Abstract
Avian leukosis virus (ALV) induces B-cell lymphomas and other malignancies in chickens through insertional activation of oncogenes, and c-myc activation has been commonly identified in ALV-induced tumors. Using ALV-transformed B-lymphoma-derived HP45 cell line, we applied in situ CRISPR-Cas9 editing of integrated proviral long terminal repeat (LTR) to examine the effects on gene expression and cell proliferation. Targeted deletion of LTR resulted in significant reduction in expression of a number of LTR-regulated genes including c-myc. LTR deletion also induced apoptosis of HP45 cells, affecting their proliferation, demonstrating the significance of LTR-mediated regulation of critical genes. Compared to the global effects on expression and functions of multiple genes in LTR-deleted cells, deletion of c-myc had a major effect on the HP45 cells proliferation with the phenotype similar to the LTR deletion, demonstrating the significance of c-myc expression in ALV-induced lymphomagenesis. Overall, our studies have not only shown the potential of targeted editing of the LTR for the global inhibition of retrovirus-induced transformation, but also have provided insights into the roles of LTR-regulated genes in ALV-induced neoplastic transformation.
1. Introduction
Avian leukosis viruses (ALV), belonging to different envelope subgroups of avian retroviruses, are major pathogens associated with different types of neoplastic diseases in poultry [1]. Similar to other retroviruses, ALV integrates into the host genome to induce tumorigenesis by modifying the expression of host genes. ALV long terminal repeat (LTR), with strong promoter-enhancer and other regulatory elements, drive the expression of genes adjacent to the insertion site through the process referred to as insertional activation, first demonstrated with the c-myc oncogene [2]. In addition to the ALV-LTR-induced enhanced expression of the c-myc, insertional activation of other genes such as c-bic resulting in synergistic enhancement of lymphomas, particularly in the late stages of tumor development, has also been reported [3]. Although the functional nature of the c-bic locus was not clear at the time of its discovery, subsequent studies have identified the multifunctional miR-155 as the gene product [4,5], and detailed studies highlighting the direct role of miR-155 in many cancers have recently been reviewed [6,7]. Retroviral integrations near host genes can result in deregulation of gene expression through different pathways, including enhancer or promoter insertion or mRNA 3’ end substitution, as well as through insertional disruption [8]. ALV-mediated induction of lymphoma through c-myc occurs mainly by direct transcriptional activation as well as through transduction of the v-myc oncogene by acutely transforming viruses such as MC29 [9]. Members of the Myc family, which include c-myc, L-myc, and N-myc, are powerful transcription factors that regulate cellular growth and proliferation [10]. This family of oncogenes is also actively associated with the development of multiple cancers [11,12]. The direct role of c-myc as an oncogene in the induction of cancer was first demonstrated by the insertional activation of the c-myc promoter in ALV-induced B-cell lymphomas [2] and subsequently confirmed in other tumors [13]. Myc transcriptional regulator function occurs through the specific interaction with the palindromic DNA sequence (CACGTG), called ‘E-Box’, with the help of myc-associated factor X (MAX) as a heterodimer [10].
While c-myc is indeed one of the most common integration sites in ALV-induced tumors [13], several other integration sites in genes such as CTDSPL, CTDSPL2, MYB, TERT, MET, EGFR, TNFRSF1A, MEF2C, CTDSPL, TAB2, RUNX1, MLL5, CXorf57, and BACH2 have been also identified in ALV-induced B-cell lymphomas [14,15,16,17]. Comprehensive identification of the integration sites in ALV-transformed cells is important in understanding the global changes in gene expression directly associated with retroviral integration. Several methods have been used for the identification of retroviral integration sites in the genome [18], including inverse PCR [19], splinkerette PCR [20] and Cas9 enrichment [21] together with the next generation sequencing (NGS) for high throughput identification of integration sites. In order to examine the direct effect of the ALV-LTR-induced gene regulation on the transformation phenotype of the HP45 cell line, we carried out CRISPR-Cas9-mediated targeted knockdown (KD) of a region of the ALV-LTR. Our studies showed that targeted editing of the ALV-LTR had a dramatic effect on the proliferation of the HP45 cell line as well as the dysregulation of the expression of LTR-regulated genes, demonstrating the broad role of the integrated proviral ALV-LTR on the transformed phenotype of the HP45 cell line. Targeted editing of c-myc showed that c-myc is a major contributor of the observed phenotypical changes induced by the ALV-LTR deletion.
