Germline Variants in 32 Cancer-Related Genes among 700 Chinese Breast Cancer Patients by Next-Generation Sequencing: A Clinic-Based, Observational Study
Abstract
1. Introduction
2. Results
2.1. Patients Characteristics
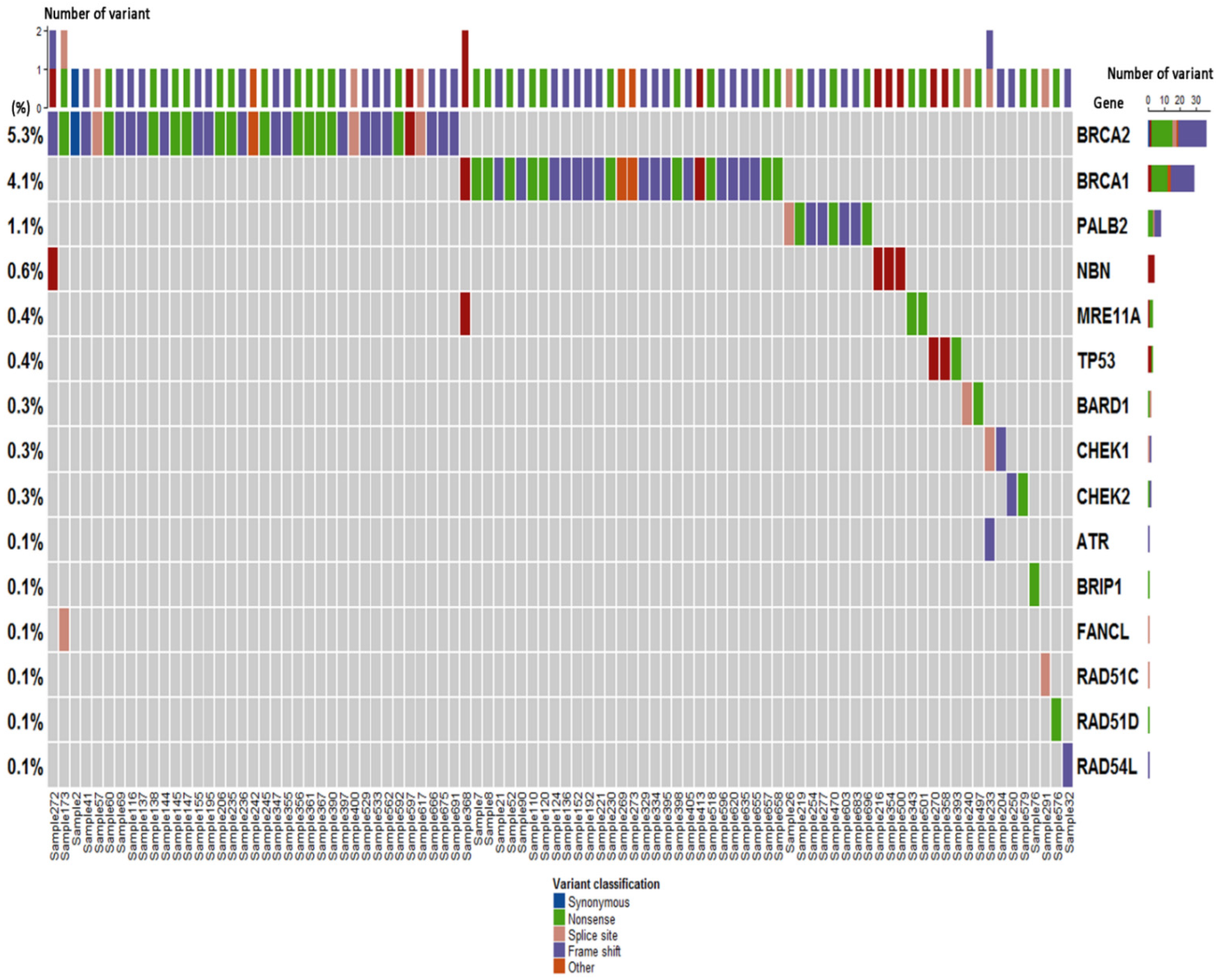
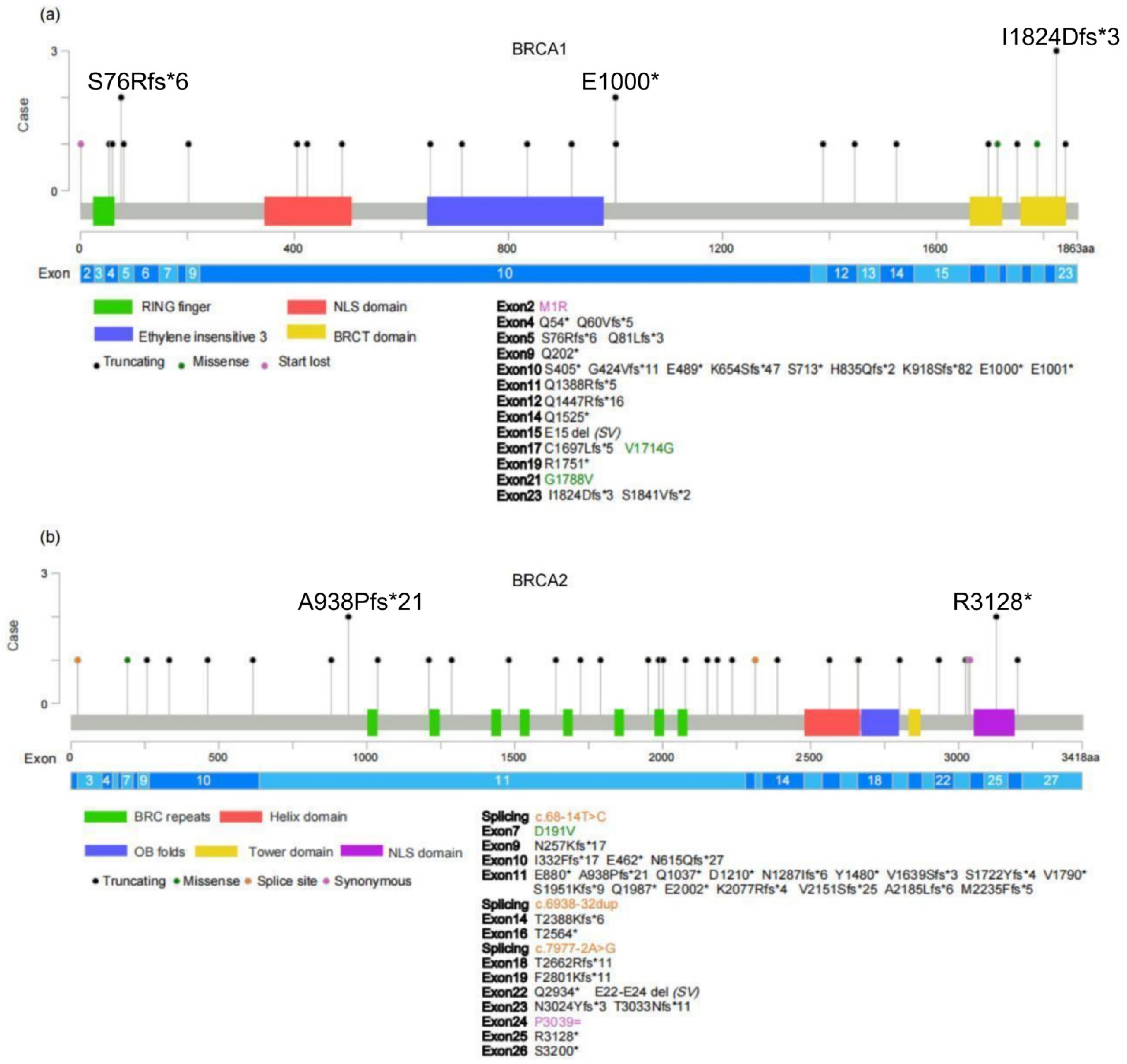
2.2. Germline Variant Spectrum
2.3. Association between Deleterious Germline Variants and Clinicopathological Characteristics
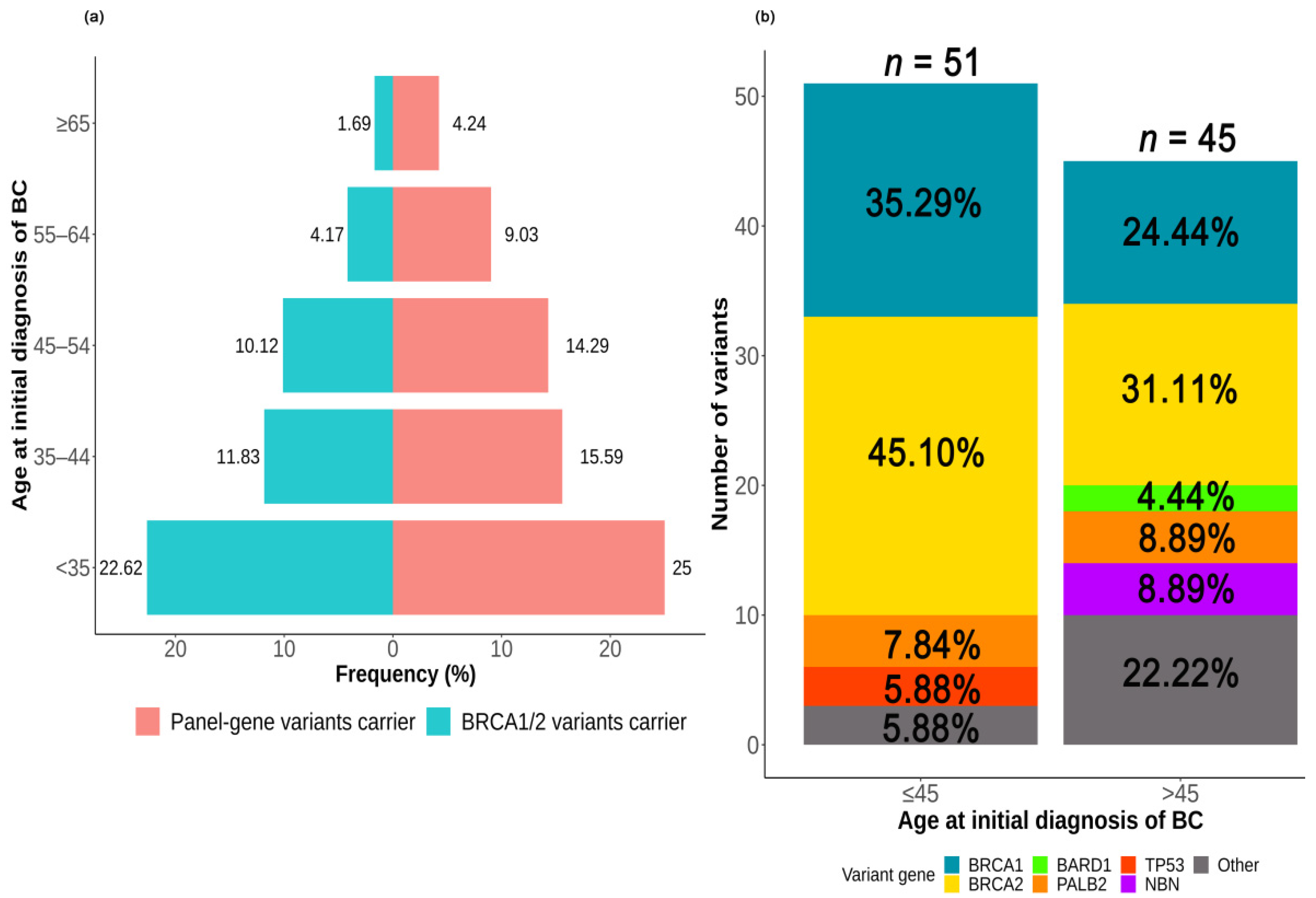
2.3.1. Association between Deleterious Germline Variants and Age at the Initial Diagnosis of BC
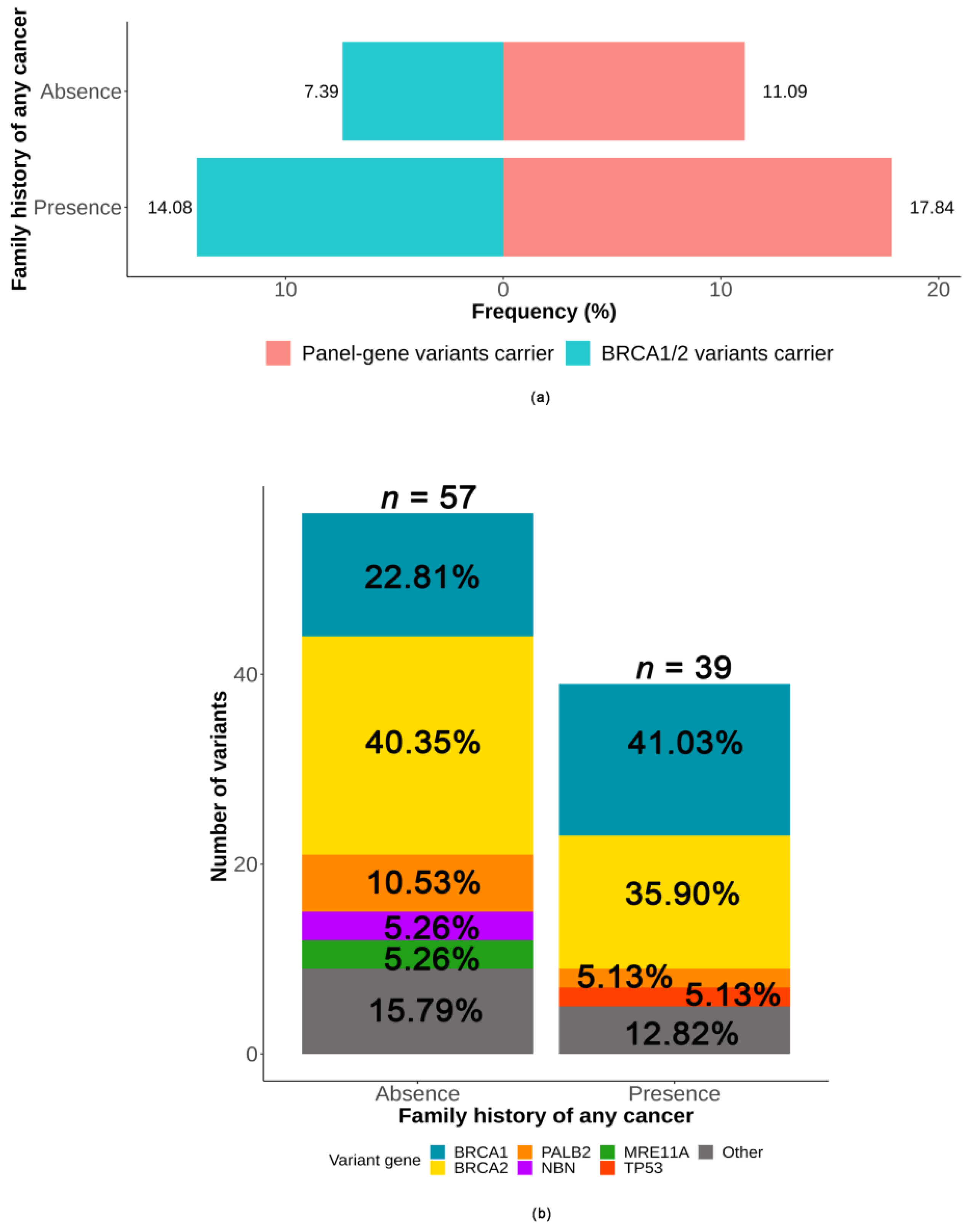
2.3.2. Association between Deleterious Germline Variants and Family History of Cancer
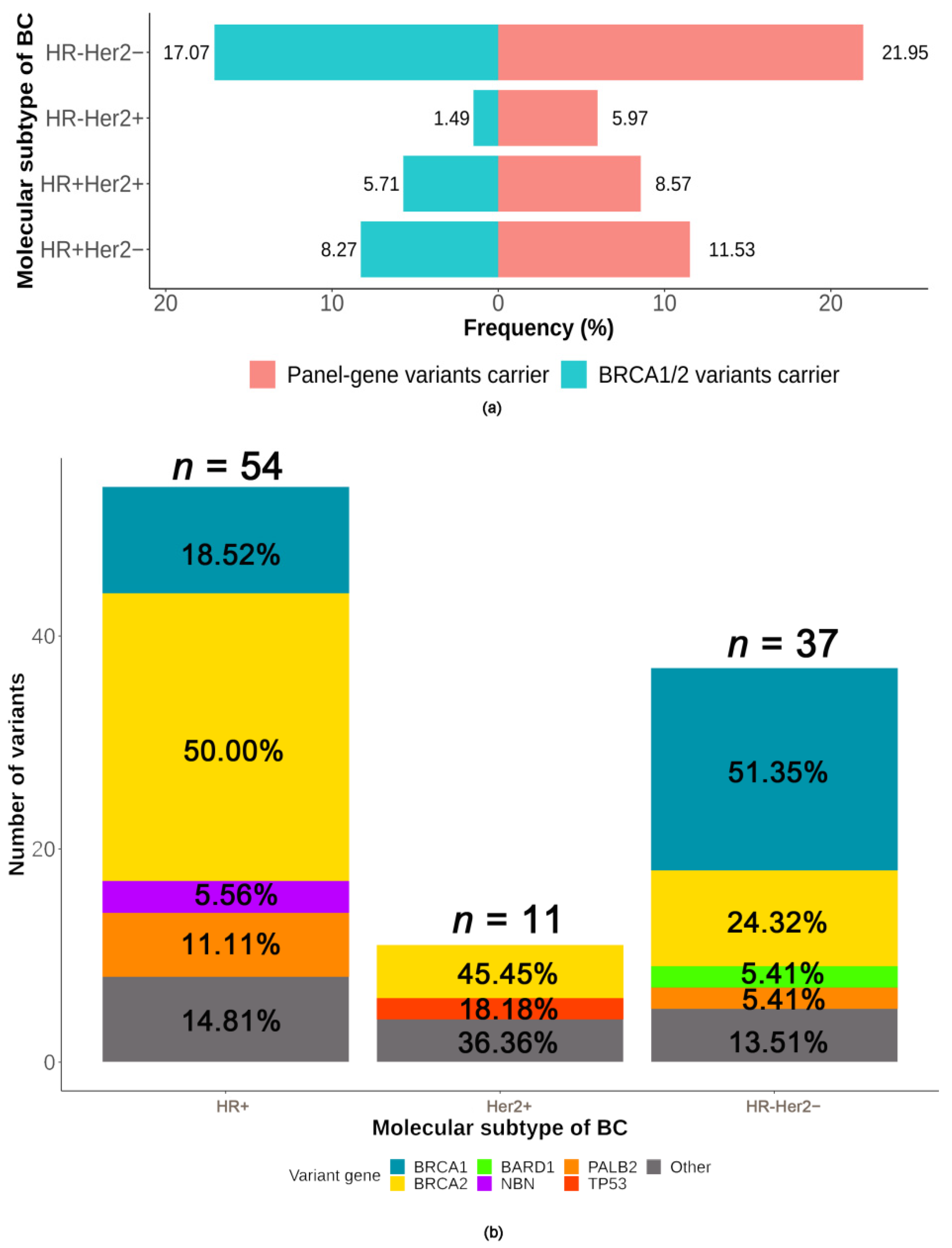
2.3.3. Association between Deleterious Germline Variants and Molecular Subtype of BC
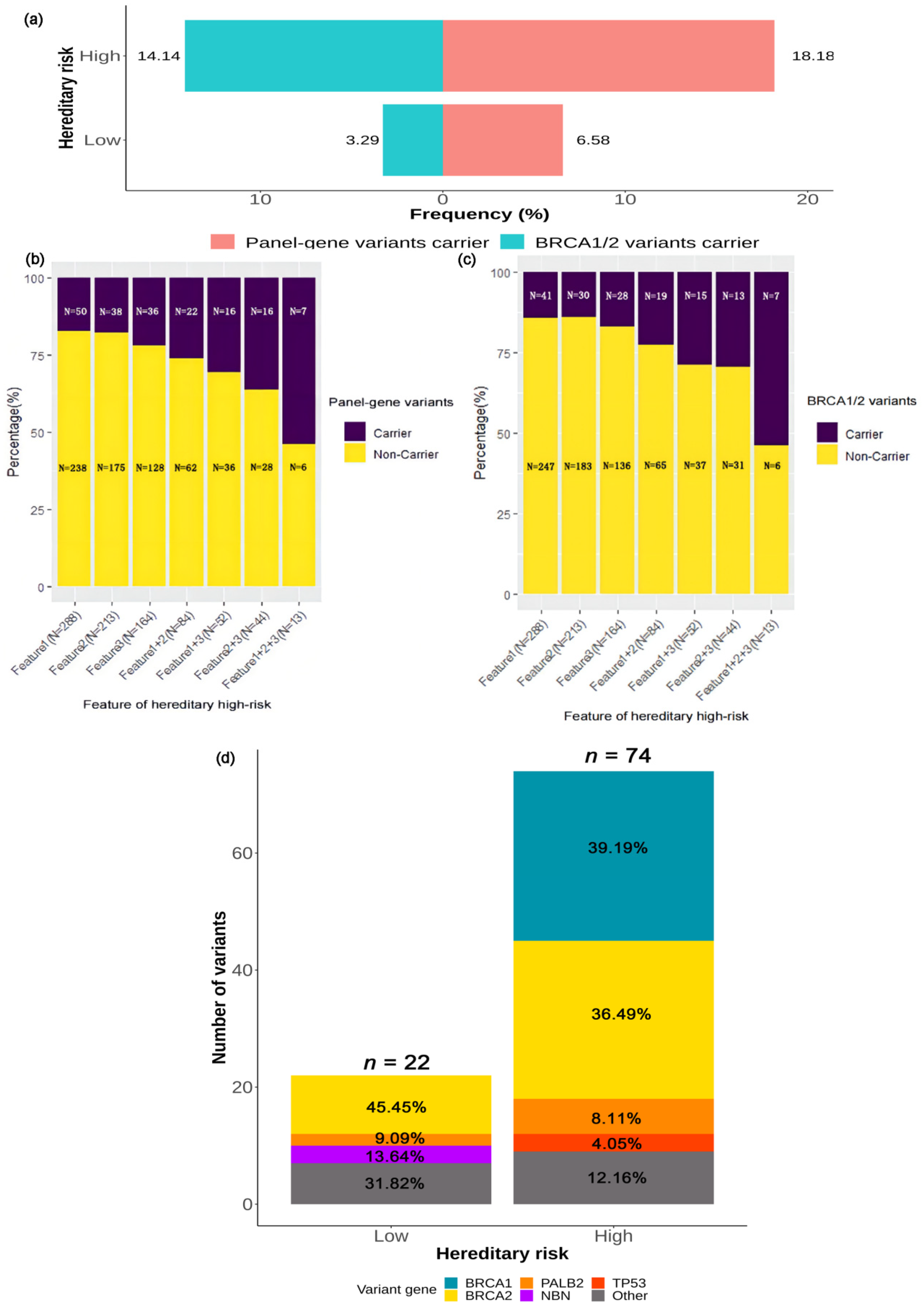
2.3.4. Association between Deleterious Germline Variants and Hereditary Risk
3. Discussion
4. Materials and Methods
4.1. Patients and Clinicopathological Factors
4.2. Hereditary High Risk Assessment
4.3. DNA Extraction
4.4. NGS Assay and Variant Classification
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Lin, N.U.; Kidd, J.; Allen, B.A.; Singh, N.; Wenstrup, R.J.; Hartman, A.R.; Winer, E.P.; Garber, J.E. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J. Clin. Oncol. 2016, 34, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Meng, H.; Yao, L.; Lv, M.; Bai, J.; Zhang, J.; Wang, L.; Ouyang, T.; Li, J.; Wang, T.; et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin. Cancer Res. 2017, 23, 6113–6119. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Jing, R.; Wei, H.; Wang, M.; Xiaowei, Q.; Liu, H.; Jian, L.; Ou, J.H.; Jiang, W.H.; Tian, F.G.; et al. Germline mutations in 40 cancer susceptibility genes among Chinese patients with high hereditary risk breast cancer. Int. J. Cancer 2019, 144, 281–289. [Google Scholar] [CrossRef]
- Breast Cancer Association Consortium; Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Luccarini, C.; Wahlström, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; et al. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Pharoah, P.D.; Antoniou, A.C.; Tischkowitz, M.; Tavtigian, S.V.; Nathanson, K.L.; Devilee, P.; Meindl, A.; Couch, F.J.; Southey, M.; et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015, 372, 2243–2257. