Abstract
Assessing tumor EGFR mutation status is necessary for the proper management of patients with advanced non–small cell lung cancer (NSCLC). We evaluated the impact of dynamic analyses of the plasma and tissue EGFR mutation using ultra-sensitive droplet digital PCR (ddPCR) assays to manage NSCLC patients treated with EGFR tyrosine kinase inhibitors (EGFR-TKIs). Paired tumor tissues and plasma samples from 137 EGFR-mutated lung adenocarcinoma patients prior to the first-line EGFR-TKIs treatment (at baseline) and at disease progression were subjected to EGFR mutation analysis using ddPCR, together with the analyses of the clinicopathological characteristics and treatment outcomes. Patients with EGFR-activating mutations detected in baseline plasma were associated with bone metastasis (p = 0.002) and had shorter progression-free survival (12.9 vs. 17.7 months, p = 0.02) and overall survival (24.0 vs. 39.4 months, p = 0.02) compared to those without. Pre-treatment EGFR T790M mutation found in baseline tumor tissues of 28 patients (20.4%; 28/137) was significantly associated with brain metastasis (p = 0.005) and a shorter brain metastasis-free survival (p = 0.001). The presence of EGFR T790M mutations in baseline tumor tissues did not correlate with the emergence of acquired EGFR T790M mutations detected at progression. At disease progression, acquired EGFR T790M mutations were detected in 26.6% (21/79) of the plasma samples and 42.9% (15/35) of the rebiopsy tissues, with a concordance rate of 71.4% (25/35). The dynamic monitoring of tissue and plasma EGFR mutation status at baseline and progression using ddPCR has a clinical impact on the evaluation of EGFR-TKIs treatment efficacy and patient outcomes, as well as the emergence of resistance in NSCLC.
1. Introduction
Lung cancer is the leading cause of cancer-related death around the world. Nearly 85% of lung cancer patients have non–small cell lung cancer (NSCLC), with an overall 5-year survival rate of less than 15% [1,2]. Epidermal growth factor receptor (EGFR) mutations are the most frequent oncogenic driver mutations found in NSCLC. Mutations of the EGFR gene that occur in exons 18 to 21 within the kinase domain, leading to the activation of the kinase activity by a ligand-independent mechanism, are referred to as “activating mutations”. EGFR-activating mutations are highly associated with tumor responses to EGFR-TKIs. EGFR exon 19 deletions and L858R mutation are the two most common activating mutations in NSCLC [3]. More than 50% of Asian NSCLC patients were found to harbor EGFR-activating mutations, which are more prevalent in female and never-smokers [4]. EGFR tyrosine kinase inhibitors (EGFR-TKIs) treatment, demonstrating superior progression-free survival, higher tumor response rates and a better quality of life, are now recommended as the primary therapy for advanced-stage NSCLC patients with EGFR-activating mutations [5,6,7,8,9]. However, most patients who initially respond to first- or second-generation EGFR-TKIs will eventually undergo disease progression after a median of 9 to 14 months due to acquired resistance [6,7,8,10,11]. Approximately 50 to 60% of cases of acquired resistance for first- and second-generation EGFR-TKIs are due to the emergence of secondary EGFR T790M mutation that affects the “gatekeeper residue” in the catalytic domain of the kinase, leading to steric hindrance of EGFR-TKI binding due to the presence of the bulkier methionine side chain in the ATP-kinase-binding pocket and subsequently abolishing the potency of ATP-competitive TKIs [3,12]. Recently, the third-generation EGFR-TKI, osimertinib, targeting both EGFR-activating mutations as well as the gatekeeper EGFR T790M mutation, has been approved as a first-line treatment for patients with EGFR-mutant metastatic NSCLC and as a second-line therapy for patients who have developed EGFR T790M mutation after failure of the first- and/or second-generation EGFR-TKI treatment [13,14]. Therefore, the continuous assessment of tumor EGFR mutation status during the course of disease is necessary for the better management of NSCLC patients, particularly for the early identification of the resistance mechanisms.
Currently, the gold standard for assessing EGFR mutations is to analyze the tumor tissues. However, obtaining tumor samples through tumor biopsies has certain limitations [15,16]. First, tumor biopsy is an invasive and potentially risky procedure. Second, a small amount of tumor tissue obtained from a single-site biopsy has a high failure rate in molecular testing and is further affected by intra-tumoral heterogeneity and sampling bias [17]. Recently, testing plasma circulating cell-free DNA (cfDNA), commonly known as liquid biopsy, has been highlighted as a promising alternative. Unlike traditional tumor biopsy, liquid biopsy is less invasive and allows for frequent sampling during follow-up. Liquid biopsy contains tumor DNA released from both the primary and/or metastatic tumor sites, which can better reflect the molecular heterogeneity of tumors [18,19].