2. Results
2.1. ALV-LTR Editing Downregulates Expression of Insertionally Activated Genes
Using an ALV-transformed B-cell line with the proviral integration in the c-myc locus, we first examined the c-myc expression in the HP45 cell line by RT-qPCR with myc-specific primers (Table 1). Primary B cells (isolated from the chicken bursa of Fabricius) expressing physiological levels of c-myc were used as a control. Analysis of the RT-qPCR data, relative to the GAPDH [22], showed that c-myc expression was almost 28-fold higher in the HP45 cells compared to the primary B cells (Supplementary Figure S1A). This suggested that insertional activation of c-myc through the LTR of the integrated ALV provirus contributes to the enhanced c-myc gene expression in the transformed HP45 cell line.

Table 1.
Primers used in PCR and RT-qPCR.
After demonstrating the enhanced expression of c-myc in the HP45 cells, we investigated whether insertional activation through the strong LTR promoter was driving the overexpression of c-myc in these cells. We applied targeted CRISPR/Cas9 editing of the integrated proviral LTR using the gRNAs listed in Table 2. The efficiency of the LTR editing was analyzed by PCR, using specific forward and reverse primers (Table 1) on genomic DNA from the edited HP45 cell population at different time points post transfection. A reduction in the size of the amplified DNA, compared to that seen in the non-targeting SgA control (SgA-NT), demonstrated the targeted editing of the LTR (Supplementary Figure S1B, top panel). Sequence analysis of the purified PCR products further confirmed the deletion of the target region compared with that of wild-type LTR sequence (Supplementary Figure S1C).

Table 2.
ALV-LTR and c-Myc guide-RNA sequence.
As one of the most important genes regulated by insertional activation, we first examined the effect of LTR editing on the expression of c-myc by RT-qPCR. The data were plotted as 2−ΔΔCT, and the gene expression was compared with the expression levels relative to SgA-NT guide-RNA, using GAPDH as an internal control. We observed a highly significant reduction in c-myc expression in LTR-edited cells at different time points (Figure 1A). We also examined the expression of other LTR-regulated genes identified by Targeted locus amplification (TLA) analysis or integration junction PCR (manuscript in preparation). Targeted editing of LTR in HP45 cells resulted in downregulation of LTR-regulated genes such as BATF (basic leucine zipper ATF-like transcription factor), TP63 (Tumor protein 63), EP400 (E1A Binding protein P400), c-Rel, and miR-155 expressed from the c-Bic. However, the expression of B2M (Beta-2-microglobulin), one of the stably expressed reference genes [23] and not targeted by ALV-LTR, has not been affected (Figure 1A).
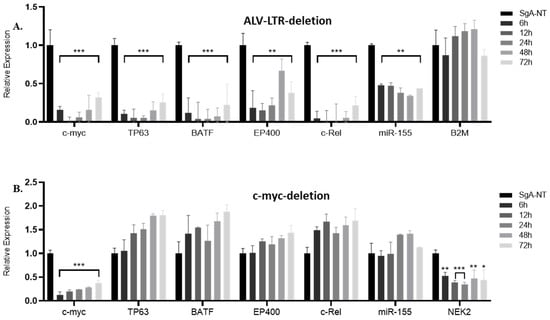
Figure 1.
Expression of ALV-LTR-activated genes following ALV-LTR or c-myc deletion in HP45. RT-qPCR analysis of c-myc, TP63, BATF, EP400, c-Rel gene and miR-155 expression in ALV-LTR deleted (A) or c-myc deleted (B) cells and the control cells. Cas9 expressing HP45 cells were transfected with non-targeting control gRNA SgA-NT or ALV-LTR-gRNAs/c-myc-gRNAs. For the ALV-LTR-gRNAs/c-myc-gRNAs transfections, the transfected cells were harvested at 6, 12, 24, 48, and 72 h post transfection, and RNA was extracted, followed by RT-qPCR analysis using gene-specific primers and SYBR green reagents. Relative expression of miR-155 was measured using miR155-specific TaqMan probe and primers (Applied Bioscience). The data were plotted as 2−ΔΔCT where transcript levels were normalized against GAPDH for c-myc, TP63, BATF, EP400, and c-Rel and against U6-small nuclear RNA (U6-SnRNA) for miR-155. The value corresponding to the level of each gene/miRNA in SgA-NT transfected cells harvested at 72 h post transfection was set as 1. All data are representative of three independent experiments and the statistical significance was measured by t-test using GraphPad software, from 3 independent experiments (* p < 0.05; ** p < 0.01; *** p < 0.001).