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for metastatic breast cancer inpatients with a germline BRCA mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Tutt, A.N.J.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; de Azambuja, E.; Fielding, A.; Balmaña, J.; et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. N. Engl. J. Med. 2021, 384, 2394–2405. [Google Scholar] [CrossRef]
- Tutt, A.; Tovey, H.; Cheang, M.C.U.; Kernaghan, S.; Kilburn, L.; Gazinska, P.; Owen, J.; Abraham, J.; Barrett, S.; Barrett-Lee, P.; et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: The TNT Trial. Nat. Med. 2018, 24, 628–637. [Google Scholar] [CrossRef]
- Daly, M.B.; Pal, T.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Goggins, M.; Hutton, M.L.; et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2021, 19, 77–102. [Google Scholar] [CrossRef]
- Kurian, A.W.; Hare, E.E.; Mills, M.A.; Kingham, K.E.; McPherson, L.; Whittemore, A.S.; McGuire, V.; Ladabaum, U.; Kobayashi, Y.; Lincoln, S.E.; et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J. Clin. Oncol. 2014, 32, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Zhuo, Z.L.; Xie, F.; Wang, S.; Zhao, X.T. Four novel BRCA variants found in Chinese hereditary breast cancer patients by next-generation sequencing. Clin. Chim. Acta 2021, 516, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Yao, L.; Yuan, H.; Xu, Y.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; Lin, B.; et al. BRCA1 c.5470_5477del, a founder mutation in Chinese Han breast cancer patients. Int. J. Cancer 2020, 146, 3044–3052. [Google Scholar] [CrossRef]
- Li, J.; Han, S.; Zhang, C.; Luo, Y.; Wang, L.; Wang, P.; Wang, Y.; Xia, Q.; Wang, X.; Wei, B.; et al. Identification of BRCA1:c.5470_5477del as a Founder Mutation in Chinese Ovarian Cancer Patients. Front Oncol. 2021, 11, 655709. [Google Scholar] [CrossRef] [PubMed]
- Salazar, R.; Cruz-Hernandez, J.J.; Sanchez-Valdivieso, E.; Rodriguez, C.A.; Gomez-Bernal, A.; Barco, E.; Fonseca, E.; Portugal, T.; Gonzalez-Sarmiento, R. BRCA1-2 mutations in breast cancer: Identification of nine new variants of BRCA1-2 genes in a population from central Western Spain. Cancer Lett. 2006, 233, 172–177. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, G.; Li, X.; Ren, C.; Wang, Y.; Li, K.; Mok, H.; Cao, L.; Wen, L.; Jia, M.; et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging 2020, 12, 3140–3155. [Google Scholar] [CrossRef]