Given that the tumor tissue is extremely heterogeneous, and the concentration of tumor DNA in peripheral blood is extremely low and varies between patients, molecular methods with a high detection sensitivity are required for more accurate diagnostics. The cobas EGFR Mutation Test is a real-time polymerase chain reaction (PCR) test that can qualitatively identify 42 mutations in exons 18, 19, 20 and 21 of the EGFR gene, including the T790M resistance mutation, by using allele-specific PCR primers specifically amplifying the targeted mutant sequences rather than wild-type sequences and/or other human genomic DNA. It has been clinically validated as a companion diagnostic (CDx) for EGFR-TKIs therapy in patients with advanced NSCLC with a detection sensitivity of around 5% for tissue-derived DNA [20]. Droplet digital PCR (ddPCR) is an ultra-sensitive assay combining microfluidics technology with TaqMan-based quantitative PCR (qPCR) to measure absolute mutation alleles through clonal amplification and fluorescence detection of tens of thousands of individual template molecules in a single reaction. Unlike classic qPCR, which depends on calibration curves for target sequence quantification, ddPCR collects fluorescence signals via end-point measurements and utilizes Poisson statistics to calculate the target concentrations, which could avoid the pitfalls associated with the variations in reaction efficiencies. Therefore, ddPCR has emerged as a promising tool for the detection and absolute quantification of gene mutations below 1% [21,22,23,24]. As ddPCR is increasingly implemented in clinical practice for detecting somatic mutations that are present at low frequencies in tumors or circulating cell-free DNA, the current study aimed to evaluate the clinical utility of assessing both plasma and tissue EGFR mutations using ultra-sensitive ddPCR assays prior to the treatment and at disease progression in monitoring the efficacies and outcomes of NSCLC patients treated with EGFR-TKIs.
2. Results
2.1. Patient Characteristics
A total of 137 lung adenocarcinoma patients with EGFR-activating mutations identified from baseline tumor biopsies using a routine cobas EGFR Mutation Test were enrolled in this study. The majority of patients (98.5%; 135/137) presented at stage IV, and the rest were at stage IIIB. There were 76 (55.5%; 76/137) females and 61 (44.5%; 61/137) males, with a median age of 64.3 years. Among these patients, 101 (73.7%; 101/137) were never smokers, whereas all others were smokers. According to the baseline tissue testing results, 77 (56.2%; 77/137) patients had L858R mutations, and 60 (43.8%; 60/137) had exon 19 deletions. All these patients received first- or second-generation EGFR-TKIs as their first-line therapy; 74 (54.0%; 74/137) were treated with afatinib, 41 (29.9%; 41/137) were treated with erlotinib and 22 (16.1%; 22/137) were treated with gefitinib. Gefitinib and erlotinib are the first-generation EGFR-TKIs that reversibly bind to EGFR and inhibit EGFR signaling, while afatinib is a second-generation EGFR-TKI that irreversibly blocks signaling from all members of the ErbB receptor family [25]. For distant metastasis, there were 56 (40.9%; 56/137) patients with brain metastases, 71 (51.8%; 71/137) with bone metastases, 34 (24.8%; 34/137) with pleura metastases and 13 (9.5%; 13/137) with liver metastases. Brain metastases were presented in 37 (66.1%; 37/56) of the cases at the time of accrual, bone metastases were present in 45 (63.4%; 45/71), pleura metastases were present in 28 (82.4%; 28/34) and liver metastases were present in 7 (53.8%; 7/13).
2.2. Detection of Tissue and Plasma EGFR Mutations at Baseline Using the ddPCR Platform
The baseline tissue and plasma samples were subjected to EGFR mutation analysis using the ddPCR platform. The baseline tissue analysis revealed that 61 (44.5%; 61/137) patients were positive for L858R mutations, 16 (11.7%; 16/137) were positive for L858R combined with T790M mutations, 48 (35.0%; 48/137) were positive for exon 19 deletions and 12 (8.8%; 12/137) were positive for exon 19 deletions combined with T790M mutations. The concordance rate between the cobas EGFR Mutation Test and the ddPCR platform for baseline tissue testing was 79.6% (109/137) for overall EGFR mutations, including activating and T790M mutations, and it was 100% (137/137) for activating mutations only (Table 1). Remarkably, 20.4% (28/137) of the baseline cases were found to harbor pre-treatment EGFR T790M mutation by the ddPCR platform, but none of them were detected by the cobas EGFR Mutation Test. As determined by baseline plasma analysis using ddPCR, 89 (65.0%; 89/137) patients were positive for EGFR mutations, including 47 (34.3%; 47/137) positive for L858R, 2 (1.5%; 2/137) positive for L858R combined with T790M mutations, 38 (27.7%; 38/137) positive for exon 19 deletions and 2 (1.5%; 2/137) positive for exon 19 deletions combined with T790M mutations. A total of 2.9% (4/137) of patients had pre-treatment EGFR T790M mutation in the baseline plasma samples. The concordance rates between baseline tissue and plasma testing using the ddPCR platform were 51.8% (71/137) for overall EGFR mutations and 65.0% (89/137) for activating mutations only (Table 2).

Table 1.
Comparison of EGFR mutation status determined from baseline tumor tissues using the cobas EGFR Mutation Test and the ddPCR platform (N = 137).

Table 2.
Comparison of EGFR mutation status determined from baseline tumor tissues and plasma samples using the ddPCR platform (N = 137).
2.3. Clinical Characteristics of Patients with Pre-Treatment Tissue EGFR T790M and Plasma EGFR-Activating Mutations
To investigate the association between the clinical characteristics of patients and pre-treatment tissue EGFR T790M mutation or plasma EGFR-activating mutations, Chi-square or Fisher’s exact test was used for analyses, and the results are shown in Table 3. There were no statistical differences for age, gender, smoking history, first-line therapies, primary tumor sizes, TNM stages and EGFR mutation types between these two groups. However, we found that patients with pre-treatment EGFR T790M mutation were significantly associated with the development of brain metastasis (p = 0.005) but not bone, pleura or liver metastasis. To further evaluate if these clinical parameters are independently associated with the mutation status, a multivariate analysis was also performed (Supplemental Table S1). Kaplan–Meier survival analysis was used to estimate the progression-free survival (PFS), overall survival (OS) and brain metastasis-free survival (BMFS). The results showed that the PFS and OS were 13.8 months and 30.8 months, respectively, for patients with pre-treatment EGFR T790M mutation and 16.7 months and 27.8 months, respectively, for those without pre-treatment EGFR T790M mutation. No significant difference regarding patient survival was found between these two groups (p = 0.72 for PFS; p = 0.87 for OS) (Figure 1A,B).