2.2. Knockdown of c-myc Did Not Affect Expression of Other LTR-Regulated Genes
Editing of the LTR had a downward effect on the expression of multiple genes regulated by LTR. To confirm that the changes in gene expression were not an indirect effect of the reduced expression of key LTR-regulated genes, such as c-myc, we carried out targeted editing of c-myc using specific gRNAs (Table 2, Supplementary Figure S1B, bottom panel and S1–D). The expression levels of different LTR-regulated genes were then measured by RT-qPCR using SYBR green master mix and gene-specific primers (Table 1). Targeted editing of the c-myc resulted in a significant reduction in c-myc expression at different time points (Figure 1B). However, editing of c-myc did not affect the expression of any of the other LTR-regulated genes, including miR-155 (Figure 1B), confirming that the changes in LTR-regulated gene expression were indeed a direct effect of LTR deletion and not c-myc downregulation. Interestingly, the expression of the NEK2 gene, one of the c-myc target genes identified by the c-myc ChIP-Seq assay of HP45 (manuscript in preparation), is downregulated after c-myc deletion, further confirming that the direct effect of LTR deletion on its integrated genes is not due to c-myc downregulation.
2.3. LTR Editing Inhibits Proliferation of HP45 Cells
During LTR editing of HP45 cells, we observed that LTR-gRNA electroporated cells did not increase in numbers when compared to the SgA-NT electroporated control cells. This suggested that LTR editing also impacted cell proliferation. To examine this further, edited cell populations were subjected to a proliferation assay using IncuCyte® NucLight Rapid Red Reagent. Dynamic changes in the proliferation of HP45-Cas9 cells electroporated with LTR-gRNAs, or SgA-NT control gRNA, were continuously monitored in IncuCyte S3 and analyzed using IncuCyte S3 2018C software. Data plotted as percentages of phase object confluency (Figure 2A upper panel) and red object confluence (Figure 2B upper panel) confirmed the reduced proliferation of the LTR-edited cells from the early time points, with highly significant differences (p < 0.001) observed at later time points. The flat growth curve of the LTR-edited cell population, compared to the rapidly proliferating control cells, was also reflected in the images taken at 72 h time point for phase (Figure 2A, lower panel) and red fluorescence (Figure 2B, lower panel) channel in the IncuCyte S3.
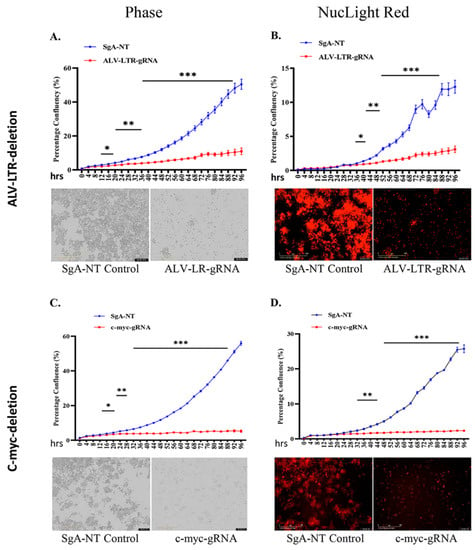
Figure 2.
Proliferation of gRNA-transfected cells monitored in real time using the IncuCyte S3 live imaging system. gRNAs targeting ALV-LTR (A,B) or c-myc (C,D) transfected cells were subjected to proliferation assay using NucLight Rapid Red Reagent in IncuCyte S3 imaging system. Phase (A,C) and red (B,D) object confluency of each cell population were determined every 4 h for 96 h from five separate regions per well and four wells per sample by IncuCyte S3 and compared with SgA-NT control. Growth curves (upper panels) are shown as mean ± standard error (SE) representative of three independent experiments. Asterisk (*) indicates statistically significant differences between gRNA-edited cells and SgA-NT cells at different time points. * p <0.05; ** p < 0.01; *** p < 0.001. Phase and red object images (bottom panels) are representative of 72 h time points. The scale bar, 400 μm.