- Shimelis, H.; LaDuca, H.; Hu, C.; Hart, S.N.; Na, J.; Thomas, A.; Akinhanmi, M.; Moore, R.M.; Brauch, H.; Cox, A.; et al. Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J. Natl. Cancer Inst. 2018, 110, 855–862. [Google Scholar] [CrossRef]
- Lang, G.T.; Shi, J.X.; Hu, X.; Zhang, C.H.; Shan, L.; Song, C.G.; Zhuang, Z.G.; Cao, A.Y.; Ling, H.; Yu, K.D.; et al. The spectrum of BRCA mutations and characteristics of BRCA-associated breast cancers in China: Screening of 2991 patients and 1043 controls by next-generation sequencing. Int. J. Cancer 2017, 141, 129–142. [Google Scholar] [CrossRef]
- Deng, M.; Chen, H.H.; Zhu, X.; Luo, M.; Zhang, K.; Xu, C.J.; Hu, K.M.; Cheng, P.; Zhou, J.J.; Zheng, S.; et al. Prevalence and clinical outcomes of germline mutations in BRCA1/2 and PALB2 genes in 2769 unselected breast cancer patients in China. Int. J. Cancer 2019, 145, 1517–1528. [Google Scholar] [CrossRef]
- Kast, K.; Rhiem, K.; Wappenschmidt, B.; Hahnen, E.; Hauke, J.; Bluemcke, B.; Zarghooni, V.; Herold, N.; Ditsch, N.; Kiechle, M.; et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J. Med. Genet. 2016, 53, 465–471. [Google Scholar] [CrossRef]
- Santonocito, C.; Rizza, R.; Paris, I.; Marchis, L.; Paolillo, C.; Tiberi, G.; Scambia, G.; Capoluongo, E. Spectrum of Germline BRCA1 and BRCA2 Variants Identified in 2351 Ovarian and Breast Cancer Patients Referring to a Reference Cancer Hospital of Rome. Cancers 2020, 12, 1286. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.J.; Reid, J.E.; Burbidge, L.A.; Pruss, D.; Deffenbaugh, A.M.; Frye, C.; Wenstrup, R.J.; Ward, B.E.; Scholl, T.A.; Noll, W.W. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer 2009, 115, 2222–2233. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Hu, Z.; Rao, N.Y.; Song, C.G.; Zhang, B.; Cao, M.Z.; Su, F.X.; Wang, Y.S.; He, P.Q.; Di, G.H.; et al. The prevalence of BRCA1 and BRCA2 germline mutations in high-risk breast cancer patients of Chinese Han nationality: Two recurrent mutations were identified. Breast Cancer Res. Treat. 2008, 110, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pei, R.; Pang, Z.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; Lin, B.; Xie, Y. Prevalence and characterization of BRCA1 and BRCA2 germline mutations in Chinese women with familial breast cancer. Breast Cancer Res. Treat. 2012, 132, 421–428. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, J.; Chen, J.; Yao, L.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; Lin, B.; et al. Comprehensive analysis of BRCA1 and BRCA2 germline mutations in a large cohort of 5931 Chinese women with breast cancer. Breast Cancer Res. Treat. 2016, 158, 455–462. [Google Scholar] [CrossRef]