Table 3.
Patient characteristics stratified by tissue EGFR T790M or plasma EGFR-activating mutation status at baseline (N = 137).
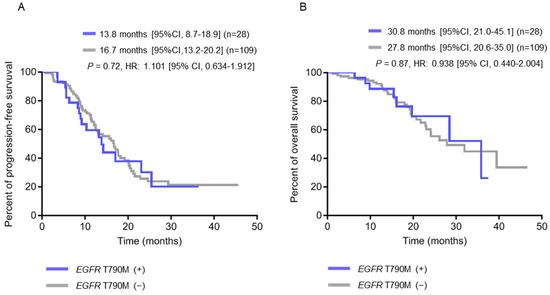
Figure 1.
Kaplan-Meier survival analysis according to pre-treatment EGFR T790M status in baseline tumor tissue (N = 137). (A) Progression-free survival (PFS) curves. (B) Overall survival (OS) curves.
Patients with pre-treatment EGFR T790M mutation had a significantly shorter BMFS compared to those without (p = 0.001) (Figure 2A). When the patients were further stratified into subgroups according to their EGFR-activating mutations, those with exon 19 deletions combined with T790M mutations had the shortest BMFS, and those with L858R mutations only had the longest BMFS (Figure 2B). On the other hand, patients with EGFR-activating mutations detected in their baseline plasma were significantly associated with larger primary tumor sizes (p = 0.004), higher node stages (p = 0.04), and the development of bone metastasis (p = 0.002) (Table 3 and Supplemental Table S1), along with shorter PFS (12.9 months vs. 17.7 months, p = 0.02) (Figure 3A) and OS (24.0 months vs. 39.4 months, p = 0.02) (Figure 3B), compared to those without EGFR-activating mutations detected.
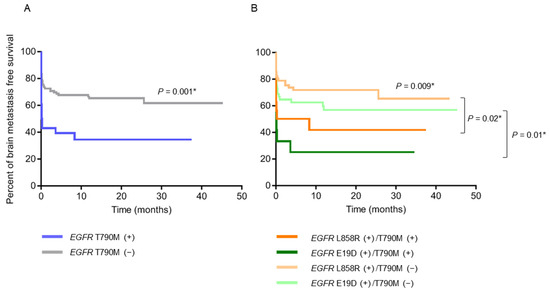
Figure 2.
Brain metastasis-free survival (BMFS) analysis (N = 137). (A) BMFS curves of patients with pre-treatment EGFR T790M (+) (N = 28) and (−) (N = 109). (B) BMFS curves of patients with pre-treatment EGFR L858R (+)/T790M (+) (N = 16), L858R (+)/T790M (−) (N = 61), E19D (+)/T790M (+) (N = 12) and E19D (+)/T790M (−) (N = 48) in baseline tumor tissue. * p < 0.05, indicating significance. Abbreviation: E19D, exon 19 deletions.
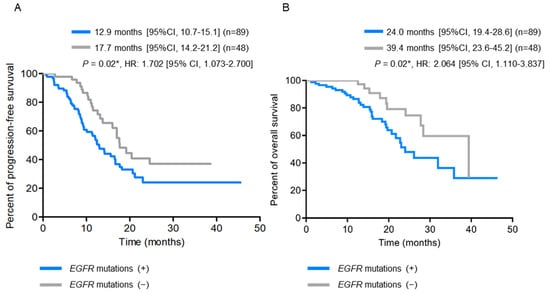
Figure 3.
Kaplan-Meier survival analysis according to baseline plasma EGFR-activating mutation status (N = 137). (A) Progression-free survival (PFS) curves. (B) Overall survival (OS) curves. * p < 0.05, indicating significance.
2.4. Detection of Tissue and Plasma EGFR Mutations at Disease Progression Using the ddPCR Platform
While all the 79 patients with tumor recurrence had plasma samples available for analysis, rebiopsy was successfully performed in only 35 patients. The plasma and tumor tissue samples at disease progression were subjected to EGFR mutation analysis using the ddPCR platform. As shown in Table 4, 57.0% (45/79) of the plasma samples were positive for EGFR mutations, including 17 (21.5%; 17/79) L858R mutations, 9 (11.4%; 9/79) L858R combined with T790M mutations, 7 (8.9%; 7/79) exon 19 deletions and 12 (15.2%; 12/79) exon 19 deletions combined with T790M mutations. In the tumor rebiopsy, 94.2% (33/35) were positive for EGFR mutations, including 8 (22.9%; 8/35) L858R mutations, 6 (17.1%; 6/35) L858R combined with T790M mutations, 10 (28.6%; 10/35) exon 19 deletions, 8 (22.9%; 8/35) exon 19 deletions combined with T790M mutations and 1 (2.9%; 1/35) T790M mutation only. Overall, acquired EGFR T790M mutation was found in 26.6% (21/79) of plasma samples and 42.9% (15/35) of rebiopsy samples. Among the 35 patients who had paired tissue and plasma results at disease progression, the concordance rate for EGFR T790M mutation was 71.4% (25/35), with 15.0% (3/20) of the patients showing negative in the tissue testing but positive in the plasma testing (Table 5). The FA of EGFR-activating and T790M mutations from plasma and tumor tissues at baseline and at disease progression is shown in Supplemental Figure S2. The FA of T790M detected from plasma or tumor tissues at disease progression was significantly higher than that at baseline; however, the FAs of L858R and exon 19 deletions in plasma or tumor tissues have no statistical difference between baseline and disease progression. Moreover, the presence of pre-treatment EGFR T790M mutation in the baseline tissue was not significantly associated with the presence of pre-treatment EGFR T790M mutation in the baseline plasma or the emergence of acquired EGFR T790M mutation in plasma or rebiopsy tumor tissues at disease progression (Table 6).