2.4. Downregulated c-myc Contributes to Proliferation Defect of LTR-Edited HP45
Having demonstrated the proliferation defect of the LTR-edited HP45 cells, we wanted to identify the LTR-regulated gene(s) that directly contributed to this phenotype. In particular, we wanted to examine whether the reduced expression of c-myc (Figure 1A) in the LTR-deleted cells contributed to the inhibition of proliferation. For this, we used the IncuCyte S3 to measure the dynamic changes in the proliferation of c-myc-knockout HP45 cells. We observed a significant defect in the proliferation of c-myc deleted HP45 cells (Figure 2C,D, phase and NucLight rapid red, respectively) in a pattern very similar to LTR-deleted cells. Control cells transfected with SgA-NT showed no defect in proliferation. This indicates that c-myc is a key contributor to proliferation defects in HP45 with LTR deletion.
2.5. Knockdown of LTR and c-myc Induces Apoptosis in HP45 Cells
Having demonstrated a significant defect in the proliferation rates of HP45 cells with deletions in the LTR and c-myc, we explored the possible mechanisms. During the early stage of the experiments, we observed that the gene-deleted cell populations were not only defective in proliferation but also dying in significant numbers, especially in the case of ALV-LTR-edited cells. In order to quantify the percentage of dying cells in the knockout populations, these cells were analyzed for apoptosis using FITC Annexin-V Apoptosis Detection Kit with 7-AAD. HP45 cells, harvested at 48 h and 72 h post electroporation with LTR- or c-myc-specific gRNAs or SgA-NT gRNA, were stained according to the manufacturer’s protocol, and data were collected using DIVA 8 software and a BD biosciences LSR Fortessa. ALV-LTR-treated cells induced a marked increase in early and late apoptosis compared to control cells. The percentage of cells that were apoptotic at 48 and 72 h post electroporation with LTR-specific gRNAs was 62% (Figure 3A, top panel) and 67.6% (Figure 3A, bottom panel), respectively. In comparison, control cells showed only 9.3% and 7.2% apoptotic cells at the same time points. These results showed that targeting ALV-LTR not only slows down the proliferation rate but also increases the apoptosis of the cell population. Targeted editing of c-myc also induced apoptosis, but at a lower rate when compared to LTR-deleted cells. The percentage of apoptotic cells detected in c-myc knockdown were 25.5% and 43.8%, compared to control cells with 6.5% and 7.2% apoptotic cells at 48 and 72 h, respectively (Figure 3B).
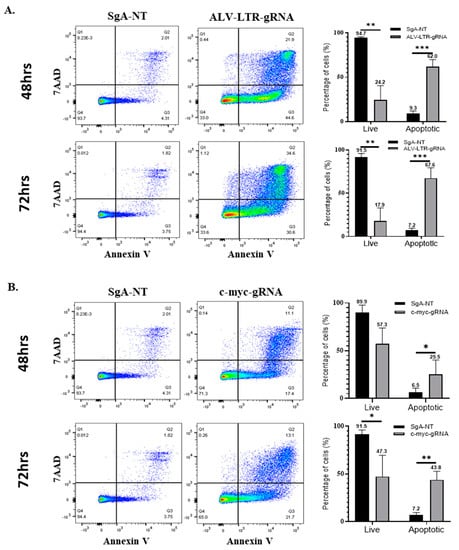
Figure 3.
Apoptosis Assay and viability assay of gRNA-transfected cells. SgA-NT (left panels) or gRNAs targeting ALV-LTR (A) and c-myc (B) (middle panels) transfected cells were stained with Annexin-V-FITC and 7AAD at 48 h (upper panels) and 72 h (lower panels) post transfection and analyzed in FlowjoX with data collected on a BD Bioscience LSR Fortessa. The histogram (right panels) shows the percentage of live and apoptotic cells calculated from the flow cytometry data. Each experiment has been performed a minimum of three times, multiple t-tests have been performed, and the statistical significance was calculated using the Holm-Sidak method. Asterisk (*) indicates statistically significant differences between gRNA-edited cells and SgA-NT cells at different time points. * p <0.05; ** p < 0.01; *** p < 0.001.