- Antoniou, A.C.; Casadei, S.; Heikkinen, T.; Barrowdale, D.; Pylkäs, K.; Roberts, J.; Lee, A.; Subramanian, D.; De Leeneer, K.; Fostira, F.; et al. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014, 371, 497–506. [Google Scholar] [CrossRef]
- Erkko, H.; Xia, B.; Nikkilä, J.; Schleutker, J.; Syrjäkoski, K.; Mannermaa, A.; Kallioniemi, A.; Pylkäs, K.; Karppinen, S.M.; Rapakko, K.; et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 2007, 446, 316–319. [Google Scholar] [CrossRef]
- Hsu, H.M.; Wang, H.C.; Chen, S.T.; Hsu, G.C.; Shen, C.Y.; Yu, J.C. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2024–2032. [Google Scholar] [CrossRef]
- Roznowski, K.; Januszkiewicz-Lewandowska, D.; Mosor, M.; Pernak, M.; Litwiniuk, M.; Nowak, J. I171V germline mutation in the NBS1 gene significantly increases risk of breast cancer. Breast Cancer Res. Treat. 2008, 110, 343–348. [Google Scholar] [CrossRef]
- Wang, Y.; Hong, Y.; Li, M.; Long, J.; Zhao, Y.P.; Zhang, J.X.; Li, Q.; You, H.; Tong, W.M.; Jia, J.D.; et al. Mutation inactivation of Nijmegen breakage syndrome gene (NBS1) in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. PLoS ONE 2013, 8, e82426. [Google Scholar] [CrossRef]
- Bougeard, G.; Renaux-Petel, M.; Flaman, J.M.; Charbonnier, C.; Fermey, P.; Belotti, M.; Gauthier-Villars, M.; Stoppa-Lyonnet, D.; Consolino, E.; Brugières, L.; et al. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J. Clin. Oncol. 2015, 33, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Polley, E.C.; Yadav, S.; Lilyquist, J.; Shimelis, H.; Na, J.; Hart, S.N.; Goldgar, D.E.; Shah, S.; Pesaran, T.; et al. The Contribution of Germline Predisposition Gene Mutations to Clinical Subtypes of Invasive Breast Cancer From a Clinical Genetic Testing Cohort. J. Natl. Cancer Inst. 2020, 112, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Näslund-Koch, C.; Nordestgaard, B.G.; Bojesen, S.E. Increased Risk for Other Cancers in Addition to Breast Cancer for CHEK2*1100delC Heterozygotes Estimated From the Copenhagen General Population Study. J. Clin. Oncol. 2016, 34, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Chen, S.Y.; Ren, J.X.; Pei, Y.C.; Jiang, C.W.; Zhao, S.; Xiao, Y.; Xu, X.E.; Liu, G.Y.; Hu, X.; et al. Molecular Features and Functional Implications of Germline Variants in Triple-Negative Breast Cancer. J. Natl. Cancer Inst. 2021, 113, 884–892. [Google Scholar] [CrossRef]