Table 4.
EGFR mutation status determined from the plasma and tumor rebiopsy samples using the ddPCR platform at disease progression.

Table 5.
Comparison of EGFR T790M mutation status determined from the plasma and tumor rebiopsy samples at disease progression (N = 35).

Table 6.
Summary of EGFR T790M mutation status in tissue and plasma tested at baseline and disease progression (N = 137).
3. Discussion
Accurately identifying tumors that harbor EGFR-activating as well as T790M resistance mutations is critical for the precision management of NSCLC [26]. In this study, through the analyses of EGFR mutation status using ddPCR performed in paired tumor and plasma samples obtained from 137 patients with advanced NSCLC prior to their EGFR-TKI treatment and at disease progression, we demonstrated that (1) baseline plasma EGFR-activating mutation status can be used as a predictive marker for EGFR-TKI therapy; (2) EGFR T790M mutation pre-existing as a minor subpopulation prior to EGFR-TKI treatment is not associated with the emergence of acquired resistance at disease progression; (3) Tissue testing for EGFR T790M mutation at disease progression cannot be used as a stand-alone assay, and the analysis of plasma should also be used to more precisely identify patients who might benefit from the subsequent third-generation EGFR-TKI treatment.
By using ddPCR, we found that plasma EGFR-activating mutations were detected in 65% (89/137) of patients prior to EGFR-TKI treatment. The positivity of EGFR-activating mutation in baseline plasma has been addressed in several previous studies, with a range from 20% to 73% [27]. The variability among studies may result from the differences in the assay sensitivity and disease stages of the enrolled patients. Our results showed that patients with detectable baseline plasma EGFR-activating mutations had a shorter PFS and OS compared to those without, suggesting that the absence of detectable EGFR-activating mutations in baseline plasma might be used as an indicator for low distant spreading activities and low systemic tumor burden in NSCLC. Our observation of an improved PFS and/or OS to EGFR-TKIs treatment in patients without EGFR-activating mutations detected in baseline plasma is also consistent with the previous findings [28,29,30].
Baseline plasma EGFR mutation testing is now recommended in the College of American Pathologists (CAP)/International Association for the Study of Lung Cancer (IASLC)/Association for Molecular Pathology (AMP) guidelines for the molecular testing of patients with NSCLC as an alternative for a diagnostic tissue biopsy in cases with insufficient tumor tissue specimens or where tissue specimens are not obtainable; however, its prognostic value for predicting EGFR-TKI outcomes, which has been demonstrated in previous studies, has not yet been applied to clinical practice [31]. One major obstacle for translating plasma EGFR mutation testing into routine clinical practice is the lack of standardization of methods for assessing tumor mutations. Therefore, the further evaluation of larger groups of NSCLC patients in prospective studies with respect to diagnosis and prognosis may be warranted for the wide implementation of liquid biopsy.
The seminal mechanism of acquired resistance to first- or second-generation EGFR-TKIs is known to be the emergence of EGFR T790M mutation. It is not clear whether EGFR T790M mutation in NSCLC patients who have relapsed from first- or second-generation EGFR-TKIs treatment is acquired during disease progression or develops from the pre-existing EGFR T790M clones in treatment-naïve patients as a minor population. The presence of pre-treatment EGFR T790M mutation in NSCLC has been discovered in several studies using highly sensitive detection methods, such as mass spectrometry, the scorpion amplification refractory mutation system, colony hybridization assays, etc.; the reported prevalence ranges from 20 to 80% [32,33,34,35].
In the present study, EGFR T790M mutation in 20.4% (28/137) of the EGFR-mutated, treatment-naïve NSCLC tumors was only detected by using the ddPCR platform but not the cobas EGFR mutation Test, and the FA of EGFR T790M is lower than that of EGFR-activating mutations in baseline tissue and plasma samples, showing that EGFR T790M pre-existing as a minor subpopulation in treatment-naïve, EGFR-mutated NSCLC has undergone clonal expansion in response to the selection pressure by the first- or second-generation EGFR-TKIs treatment. However, through analyses of EGFR mutations in plasma and tissue at baseline and disease progression, we found that the presence of EGFR T790M mutation at baseline was not statistically associated with the emergence of acquired EGFR T790M resistance at disease progression. One possible explanation is that the intratumoral heterogeneity of EGFR T790M mutation in tumor tissues results in sampling bias during the tumor biopsy procedures, which contributes to underestimating the incidence of this mutation and leads to the lack of statistical differences found in the present study. Indeed, a study evaluating EGFR T790M mutation in the sequential rebiopsies, along with the course of first-generation EGFR-TKI treatment, found that some patients who were EGFR T790M-positive at the first post-TKI biopsy became EGFR T790M-negative in later post-TKI rebiopsies, and vice versa, which is suggestive of the intratumoral heterogeneity of the mutation [36]. Moreover, the involvement of other molecular changes in the resistance mechanisms may also contribute to the results. In the present study, the concordance rate for EGFR T790M mutation was only 71.4% (25/35) between paired tissue and plasma samples at disease progression, with some being EGFR T790M-positive in the plasma and negative in the tissue, and vice versa. These findings indicate that tissue or plasma testing for EGFR T790M mutation should not be used as a stand-alone assay for selecting patients that are eligible for the subsequent osimertinib treatment, and the repeat or combined testing should be considered.