3. Discussion
It has been reported that there is a strong association between retroviral integration, oncogenic activation, and tumorigenesis, not only in humans but in other species as well. Integration of ALV into various cellular proto-oncogenes such as c-myc, MYB, TERT, and RUNX1 triggers the induction of tumors causing lymphoid, myeloid, or erythroid tumors, depending on the cell types affected [1,13,16]. The most common site of ALV integration in bursal lymphomas and derivative cell lines corresponds to the 5’ flanking region of the c-myc gene, with the proviral integration occurring mainly within the 5 kb region of the transcriptional start site [24]. In many of these lymphomas, there is also second integration in the c-bic oncogene that encodes the oncogenic microRNA miR-155 [4,25,26,27,28,29].
In this study, we have used CRISPR-Cas9-based targeted editing of the integrated ALV proviral genome to demonstrate the broad effect of the LTR on the expression of ALV-LTR integration-associated genes. We have examined c-myc, TP63, BATF, EP400, c-REL, and miR155 gene expression levels in HP45 cells where the LTR region was targeted using specific guide-RNAs. Among these genes, the roles of c-myc and TP63 on cell proliferation or growth have been published [30,31]. In this study, we investigated the effect of knockdown of ALV-LTR and c-myc on HP45 cell proliferation using the IncuCyte S3 live cell analysis system. These studies showed significant proliferation defects in both ALV-LTR and c-myc KD cells compared to control cells, indicating that c-myc is a major contributor to the observed proliferation defect after the ALV-LTR deletion. ALV-LTR KD cells exhibited high levels of cell death when viewed under the microscope. Although c-myc has a prominent role in driving apoptosis [32], it has also been reported to mediate the inhibition of apoptosis [33]. Our data have shown that KD of c-myc also induced apoptosis of HP45 cells. Interestingly, the apoptosis levels were significantly higher when targeting the LTR region compared to the cells where c-myc was targeted. This suggests that in addition to c-myc, other LTR-activated genes, such as TP63, may also contribute to the apoptosis induced by ALV-LTR editing.
The oncogene c-myc has been implicated in driving neoplastic transformation by other oncogenic viruses. Epstein-Barr Virus (EBV) is highly pathogenic and causes a number of B-cell malignancies. It has been reported that a high level of c-myc expression reduces the latent membrane protein 1 (LMP1) expression, which is essential for the EBV infected B-cell immortalization; thus, the effect of EBV infection in primary B cells in the first few weeks is not evident [34]. Recent data has also demonstrated that c-myc controls the EBV lytic switch by remodeling the viral genome [35].
In summary, we have demonstrated the successful application of CRISPR/Cas9-based targeted in situ editing of the LTR of the integrated retroviral genome as well as the insertionally activated major oncogene c-myc in an avian B-cell lymphoma-derived tumor cell line. Overall, our data suggest that KD of ALV-LTR has a broad effect on the ALV integration-related gene expression, proliferation, and apoptosis. KD of c-myc also had a similar functional effect indicating its importance among the multiple insertionally activated target genes. The lower level of apoptosis induced by c-myc deletion compared to ALV-LTR deletion suggested that other ALV-LTR insertionally activated genes may also contribute to the overall apoptosis effect with ALV-LTR deletion. The exact role and degree of contribution by other integrated genes need further investigation. While the targeted editing of genes in transformed cell lines in situ can provide insights into the gene functions and downstream regulatory pathways, these approaches could also be utilized in developing a therapeutic target by directly aiming to inactivate the LTR region and indirectly targeting many genes, thus reducing the proliferation and growth of the cancerous tissues.