- WHO Classification of Tumors Editorial Board. Breast Tumors; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- Giuliano, A.E.; Edge, S.B.; Hortobagyi, G.N. Eighth Edition of the AJCC Cancer Staging Manual: Breast Cancer. Ann Surg Oncol. 2018, 25, 1783–1785. [Google Scholar] [CrossRef]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Guideline Update. Arch. Pathol. Lab. Med. 2020, 144, 545–563. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Rannala, B. Phylogenetic methods come of age: Testing hypotheses in an evolutionary context. Science 1997, 276, 227–232. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic. Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Panel Members of China Expert Consensus on BRCA Variant Interpretation. China expert consensus on BRCA variant interpretation. Zhonghua Bing Li Xue Za Zhi 2017, 46, 293–297. [Google Scholar] [CrossRef]
| Characteristics | No. | % |
|---|---|---|
| Gender | ||
| Female | 695 | 99.3 |
| Male | 5 | 0.7 |
| Nationality | ||
| Han | 696 | 99.4 |
| Mongolian | 2 | 0.3 |
| Uyghur | 2 | 0.3 |
| Age at the initial diagnosis of BC | ||
| <35 | 84 | 12.0 |
| 35–44 | 186 | 26.6 |
| 45–54 | 168 | 24.0 |
| 55–64 | 144 | 20.6 |
| ≥65 | 118 | 16.9 |
| Family history 1 | ||
| No | 487 | 69.6 |
| Yes | 213 | 30.4 |
| Personal history of cancer 2 | ||
| No | 675 | 96.4 |
| Yes | 25 | 3.6 |
| Hereditary risk 3 | ||
| Low | 304 | 43.4 |
| High | 396 | 56.6 |
| Laterality of BC | ||
| Unilateral | 673 | 96.1 |
| Bilateral | 27 | 3.9 |
| Histology type | ||
| DCIS | 59 | 8.4 |
| Ductal | 534 | 76.3 |
| Lobular | 19 | 2.7 |
| Mixed | 57 | 8.1 |
| Other 4 | 31 | 4.4 |
| Histology grade | ||
| I | 45 | 6.4 |
| II | 390 | 55.7 |
| III | 185 | 26.4 |
| UNK | 80 | 11.4 |
| Tumor stage | ||
| Tis | 59 | 8.4 |
| T1 | 327 | 46.7 |
| T2 | 266 | 38.0 |
| T3–T4 | 48 | 6.9 |
| Nodal status | ||
| Negative | 419 | 59.9 |
| Positive | 281 | 40.1 |
| TNM stage | ||
| 0 | 59 | 8.4 |
| I | 222 | 31.7 |
| II | 301 | 43.0 |
| III | 93 | 13.3 |
| IV | 25 | 3.6 |
| Molecular subtype | ||
| HR+Her2− | 399 | 57.0 |
| HR+Her2+ | 70 | 10.0 |