Our findings that pre-treatment EGFR T790M mutation was significantly associated with brain metastasis in patients with EGFR-mutated tumors receiving first- or second-generation EGFR-TKIs suggested that therapeutically targeting the pre-existing minor subpopulation harboring T790M mutation may have the advantage of preventing the development of brain metastasis in NSCLC. In the FLAURA trial, an analysis of a subset of treatment-naïve patients with EGFR-mutated advanced NSCLC and CNS metastases showed that the PFS was longer for patients receiving osimertinib compared to those receiving either gefitinib or erlotinib (15.2 versus 9.6 months; HR 0.47, 95% CI 0.30–0.74), indicating that osimertinib has a higher CNS efficacy in patients with untreated EGFR-mutated NSCLC [14]. Ballard et al. has further validated the CNS activity of osimertinib in a mouse model [37]. The findings in our current study of an association between pre-treatment EGFR T790M and brain metastasis might also support the use of osimertinib as a front-line EGFR-TKI treatment in preventing CNS progression.
The prognostic value of pre-treatment EGFR T790M mutation in advanced NSCLC patients treated with TKIs remains inconclusive. Some studies showed that patients with detectable EGFR T790M mutation had a shorter PFS, but this had no impact on OS, while others found that the presence of pre-treatment EGFR T790M mutation indicated favorable outcomes in advanced NSCLC patients treated with EGFR-TKIs [32,33,34,35]. In our study, we found no significant difference in patient survival between those with and without pre-treatment EGFR T790M. Possible explanations for the discordance include the differences in assay methodologies, with different sensitivities used among studies, the source of tumor samples (e.g., surgical resection or biopsies) used for analysis and the first- or second-generation EGFR TKIs chosen for treatment. Noteworthily, most patients enrolled in other studies were treated with first-generation EGFR-TKIs, such as gefitinib or erlotinib, but, in our present study, approximately 50% of patients were treated with the second-generation EGFR-TKI afatinib as the first-line therapy. In the phase IIB LUX-Lung 7 trial comparing afatinib and gefitinib as the first-line treatment for EGFR-mutated NSCLC patients, the researchers demonstrated that afatinib significantly improved PFS and time-to-treatment failure compared with gefitinib [38]. In addition, afatinib has been shown to inhibit the growth of gefitinib-resistant lung cancer cells harboring low levels of EGFR T790M mutation, but not those with high levels of EGFR T790M mutation [25]. Indeed, in this study, we did find that the frequency of developing acquired EGFR T790M mutation in patients treated with gefitinib or erlotinib was higher than those treated with afatinib (32.6% vs. 19.4%). In addition, our results showed that patients with detectable baseline EGFR-activating mutations are significantly associated with bone metastasis. Previous studies have found that the incidence of EGFR-activating mutations in bone metastasis is higher than that in primary adenocarcinomas or metastases to other organs. A study by Furugaki et al. reported that the inhibition of EGFR signaling by erlotinib prevents the tumor-induced osteolytic invasion of NCI-H292 cell lines [39]. These findings suggest that the activation of EGFR signaling promotes the osteolytic invasion and metastasis of tumor cells and provides supportive evidence to our result that baseline plasma EGFR-activating mutation status might serve as a predictive marker for the development of bone metastasis in NSCLC.
There were limitations in our current study. First, it was a single-center study with a small sample size due to the difficulty in longitudinally collecting the tissue and plasma samples in advanced NSCLC patients prior to treatment and at disease progression, and further investigation in a larger cohort may be required to confirm the findings. Second, the assay sensitivity and specificity towards EGFR mutation testing using the ddPCR platform cannot be accurately evaluated in this study because of a lack of enrolled EGFR mutation-negative patients. Third, patients enrolled in this study were at advanced stages, and their tumor tissues were obtained via tumor biopsy. The intratumor heterogeneity of EGFR T790M mutation may lead to an underestimation of the prevalence due to sampling bias, which may, in turn, influence its clinical impact.
In conclusion, our study demonstrated the significance of using the ultra-sensitive ddPCR platform for the dynamic assessment of tissue and plasma EGFR mutation status at baseline and disease progression in EGFR mutant NSCLC treated with EGFR-TKIs. In addition to evaluating the EGFR-TKI treatment efficacy and the emergence of resistance mechanisms, the use of the ddPCR method for EGFR mutation detection is proven to be valuable in predicting tumor metastasis and patient outcomes.
4. Materials and Methods
4.1. Study Population
This was a single-center prospective observational study. Patients who were pathologically diagnosed with lung adenocarcinoma, were positive for EGFR-activating mutations determined by the routine cobas EGFR Mutation Test (v1 was used before 31 March 2017, and v2 was applied thereafter; Roche Molecular Systems, Inc.), had inoperable stage IIIB or IV diseases according to the 7th edition of the American Joint Committee for Cancer staging system [26] and planned to receive EGFR-TKIs as their first-line therapy with either gefitinib, erlotinib or afatinib were enrolled. Gefitinib, erlotinib and afatinib are the only first-line EGFR-TKIs that have been reimbursed by the Taiwan National Health Insurance scheme for advanced EGFR mutation-positive lung adenocarcinoma; therefore, patients with osimertinib as a first-line treatment were not available in this study. Patients were excluded if they had another active malignancy or any prior treatment that could influence the tumor burden. All enrolled patients were subjected to blood collection according to standard procedures at baseline (within 7 days before the first dose of EGFR-TKI) and at the time of disease progression. Treatment response was evaluated every 3 months with computed tomography (CT) by specialized radiologists, according to the Response Evaluation Criteria in Solid Tumors version 1.1 [40]. From June 2015 to July 2018, a total of 137 lung adenocarcinoma patients at Taipei Veterans General Hospital (Taipei city, Taiwan) were enrolled in this study (Figure 4). The prior-to-treatment (at baseline) and rebiopsy (at disease progression) tumor tissues and plasma samples were analyzed for EGFR mutation status using ddPCR. Clinical characteristics, including the patients’ age, gender, smoking history, distant organ metastasis, first-line EGFR-TKIs, EGFR mutation status, date of initial diagnosis, treatment start date, time to disease progression, date of death or last follow-up, etc., were collected. Never-smokers were defined as patients who had never smoked cigarettes, whereas smokers were defined as those who were current or former smokers. The study protocol was approved by the institutional review board (IRB) of Taipei Veterans General Hospital.