4. Materials and Methods
4.1. Cell Culture and Generation of HP45-Cas9 Cell Line
ALV-transformed HP45 cells [36] were cultured in RPMI 1640 (Sigma-Aldrich, Burlington, MA, USA) supplemented with 10% tryptose phosphate broth (TPB), 10% heat inactivated FBS (Sigma-Aldrich, Burlington, MA, USA), 10 mM Sodium pyruvate (Sigma-Aldrich, Burlington, MA, USA), 50 mM 2-mercaptoethanol (GIBCO, Waltham, MA, USA) and 100 units/mL penicillin and streptomycin (GIBCO, Waltham, MA, USA), kept at 38.5 °C incubator with 5% CO2 and humidity.
To generate the stable HP45 cell line expressing Cas9, 1 × 106 of HP45 cells were resuspended in 96 µL of Opti-MEM medium (GIBCO, Waltham, MA, USA) and electroporated with 10 µg of pX459-V2.0 plasmid (Addgene, Watertown, MA, USA) using a NEPA21 electroporator (Sonidel Ltd., Dublin, Ireland). After selection with 1 µg/mL puromycin (Sigma-Aldrich, Burlington, MA, USA) for 7 days, single cell clones were isolated using fluorescence-activated cell sorting using FACSAria IIu (BD Bioscience, Wokingham, Berkshire, UK). The single cell clone with Cas9 expression detected by Western blotting with anti-Flag antibody (Supplementary Figure S2) was grown and stored in liquid nitrogen for further studies.
4.2. Guide-RNA Design
The gRNAs targeting ALV-LTR and c-myc were designed using http://crispor.tefor.net/, accessed on 8 March 2019, and the MIT specificity score was considered not less than 95%. A two-part guide-RNA system containing crRNA and tracrRNA guide complex was used for gene editing. Sequences of ALV-LTR [37] and c-myc [38] from HP45 genomic DNA were confirmed by sequencing of the PCR products. The gRNAs designed to target ALV-LTR and c-myc (Table 2) were produced by Integrated DNA Technologies (IDT, Diego, CA, USA). Aliquots of lyophilized crRNA and tracrRNA pellets were resuspended in duplex buffer (IDT) at 200 µM concentration and stored at −80 °C.
4.3. Genomic DNA Isolation
Edited HP45-Cas9 cells and SgA-NT transfected control cells were harvested after electroporation at different time points. Cell pellets were washed twice with PBS and resuspended in 100 µL of 1× Proteinase K-based DNA extraction buffer (10 mM Tris-HCL, 1 mM EDTA, 25 mM NaCl, and 200 µg/mL Proteinase K) and incubated at 65 ℃ for 30 min followed by proteinase K inactivation at 95 ℃ for 2 min and stored at −20 ℃. A total of 1 µL of genomic DNA was used for PCR.
4.4. Electroporation of gRNAs into HP45-Cas9 Cell Line for Targeted Editing
To knockout the ALV-LTR region or c-myc gene, 2 × 106 of HP45-Cas9 cells were resuspended in 96 µL Opti-MEM medium. ALV-LTR-gRNAs 1-3 or c-myc-gRNAs 1-2 were mixed with equimolar of tracrRNA to reach a final duplex concentration of 100 µM. The mix was incubated at 95 °C for 5 min and allowed to cool to room temperature until use. The duplex was mixed with the cell suspension in a total volume of 100 µL and electroporated using a NEPA21 electroporator (Nepa Gene Co., Ltd., Ichikawa, Japan). Electroporated cells were immediately transferred into a 6-well plate containing 2 mL of complete growth media. Cells were harvested at 6 h, 12 h, 24 h, 48 h, and 72 h intervals for RT-qPCR and PCR analysis. The proliferation assay was set up immediately following the electroporation process. The cells were harvested at 48 h and 72 h time points and analyzed in an apoptosis assay.
4.5. RNA Extraction, cDNA Synthesis, and RT-qPCR
Total RNA was extracted using TRIzol Reagent (Life Technologies, Paisley, UK) following the manufacturer’s protocol. The RNA was quantified using a Nanodrop (ThermoFisher Scientific, Gloucester, UK) and diluted to 500 ng/µL with RNase-free water for cDNA synthesis. c-DNAs were synthesized using High-Capacity RNA-to-cDNA Kit (Applied BiosystemTM, Waltham, MA, USA), and 500 ng RNA was used for each reaction. qPCR was performed using Luna®® Universal qPCR Master Mix (New England Biolabs, Herts, UK), which uses real-time fluorescence of a double-stranded DNA (dsDNA) binding dye, commonly SYBR Green, and 10 µM forward and reverse primer specific to each gene (Table 1). The transcripts were normalized to endogenous GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and to the experimental SgA-NT control and plotted as 2−ΔΔCt [22].