| HR−Her2+ | 67 | 9.6 |
| HR−Her2− | 164 | 23.4 |
| Ki-67 Index | ||
| ≤20% | 374 | 53.4 |
| >20% | 326 | 46.6 |
| Criteria of Hereditary High Risk | No. (%) | |
|---|---|---|
| C.1 | Diagnosed with breast cancer at age ≤ 45 years. | 288 (41.1) |
| C.2 | Diagnosed with breast cancer at age 46–50 years with one of the following: | 12 (1.7) |
| (1) A second breast cancer diagnosed at any age; | ||
| (2) ≥1 close blood relative with BRCA-related cancer 1 at any age. | ||
| C.3 | ≥1 close blood relative diagnosed with BRCA-related cancer 1 at age ≤ 50 years. | 54 (7.7) |
| C.4 | ≥2 Non-close blood relatives diagnosed with BRCA-related 1 cancer at any age. | 9 (1.3) |
| C.5 | Diagnosed at age ≤ 60 years with triple-negative breast cancer. | 120 (17.1) |
| C.6 | Diagnosed at any age with male breast cancer. | 5 (0.7) |
| C.7 | Diagnosed at any age with BRCA-related cancer 1. | 5 (0.7) |
| Characteristics | No. | Panel-Gene Variants | HRR-Gene Variants | BRCA1/2 Variants | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Carrier n = 92 | Non-Carrier n = 608 | p | Carrier n = 89 | Non-Carrier n = 611 | p | Carrier n = 66 | Non-Carrier n = 634 | p | ||
| Gender (%) | ||||||||||
| Female | 695 | 91 (98.9) | 604 (99.3) | 1.000 | 88 (98.9) | 607 (99.3) | 1.000 | 66 (100.0) | 629 (99.2) | 1.000 |
| Male | 5 | 1 (1.1) | 4 (0.7) | 1 (1.1) | 4 (0.7) | 0 (0.0) | 5 (0.8) | |||
| Nationality (%) | ||||||||||
| Han | 696 | 92 (100.0) | 604 (99.3) | NA | 89 (100.0) | 607 (99.3) | NA | 66 (100.0) | 630 (99.4) | NA |
| Mongolian | 2 | 0 (0.0) | 2 (0.3) | 0 (0.0) | 2 (0.3) | 0 (0.0) | 2 (0.3) | |||
| Uyghur | 2 | 0 (0.0) | 2 (0.3) | 0 (0.0) | 2 (0.3) | 0 (0.0) | 2(0.3) | |||
| Age (%) | ||||||||||
| <35 | 84 | 21 (22.8) | 63 (10.4) | <0.001 | 21 (23.6) | 63 (10.3) | <0.001 | 19 (28.8) | 65 (10.3) | <0.001 |
| 35–44 | 186 | 29 (31.5) | 157 (25.8) | 26 (29.2) | 160 (26.2) | 22 (33.3) | 164 (25.9) | |||
| 45–54 | 168 | 24 (26.1) | 144 (23.7) | 24 (27.0) | 144 (23.6) | 17 (25.8) | 151 (23.8) | |||
| 55–64 | 144 | 13 (14.1) | 131 (21.5) | 13 (14.6) | 131 (21.4) | 6 (9.1) | 138 (21.8) | |||
| ≥65 | 118 | 5 (5.4) | 113 (18.6) | 5 (5.6) | 113 (18.5) | 2 (3.0) | 116 (18.3) | |||
| Family history 1 (%) | ||||||||||
| No | 487 | 54 (58.7) | 433 (71.2) | 0.021 | 53 (59.6) | 434 (71.0) | 0.038 | 36 (54.5) | 451 (71.1) | 0.008 |
| Yes | 213 | 38 (41.3) | 175 (28.8) | 36 (40.4) | 177 (29.0) | 30 (45.5) | 183 (28.9) | |||
