, 4553–4561. [Google Scholar] [CrossRef] [PubMed]
- Seong, G.H.; Yanagida, Y.; Aizawa, M.; Kobatake, E. Atomic Force Microscopy Identification of Transcription Factor NFkappaB Bound to Streptavidin-Pin-Holding DNA Probe. Anal. Biochem. 2002, 309, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Buzio, R.; Repetto, L.; Giacopelli, F.; Ravazzolo, R.; Valbusa, U. Label-Free, Atomic Force Microscopy-Based Mapping of DNA Intrinsic Curvature for the Nanoscale Comparative Analysis of Bent Duplexes. Nucleic Acids Res. 2012, 40, e84. [Google Scholar] [CrossRef] [PubMed]
- Meir, A.; Helppolainen, S.H.; Podoly, E.; Nordlund, H.R.; Hytönen, V.P.; Määttä, J.A.; Wilchek, M.; Bayer, E.A.; Kulomaa, M.S.; Livnah, O. Crystal Structure of Rhizavidin: Insights into the Enigmatic High-Affinity Interaction of an Innate Biotin-Binding Protein Dimer. J. Mol. Biol. 2009, 386, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Helppolainen, S.H.; Nurminen, K.P.; Määttä, J.A.E.; Halling, K.K.; Slotte, J.P.; Huhtala, T.; Liimatainen, T.; Ylä-Herttuala, S.; Airenne, K.J.; Närvänen, A.; et al. Rhizavidin from Rhizobium Etli: The First Natural Dimer in the Avidin Protein Family. Biochem. J. 2007, 405, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Stormberg, T.; Vemulapalli, S.; Filliaux, S.; Lyubchenko, Y.L. Effect of Histone H4 Tail on Nucleosome Stability and Internucleosomal Interactions. Sci. Rep. 2021, 11, 24086. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J. Electron Microscopy Methods and Protocols, 3rd ed.; Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 1117. [Google Scholar]
- Shlyakhtenko, L.S.; Lushnikov, A.J.; Li, M.; Harris, R.S.; Lyubchenko, Y.L. Interaction of APOBEC3A with DNA Assessed by Atomic Force Microscopy. PLoS ONE 2014, 9, e99354. [Google Scholar] [CrossRef]
- Shlyakhtenko, L.S.; Lushnikov, A.Y.; Miyagi, A.; Lyubchenko, Y.L. Specificity of Binding of Single-Stranded DNA-Binding Protein to Its Target. Biochemistry 2012, 51, 1500–1509. [Google Scholar] [CrossRef][Green Version]
- Hart, D.J.; Speight, R.E.; Cooper, M.A.; Sutherland, J.D.; Blackburn, J.M. The Salt Dependence of DNA Recognition by N-ΚB P50: A Detailed Kinetic Analysis of the Effects on Affinity and Specificity. Nucleic Acids Res. 1999, 27, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.A.; Honig, B. Salt Effects on Nucleic Acids. Curr. Opin. Struct. Biol. 1995, 5, 323–328. [Google Scholar] [CrossRef]
- Jensen, J. Calculating PH and Salt Dependence of Protein-Protein Binding. Curr. Pharm. Biotechnol. 2008, 9, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Waldron, T.T.; Schrift, G.L.; Murphy, K.P. The Salt-Dependence of a Protein–Ligand Interaction: Ion–Protein Binding Energetics. J. Mol. Biol. 2005, 346, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Yager, T.D.; Van Holde, K.E. Dynamics and Equilibria of Nucleosomes at Elevated Ionic Strength. J. Biol. Chem. 1984, 259, 4212–4222. [Google Scholar] [CrossRef]