To determine the microRNA (miRNA) expression level, total RNA was extracted using a miRNeasy Mini Kit (Qiagen, Hilden, North Rhine-Westphalia, Germany), following the manufacturer’s protocol. The RNA was quantified using a Nanodrop (Thermo Fisher Scientific, Paisley, Scotland, UK) and diluted to 10 ng/µL with RNase-free water for cDNA synthesis. cDNAs were synthesized using a TaqMan™ MicroRNA Reverse Transcription Kit (Applied BiosystemTM, Waltham, MA, USA). TaqMan™ MicroRNA Assay RT-hsa-miR-155 and RT-U6 snRNA primers were used with 50 ng of miRNA samples for each reaction. For qPCR, ABsolute Blue qPCR Low ROX master mix was used along with TaqMan TM-hsa-miR-155 and TM-U6 snRNA probe. For the relative quantification of miRNA155, Ct values were normalized to the expression level of endogenous U6-SnRNA and the experimental SgA-NT control and plotted as 2−ΔΔCT. The value corresponding to the level of miR-155 in SgA-NT transfected HP45-Cas9 was set as 1 for calibration.
4.6. Proliferation Assay with IncuCyte NucLight Rapid Red Reagent
After electroporation of HP45-Cas9 cells with gRNAs targeting different sequences and SgA-NT gRNAs, 5000 cells/well were plated for each sample into a 96-well plate in quadruplicates and measured for proliferation with IncuCyte NucLight Rapid Red reagent (1:500 dilution) in an IncuCyte (IncuCyte S3 Sartorius, ESSEN BioScience, Hertfordshire, UK). The plate was scanned from five separate regions per well using a 10× objective every 4 h. The phase object confluence and the red object count data were plotted using IncuCyteS3-2018C software. The IncuCyte data were analyzed by two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test using GraphPad Prism version 8 Software. The results are shown as means ± standard errors (SE) of results from four replicates, each with five separate regions per well and representative of three independent experiments. p values of <0.05 were considered to be significant.
4.7. B-Cell Isolation from Bursa of Fabricius (BF)
Freshly collected BF tissues were washed 2–4 times with PBS before digesting in 5 mL (8 mg/mL) collagenase D (Sigma-Aldrich, Burlington, MA, USA) in 1× Hanks Balanced Salt Solution (HBSS) with calcium (Thermo Fisher Scientific, Paisley, Scotland, UK) at 37 ℃ for 15–20 min. Digested tissues were passed through a 100 µM cell strainer and centrifuged at 1100 rpm for 10 min, and the pellet was resuspended in 10 mL of complete IMDM media (with 2% chicken serum, 8% FBS, β-mercaptoethanol, streptomycin, and insulin transfer). Cells were carefully layered on 5 mL Histopaque (Sigma-Aldrich, Burlington, MA, USA) and centrifuged at 2000 rpm for 20 min at 4 ℃. Cells from the interface were collected and resuspended in cold PBS followed by centrifuge at 1800 rpm for 10 min at 4 ℃. The cells were washed twice the same way and resuspended in an appropriate volume of complete IMDM media.
4.8. Apoptosis Assay
Populations of ALV-LTR and c-myc-edited and SgA-NT control cells were subjected to Annexin-V apoptosis assay using FITC Annexin-V Apoptosis Detection Kit with 7-AAD (BioLegend, London, UK), as per manufacturer’s protocols. Briefly, cells washed twice with cold buffer (BioLegend, London, UK) were resuspended in Annexin-V binding buffer (0.25–1.0 × 107 cells/mL), and 100 µL of cell suspension was mixed with 5 µL of FITC Annexin-V, followed by 5 µL of 7-AAD viability staining solution. After 15 min incubation at room temperature in the dark, Annexin-V binding buffer (400 µL) was added and analyzed by flow cytometry. Cells were identified, and data were collected using DIVA 8 software and a BD-LSR-Fortessa (BD bioscience, Wokingham, Berkshire, UK) flow cytometer, and 50,000 events were acquired for each sample. Data were analyzed using FlowJo v10.8 software (BD bioscience, Wokingham, Berkshire, UK). In brief, samples were gated on cells (SSC-A vs. FSC-A), singlets (SSC-A vs. SSC-H), and then early apoptotic cells were determined as AnnexinV-FITC-positive (blue 530/30 BP), and 7AAD-negative (YG 630/30 BP) and late apoptotic cells were determined as AnnexinV-FITC-positive and 7AAD-positive. Staurosporine (Abcam, Cambridge, UK)-treated cells were used as a positive control for early and late apoptosis.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911263/s1.