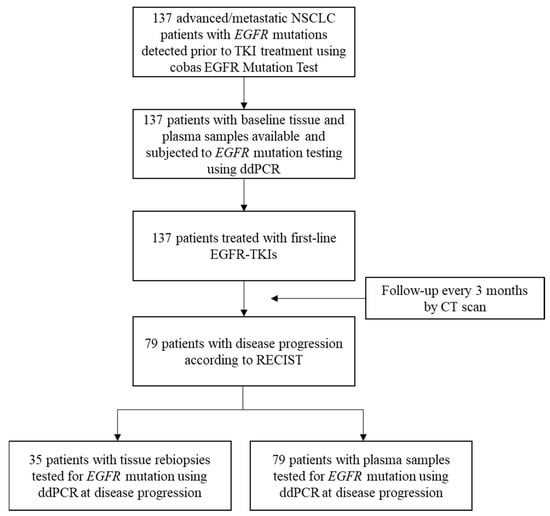
Figure 4.
Flow diagram of the study population. NSCLC, non-small cell lung cancer; ddPCR, droplet digital polymerase chain reaction; CT, computed tomography.
4.2. DNA Isolation from Plasma and Formalin-Fixed Paraffin-Embedded Tissues
For cfDNA isolation, 10 mL of peripheral blood was collected in a K2 EDTA tube and was processed within 4 h. The tubes were centrifuged at 1600× g for 15 min, and cfDNA was isolated using the cobas cfDNA Sample Preparation Kit (Roche Molecular Systems, Inc., Pleasanton, CA, USA) according to the manufacturer’s protocol. For the genomic DNA isolation from tumor tissues, formalin-fixed paraffin-embedded (FFPE) tissues were stained with Hematoxylin & Eosin (H&E) and reviewed by pathologists to select the tumor areas. The selected areas were then manually macrodissected from the corresponding ones in the consecutive tissue sections and, after deparaffinization, were subjected to genomic DNA extraction using the cobas DNA Sample Preparation Kit (Roche Molecular Systems, Inc., Pleasanton, CA, USA) according to the manufacturer’s instructions.
4.3. EGFR Mutation Analysis Using Droplet Digital PCR
This assay was performed on a QX200 Droplet Digital PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), in which a PCR sample is partitioned into approximately 20,000 droplets. The mean of the accepted droplets in the samples of this study was 14,725 with a standard deviation of 1609 droplets. After amplification, each droplet is scored as positive or negative via fluorescence detection of the target sequence. Poisson statistical analysis is used for the absolute quantification of the target sequence. For EGFR mutation analysis, the ddPCR Mutation Detection Assay Kits for the analysis of EGFR T790M Mutation and EGFR L858R Mutation and the ddPCR EGFR Exon 19 Deletions Screening Kit purchased from Bio-Rad Laboratories, Inc., (Hercules, CA, USA) were applied according to the manufacturer’s instructions. The emulsified samples were then transferred to 96-well plates for the PCR reaction. The PCR condition used was as follows: initial enzyme activation for 10 min at 95 °C, 40 cycles of 30 sec denaturation at 94 °C, annealing/extension for 1 min at 55 °C and then enzyme deactivation at 98 °C for 10 min. Subsequently, the PCR products were loaded into the QX200 droplet reader for analysis using QuantaSoft Software version 1.7.4.0917 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). A series of experimental controls were also performed in each assay, including a no template control (NTC) for monitoring environmental contamination, a negative control (the EGFR wild-type reference standard purchased from Horizon Discovery Group PLC, Cambridge, UK) for the evaluation of the false positive rates and a positive control (the EGFR mutation-specific reference standard purchased from Horizon Discovery Group PLC, Cambridge, UK) for the confirmation of the assay performance and the determination of the threshold value. In ddPCR analysis, the limit of blank (LOB) is defined as a finite number of false events of mutant droplets detected in each assay [41]. By evaluating a total of 20 normal samples, including 10 non-tumor lung tissue and 10 normal plasma samples, the LOB was determined by fitting a Poisson model to the false positive frequency distribution for each target and evaluating the 95% one-tailed upper limit of the model distribution. The number of false positive droplet events ranged from 0 to 2 for T790M and was 0 for L858R or exon 19 deletions. Therefore, the positive threshold for each target was set to at least three positive droplets detected in the assay. According to the instructions for the ddPCR Mutation Detection Assay Kits and ddPCR EGFR Exon 19 Deletions Screening Kit provided by the manufacturer (Bio-Rad Laboratories, Inc., Hercules, CA, USA), the limit of detection (LoD) for L858R and T790M was claimed to be 0.1%, and that for exon 19 deletions was 0.5%. To further verify the LoD, EGFR mutation-specific reference standards (Horizon Discovery Group PLC, Cambridge, UK) were serially diluted with the EGFR wild-type reference standard (Horizon Discovery Group PLC, Cambridge, UK) to the specified variant allele frequencies (VAFs) from 50% to 0.1%. The input DNA with varying proportions of mutant DNA mixed into wild-type DNA was subjected to ddPCR analysis. All reactions were repeated three times at each VAF. The minimum VAF that can be reliably detected (coefficient of variation (CV) < 30%) was 0.1% for L858R, exon 19 deletions and T790M (Supplemental Figure S1). Therefore, in this study, the LoD for L858R and T790M was defined as 0.1%, and for exon 19 deletions, it was 0.5%. Samples with at least three positive droplets detected and with a fractional abundance (FA; %) ≥ LoD were considered as mutation-positive cases. All specimens with pre-treatment EGFR T790M detected were independently re-tested to confirm the obtained positive results.