| Personal history of cancer 2 (%) | ||||||||||
| No | 675 | 88 (95.7) | 587 (96.5) | 0.897 | 86 (96.6) | 589 (96.4) | 1.000 | 64 (97.0) | 611 (96.4) | 1.000 |
| Yes | 25 | 4 (4.3) | 21 (3.5) | 3 (3.4) | 22 (3.6) | 2 (3.0) | 23 (3.6) | |||
| Hereditary risk 3 (%) | ||||||||||
| Low | 304 | 20 (21.7) | 284 (46.7) | <0.001 | 20 (22.5) | 284 (46.5) | <0.001 | 10 (15.2) | 294 (46.4) | <0.001 |
| High | 396 | 72 (78.3) | 324 (53.3) | 69 (77.5) | 327 (53.5) | 56 (84.8) | 340 (53.6) | |||
| Laterality (%) | ||||||||||
| Unilateral | 673 | 88 (95.7) | 585 (96.2) | 0.896 | 86 (96.6) | 587 (96.1) | 0.899 | 64 (97.0) | 609 (96.1) | 0.877 |
| Bilateral | 27 | 4 (4.3) | 23 (3.8) | 3 (3.4) | 24 (3.9) | 2 (3.0) | 25 (3.9) | |||
| Histology type (%) | ||||||||||
| DCIS | 59 | 4 (4.3) | 55 (9.0) | 0.589 | 4 (4.5) | 55 (9.0) | 0.637 | 4 (6.1) | 55 (8.7) | 0.773 |
| Ductal | 534 | 74 (80.4) | 460 (75.7) | 71 (79.8) | 463 (75.8) | 51 (77.3) | 483 (76.2) | |||
| Lobular | 19 | 3 (3.3) | 16 (2.6) | 3 (3.4) | 16 (2.6) | 3 (4.5) | 16 (2.5) | |||
| Mixed | 57 | 8 (8.7) | 49 (8.1) | 8 (9.0) | 49 (8.0) | 6 (9.1) | 51 (8.0) | |||
| Other 4 | 31 | 3 (3.3) | 28 (4.6) | 3 (3.4) | 28 (4.6) | 2 (3.0) | 29 (4.6) | |||
| Histology grade (%) | ||||||||||
| I | 45 | 2 (2.2) | 43 (7.1) | 0.115 | 2 (2.2) | 43 (7.0) | 0.144 | 1 (1.5) | 44 (6.9) | 0.131 |
| II | 390 | 53 (57.6) | 337 (55.4) | 51 (57.3) | 339 (55.5) | 37 (56.1) | 353 (55.7) | |||
| III | 185 | 30 (32.6) | 155 (25.5) | 29 (32.6) | 156 (25.5) | 23 (34.8) | 162 (25.6) | |||
| UNK | 80 | 7 (7.6) | 73 (12.0) | 7 (7.9) | 73 (11.9) | 5 (7.6) | 75 (11.8) | |||
| Tumor stage (%) | ||||||||||
| Tis | 59 | 4 (4.3) | 55 (9.0) | 0.332 | 4 (4.5) | 55 (9.0) | 0.415 | 4 (6.1) | 55 (8.7) | 0.454 |
| T1 | 327 | 41 (44.6) | 286 (47.0) | 40 (44.9) | 287 (47.0) | 27 (40.9) | 300 (47.3) | |||
| T2 | 266 | 41 (44.6) | 225 (37.0) | 39 (43.8) | 227 (37.2) | 31 (47.0) | 235 (37.1) | |||
| T3-T4 | 48 | 6 (6.5) | 42 (6.9) | 6 (6.7) | 42 (6.9) | 4 (6.1) | 44 (6.9) | |||
| Nodal status (%) | ||||||||||
| Negative | 419 | 47 (51.1) | 372 (61.2) | 0.084 | 46 (51.7) | 373 (61.0) | 0.092 | 33 (50.0) | 386 (60.9) | 0.133 |
| Positive | 281 | 45 (48.9) | 236 (38.8) | 43 (48.3) | 238 (39.0) | 33 (50.0) | 248 (39.1) | |||
| TNM stage (%) | ||||||||||
| 0 | 59 | 4 (4.3) | 55 (9.0) | 0.073 | 4 (4.5) | 55 (9.0) | 0.098 | 4 (6.1) | 55 (8.7) | 0.092 |