- Ballestar, E.; Boix-Chornet, M.; Franco, L. Conformational Changes in the Nucleosome Followed by the Selective Accessibility of Histone Glutamines in the Transglutaminase Reaction: Effects of Ionic Strength. Biochemistry 2001, 40, 1922–1929. [Google Scholar] [CrossRef]
- Bednar, J.; Horowitz, R.A.; Dubochet, J.; Woodcock, C.L. Chromatin Conformation and Salt-Induced Compaction: Three-Dimensional Structural Information from Cryoelectron Microscopy. J. Cell Biol. 1995, 131, 1365–1376. [Google Scholar] [CrossRef]
- Allahverdi, A.; Chen, Q.; Korolev, N.; Nordenskiöld, L. Chromatin Compaction under Mixed Salt Conditions: Opposite Effects of Sodium and Potassium Ions on Nucleosome Array Folding. Sci. Rep. 2015, 5, 8512. [Google Scholar] [CrossRef]
- Billingsley, D.J.; Crampton, N.; Kirkham, J.; Thomson, N.H.; Bonass, W.A. Single-Stranded Loops as End-Label Polarity Markers for Double-Stranded Linear DNA Templates in Atomic Force Microscopy. Nucleic Acids Res. 2012, 40, e99. [Google Scholar] [CrossRef] [PubMed]
- Chammas, O.; Billingsley, D.J.; Bonass, W.A.; Thomson, N.H. Single-Stranded DNA Loops as Fiducial Markers for Exploring DNA-Protein Interactions in Single Molecule Imaging. Methods 2013, 60, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtenko, L.S.; Appella, E.; Harrington, R.E.; Kutyavin, I.; Lyubchenko, Y.L. Structure of Three-Way Dna Junctions. 2. Effects of Extra Bases and Mismatches. J. Biomol. Struct. Dyn. 1994, 12, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtenko, L.S.; Rekesh, D.; Lindsay, S.M.; Kutyavin, I.; Appella, E.; Harrington, R.E.; Lyubchenko, Y.L. Structure of Three-Way Dna Junctions: 1. Non-Planar Dna Geometry. J. Biomol. Struct. Dyn. 1994, 11, 1175–1189. [Google Scholar] [CrossRef] [PubMed]
- Oussatcheva, E.A.; Shlyakhtenko, L.S.; Glass, R.; Sinden, R.R.; Lyubchenko, Y.L.; Potaman, V.N. Structure of Branched DNA Molecules: Gel Retardation and Atomic Force Microscopy Studies. J. Mol. Biol. 1999, 292, 75–86. [Google Scholar] [CrossRef]
- Yamamoto, D.; Nagura, N.; Omote, S.; Taniguchi, M.; Ando, T. Streptavidin 2D Crystal Substrates for Visualizing Biomolecular Processes by Atomic Force Microscopy. Biophys. J. 2009, 97, 2358–2367. [Google Scholar] [CrossRef]
- Woolley, A.T.; Cheung, C.L.; Hafner, J.H.; Lieber, C.M. Structural Biology with Carbon Nanotube AFM Probes. Chem. Biol. 2000, 7, R193–R204. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Kodera, N. High-Speed AFM and Applications to Biomolecular Systems. Annu. Rev. Biophys. 2013, 42, 393–414. [Google Scholar] [CrossRef]
- Miyagi, A.; Ando, T.; Lyubchenko, Y.L. Dynamics of Nucleosomes Assessed with Time-Lapse High-Speed Atomic Force Microscopy. Biochemistry 2011, 50, 7901–7908. [Google Scholar] [CrossRef]