4.4. ddPCR Data Analysis
The ddPCR data were analyzed using QuantaSoft Software version 1.7.4.0917 (Bio-Rad Laboratories, Inc., Hercules, CA, USA), by which the numbers of positive and negative droplets for each fluorophore in each sample were measured. It then fits the fraction of positive droplets to a Poisson algorithm to determine the starting concentration of the target DNA molecule in units of copies/µL input. The absolute quantification mode used for the concentration calculation of target molecules is based on the following formula:
Concentration (copies/µL) = −ln (N–neg/N–total)/V–droplet
[N–neg = the number of total negative droplets in a well; N–total = the number of total droplets in a well; V–droplet = the volume of a droplet]
Then, Fractional Abundance (FA), referring to the proportion of mutant allele frequencies by QuantaSoft, is auto-calculated by the software with the formula:
FA = Concentration of mutant allele/(Concentration of mutant allele + Concentration of wild-type allele)
4.5. Statistical Analysis
The concordance rates between the tissue and the plasma and between the different assay platforms were calculated as the number of positive and negative samples out of the total number of tested samples. The Kaplan–Meier method was used to analyze the progression-free survival (PFS), overall survival (OS) and brain metastasis-free survival (BMFS), and the log-rank test was used to compare the survival times between groups and to calculate the hazard ratio with a 95% confidence interval. The Chi-square test or Fisher’s exact test was used to analyze the associations between the EGFR mutation status and clinical characteristics of patients. The association between clinical factors and the pre-treatment EGFR T790M mutation or plasma EGFR-activating mutations was also examined using multivariate regression analyses. A p value < 0.05 was considered to indicate a statistically significant difference. All statistical analyses were conducted using SAS Statistics v.6.1 and GraphPad Prism v.8.0.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911353/s1.
Author Contributions
Conceptualization, H.-L.H., C.-M.T., C.-H.C. and T.-Y.C.; Data curation, H.-L.H., F.-Y.W. and C.-L.C.; Formal analysis, H.-L.H., F.-Y.W., C.-L.C. and C.-H.C.; Funding acquisition, H.-L.H. and T.-Y.C.; Investigation, H.-L.H., C.-M.T. and T.-Y.C.; Methodology, H.-L.H. and T.-Y.C.; Resources, F.-Y.W. and C.-L.C.; Software, F.-Y.W. and C.-L.C.; Supervision, H.-L.H., C.-M.T., C.-H.C. and T.-Y.C.; Validation, H.-L.H. and T.-Y.C.; Writing—original draft, H.-L.H. and F.-Y.W.; Writing—review & editing, H.-L.H. and T.-Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants V110C-202, V110E-001-2, V111C-053, V111C-205 and V111E-004-2 from Taipei Veterans General Hospital and grant MOST 109-2320-B-075-007-MY3 from the Ministry of Science and Technology, Taiwan.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Taipei Veterans General Hospital, Taipei, Taiwan (IRB number: 2015-03-006A/March 2015, 2019-01-002BC/January 2019, 2020-07-022AC/July 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Au, J.S.; Thongprasert, S.; Srinivasan, S.; Tsai, C.M.; Khoa, M.T.; Heeroma, K.; Itoh, Y.; Cornelio, G.; Yang, P.C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J. Thorac. Oncol. 2014, 9, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Mitsudomi, T.; Morita, S.; Yatabe, Y.; Negoro, S.; Okamoto, I.; Tsurutani, J.; Seto, T.; Satouchi, M.; Tada, H.; Hirashima, T.; et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010, 11, 121–128. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Y.L.; Chen, G.; Feng, J.; Liu, X.Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef]
- Fukuoka, M.; Wu, Y.L.; Thongprasert, S.; Sunpaweravong, P.; Leong, S.S.; Sriuranpong, V.; Chao, T.Y.; Nakagawa, K.; Chu, D.T.; Saijo, N.; et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J. Clin. Oncol. 2011, 29, 2866–2874. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Sequist, L.V.; Yang, J.C.; Yamamoto, N.; O’Byrne, K.; Hirsh, V.; Mok, T.; Geater, S.L.; Orlov, S.; Tsai, C.M.; Boyer, M.; et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 2013, 31, 3327–3334. [Google Scholar] [CrossRef] [PubMed]
- Suda, K.; Onozato, R.; Yatabe, Y.; Mitsudomi, T. EGFR T790M mutation: A double role in lung cancer cell survival? J. Thorac. Oncol. 2009, 4, 1–4. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.L.; Ahn, M.J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.; et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Pirker, R.; Herth, F.J.; Kerr, K.M.; Filipits, M.; Taron, M.; Gandara, D.; Hirsch, F.R.; Grunenwald, D.; Popper, H.; Smit, E.; et al. Consensus for EGFR mutation testing in non-small cell lung cancer: Results from a European workshop. J. Thorac. Oncol. 2010, 5, 1706–1713. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Karachaliou, N. Implications of Blood-Based T790M Genotyping and Beyond in Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3361–3362. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.M.; de Petris, G.; Lopez, J.I. Detection of Intratumor Heterogeneity in Modern Pathology: A Multisite Tumor Sampling Perspective. Front. Med. 2017, 4, 25. [Google Scholar] [CrossRef]