| I | 222 | 25 (27.2) | 197 (32.4) | 24 (27.0) | 198 (32.4) | 15 (22.7) | 207 (32.6) | |||
| II | 301 | 51 (55.4) | 250 (41.1) | 49 (55.1) | 252 (41.2) | 39 (59.1) | 262 (41.3) | |||
| III | 93 | 11 (12.0) | 82 (13.5) | 11 (12.4) | 82 (13.4) | 7 (10.6) | 86 (13.6) | |||
| IV | 25 | 1 (1.1) | 24 (3.9) | 1 (1.1) | 24 (3.9) | 1 (1.5) | 24 (3.8) | |||
| Subtype (%) | ||||||||||
| HR+Her2− | 399 | 46 (50.0) | 353 (58.1) | 0.001 | 46 (51.7) | 353 (57.8) | <0.001 | 33 (50.0) | 366 (57.7) | <0.001 |
| HR+Her2+ | 70 | 6 (6.5) | 64 (10.5) | 6 (6.7) | 64 (10.5) | 4 (6.1) | 66 (10.4) | |||
| HR−Her2+ | 67 | 4 (4.3) | 63 (10.4) | 2 (2.2) | 65 (10.6) | 1 (1.5) | 66 (10.4) | |||
| HR−Her2− | 164 | 36 (39.1) | 128 (21.1) | 35 (39.3) | 129 (21.1) | 28 (42.4) | 136 (21.5) | |||
| Ki-67 Index (%) | ||||||||||
| ≤20% | 374 | 31 (33.7) | 343 (56.4) | <0.001 | 31 (34.8) | 343 (56.1) | <0.001 | 20 (30.3) | 354 (55.8) | <0.001 |
| >20% | 326 | 61 (66.3) | 265 (43.6) | 58 (65.2) | 268 (43.9) | 46 (69.7) | 280 (44.2) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Xie, F.; Liu, C.; Zhao, J.; Hu, T.; Wu, J.; Zhao, X.; Wang, S. Germline Variants in 32 Cancer-Related Genes among 700 Chinese Breast Cancer Patients by Next-Generation Sequencing: A Clinic-Based, Observational Study. Int. J. Mol. Sci. 2022, 23, 11266. https://doi.org/10.3390/ijms231911266
Yang L, Xie F, Liu C, Zhao J, Hu T, Wu J, Zhao X, Wang S. Germline Variants in 32 Cancer-Related Genes among 700 Chinese Breast Cancer Patients by Next-Generation Sequencing: A Clinic-Based, Observational Study. International Journal of Molecular Sciences. 2022; 23(19):11266. https://doi.org/10.3390/ijms231911266
Chicago/Turabian StyleYang, Liu, Fei Xie, Chang Liu, Jin Zhao, Taobo Hu, Jinbo Wu, Xiaotao Zhao, and Shu Wang. 2022. "Germline Variants in 32 Cancer-Related Genes among 700 Chinese Breast Cancer Patients by Next-Generation Sequencing: A Clinic-Based, Observational Study" International Journal of Molecular Sciences 23, no. 19: 11266. https://doi.org/10.3390/ijms231911266
APA StyleYang, L., Xie, F., Liu, C., Zhao, J., Hu, T., Wu, J., Zhao, X., & Wang, S. (2022). Germline Variants in 32 Cancer-Related Genes among 700 Chinese Breast Cancer Patients by Next-Generation Sequencing: A Clinic-Based, Observational Study. International Journal of Molecular Sciences, 23(19), 11266. https://doi.org/10.3390/ijms231911266