- Vemulapalli, S.; Hashemi, M.; Lyubchenko, Y.L. Site-Search Process for Synaptic Protein-Dna Complexes. Int. J. Mol. Sci. 2022, 23, 212. [Google Scholar] [CrossRef]
- Zhang, Y.; Hashemi, M.; Lv, Z.; Williams, B.; Popov, K.I.; Dokholyan, N.v.; Lyubchenko, Y.L. High-Speed Atomic Force Microscopy Reveals Structural Dynamics of α-Synuclein Monomers and Dimers. J. Chem. Phys. 2018, 148, 5835–5840. [Google Scholar] [CrossRef]
- Lyubchenko, Y.L. Direct AFM Visualization of the Nanoscale Dynamics of Biomolecular Complexes. J. Phys. D Appl. Phys. 2018, 51, 403001. [Google Scholar] [CrossRef]
- Lowary, P.T.; Widom, J. New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-Directed Nucleosome Positioning. J. Mol. Biol. 1998, 276, 19–42. [Google Scholar] [CrossRef]
- Stumme-Diers, M.P.; Banerjee, S.; Hashemi, M.; Sun, Z.; Lyubchenko, Y.L. Nanoscale Dynamics of Centromere Nucleosomes and the Critical Roles of CENP-A. Nucleic Acids Res. 2018, 46, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Stormberg, T.; Filliaux, S.; Baughman, H.E.R.; Komives, E.A.; Lyubchenko, Y.L. Transcription Factor NF-ΚB Unravels Nucleosomes. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129934. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtenko, L.S.; Gall, A.A.; Lyubchenko, Y.L. Mica Functionalization for Imaging of DNA and Protein-DNA Complexes with Atomic Force Microscopy. In Cell Imaging Techniques. Methods in Molecular Biology (Methods and Protocols); Humana Press: Totowa, NJ, USA, 2012; pp. 295–312. [Google Scholar]
- Stumme-Diers, M.P.; Stormberg, T.; Sun, Z.; Lyubchenko, Y.L. Probing the Structure and Dynamics of Nucleosomes Using Atomic Force Microscopy Imaging. J. Vis. Exp. 2019, 143, e58820. [Google Scholar] [CrossRef] [PubMed]
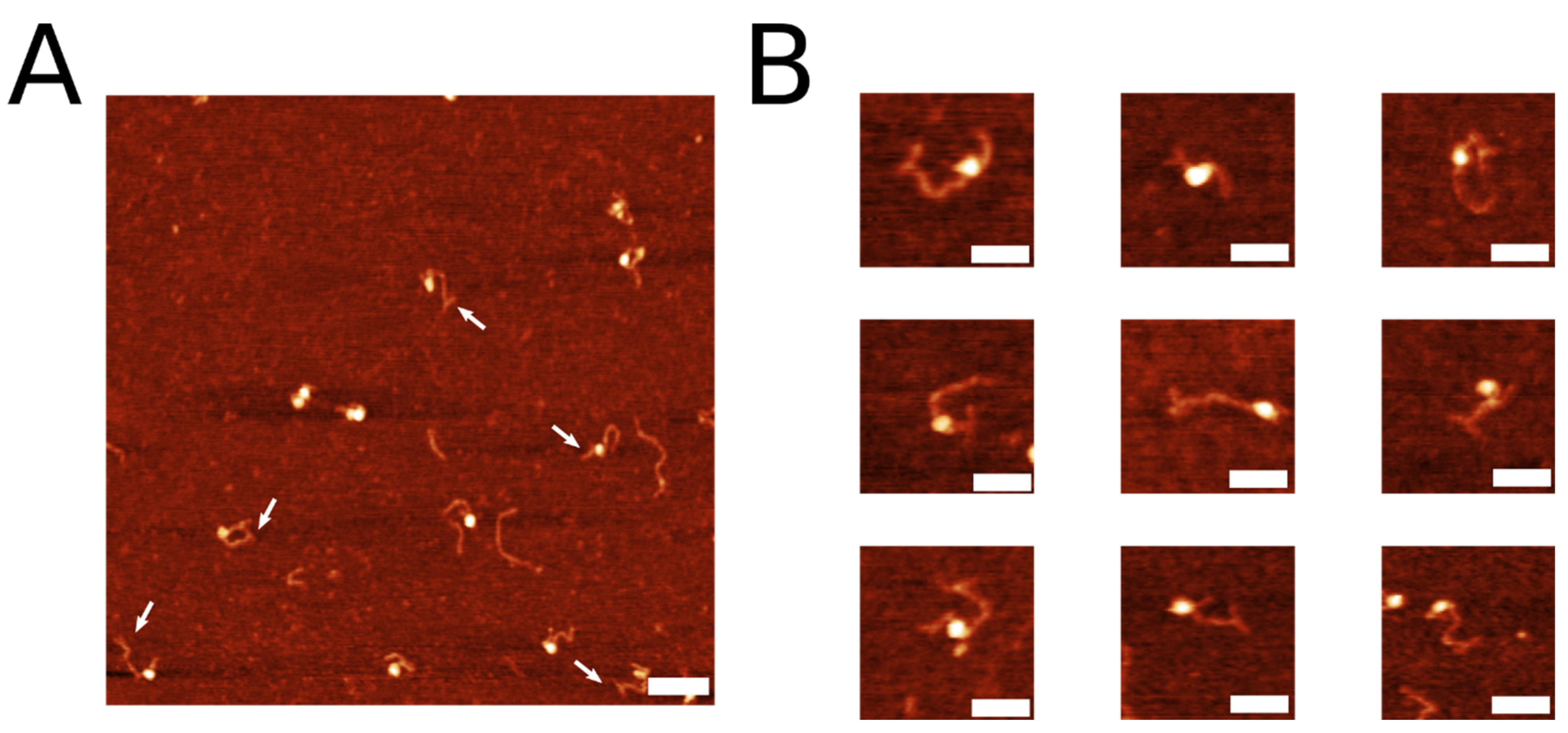

| Label Conformation | Frequency |
|---|---|
| “T” Label | 52.6% (n = 72) |
| “Y” Label | 39.4% (n = 54) |
| “Bulge” Label | 8.0% (n = 11) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Stormberg, T.; Filliaux, S.; Lyubchenko, Y.L. Three-Way DNA Junction as an End Label for DNA in Atomic Force Microscopy Studies. Int. J. Mol. Sci. 2022, 23, 11404. https://doi.org/10.3390/ijms231911404
Sun Z, Stormberg T, Filliaux S, Lyubchenko YL. Three-Way DNA Junction as an End Label for DNA in Atomic Force Microscopy Studies. International Journal of Molecular Sciences. 2022; 23(19):11404. https://doi.org/10.3390/ijms231911404
Chicago/Turabian StyleSun, Zhiqiang, Tommy Stormberg, Shaun Filliaux, and Yuri L. Lyubchenko. 2022. "Three-Way DNA Junction as an End Label for DNA in Atomic Force Microscopy Studies" International Journal of Molecular Sciences 23, no. 19: 11404. https://doi.org/10.3390/ijms231911404
APA StyleSun, Z., Stormberg, T., Filliaux, S., & Lyubchenko, Y. L. (2022). Three-Way DNA Junction as an End Label for DNA in Atomic Force Microscopy Studies. International Journal of Molecular Sciences, 23(19), 11404. https://doi.org/10.3390/ijms231911404