- Esposito, A.; Criscitiello, C.; Locatelli, M.; Milano, M.; Curigliano, G. Liquid biopsies for solid tumors: Understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol. Ther. 2016, 157, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.K.; Pandey, T.; Dey, P. Liquid biopsy: A new avenue in pathology. Cytopathology 2019, 30, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Sirera, R.; Jantus-Lewintre, E.; Reclusa, P.; Calabuig-Farinas, S.; Blasco, A.; Pisapia, P.; Rolfo, C.; Camps, C. Profile of the Roche cobas(R) EGFR mutation test v2 for non-small cell lung cancer. Expert Rev. Mol. Diagn. 2017, 17, 209–215. [Google Scholar] [CrossRef]
- Pekin, D.; Skhiri, Y.; Baret, J.C.; Le Corre, D.; Mazutis, L.; Salem, C.B.; Millot, F.; El Harrak, A.; Hutchison, J.B.; Larson, J.W.; et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 2011, 11, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.; Lu, H.; Garlan, F.; Taly, V. Droplet-Based Digital PCR: Application in Cancer Research. Adv. Clin. Chem. 2017, 79, 43–91. [Google Scholar] [PubMed]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Yoon, B.W.; Kim, J.H.; Lee, S.H.; Choi, C.M.; Rho, J.K.; Yoon, S.; Lee, D.H.; Kim, S.W.; Jang, T.W.; Lee, J.C. Comparison of T790M Acquisition Between Patients Treated with Afatinib and Gefitinib as First-Line Therapy: Retrospective Propensity Score Matching Analysis. Transl. Oncol. 2019, 12, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular Testing Guideline for the Selection of Patients with Lung Cancer for Treatment with Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [PubMed]
- Kim, H.R.; Lee, S.Y.; Hyun, D.S.; Lee, M.K.; Lee, H.K.; Choi, C.M.; Yang, S.H.; Kim, Y.C.; Lee, Y.C.; Kim, S.Y.; et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J. Exp. Clin. Cancer Res. 2013, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, T.; Saito, H.; Furuya, N.; Watanabe, K.; Sugawara, S.; Iwasawa, S.; Tsunezuka, Y.; Yamaguchi, O.; Okada, P.M.; Yoshimori, K.; et al. Evaluation of plasma EGFR mutation as an early predictor of response of erlotinib plus bevacizumab treatment in the NEJ026 study. EBioMedicine 2020, 57, 102861. [Google Scholar] [CrossRef]
- Wu, Y.L.; Lee, V.; Liam, C.K.; Lu, S.; Park, K.; Srimuninnimit, V.; Wang, J.; Zhou, C.; Appius, A.; Button, P.; et al. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer 2018, 126, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [PubMed]
- Fujita, Y.; Suda, K.; Kimura, H.; Matsumoto, K.; Arao, T.; Nagai, T.; Saijo, N.; Yatabe, Y.; Mitsudomi, T.; Nishio, K. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J. Thorac. Oncol. 2012, 7, 1640–1644. [Google Scholar] [CrossRef]
- Ma, C.; Wei, S.; Song, Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. J. Thorac. Dis. 2011, 3, 10–18. [Google Scholar] [PubMed]
- Su, K.Y.; Chen, H.Y.; Li, K.C.; Kuo, M.L.; Yang, J.C.; Chan, W.K.; Ho, B.C.; Chang, G.C.; Shih, J.Y.; Yu, S.L.; et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J. Clin. Oncol. 2012, 30, 433–440. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, J.; Yin, L.; Jiang, H.; Zhang, W.; Song, Y.; Liu, M. The prognostic role of pretreatment epidermal growth factor receptor T790M mutation in advanced non-small cell lung cancer patients treated with EGFR tyrosine kinase inhibitors. Oncotarget 2017, 8, 50941–50948. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, J.L.; Heideman, D.A.; Thunnissen, E.; Paul, M.A.; van Wijk, A.W.; Postmus, P.E.; Smit, E.F. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer 2014, 85, 19–24. [Google Scholar] [CrossRef]
- Ballard, P.; Yates, J.W.; Yang, Z.; Kim, D.W.; Yang, J.C.; Cantarini, M.; Pickup, K.; Jordan, A.; Hickey, M.; Grist, M.; et al. Preclinical Comparison of Osimertinib with Other EGFR-TKIs in EGFR-Mutant NSCLC Brain Metastases Models, and Early Evidence of Clinical Brain Metastases Activity. Clin. Cancer Res. 2016, 22, 5130–5140. [Google Scholar] [CrossRef]
- Park, K.; Tan, E.H.; O’Byrne, K.; Zhang, L.; Boyer, M.; Mok, T.; Hirsh, V.; Yang, J.C.; Lee, K.H.; Lu, S.; et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): A phase 2B, open-label, randomised controlled trial. Lancet. Oncol. 2016, 17, 577–589. [Google Scholar] [CrossRef]
- Furugaki, K.; Moriya, Y.; Iwai, T.; Yorozu, K.; Yanagisawa, M.; Kondoh, K.; Fujimoto-Ohuchi, K.; Mori, K. Erlotinib inhibits osteolytic bone invasion of human non-small-cell lung cancer cell line NCI-H292. Clin. Exp. Metastasis 2011, 28, 649–659. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 2013, 59, 1722–1731. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).