Author Contributions
Conceptualization, S.R., Y.Y. and V.N.; methodology, S.R., M.S.B., Y.Z. and K.M.; validation, S.R. and Y.Z.; formal analysis, S.R., K.M., Y.Z., Y.Y. and V.N.; investigation, S.R., Y.Z. and K.M.; writing—original draft preparation, S.R.; writing—review and editing V.N. and Y.Y.; supervision, V.N. and Y.Y.; funding acquisition, V.N. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) grants BBS/E/I/00007030, BBS/E/I/00007032, BBS/E/I/00007038, BBS/E/I/00007039 and BB/R012865/1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors report no conflict of interest.
References
- Nair, V. Leukosis/sarcoma group. In Diseases of Poultry; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., Eds.; Wiley: Hoboken, NJ, USA, 2020; Volume 1, pp. 587–625. [Google Scholar]
- Hayward, W.S.; Neel, B.G.; Astrin, S.M. Activation of a cellular onc gene by promoter insertion in alv-induced lymphoid leukosis. Nature 1981, 290, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Clurman, B.E.; Hayward, W.S. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: Evidence for stage-specific events. Mol. Cell. Biol. 1989, 9, 2657–2664. [Google Scholar]
- Tam, W.; Hughes, S.H.; Hayward, W.S.; Besmer, P. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding rna, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J. Virol. 2002, 76, 4275–4286. [Google Scholar] [CrossRef]
- Tam, W.; Dahlberg, J.E. Mir-155/bic as an oncogenic microrna. Genes Chromosomes Cancer 2006, 45, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Gulei, D.; Raduly, L.; Broseghini, E.; Ferracin, M.; Berindan-Neagoe, I. The extensive role of mir-155 in malignant and non-malignant diseases. Mol. Asp. Med. 2019, 70, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Witten, L.; Slack, F.J. Mir-155 as a novel clinical target for hematological malignancies. Carcinogenesis 2020, 41, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D. Retroviral insertional mutagenesis in humans: Evidence for four genetic mechanisms promoting expansion of cell clones. Mol. Ther. 2020, 28, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Kung, H.J.; Liu, J.L. Retroviral oncogenesis. In Viral Pathogenesis; Nathanson, N., Ed.; Lippincott-Raven Publishers: Philadelphia, PA, USA, 1997; pp. 235–266. [Google Scholar]
- Beaulieu, M.E.; Castillo, F.; Soucek, L. Structural and biophysical insights into the function of the intrinsically disordered myc oncoprotein. Cells 2020, 9, 1038. [Google Scholar] [CrossRef]
- Amjadi-Moheb, F.; Paniri, A.; Akhavan-Niaki, H. Insights into the links between myc and 3d chromatin structure and epigenetic regulation: Implications for cancer therapy. Cancer Res. 2021, 81, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.D. The myc oncogene: Its role in transformation and differentiation. Annu. Rev. Genet. 1986, 20, 361–384. [Google Scholar] [CrossRef] [PubMed]
- Justice, J.F., IV; Morgan, R.W.; Beemon, K.L. Common viral integration sites identified in avian leukosis virus-induced b-cell lymphomas. mBio 2015, 6, e01863-15. [Google Scholar] [CrossRef]
- Justice, J., IV; Malhotra, S.; Ruano, M.; Li, Y.; Zavala, G.; Lee, N.; Morgan, R.; Beemon, K. The met gene is a common integration target in avian leukosis virus subgroup j-induced chicken hemangiomas. J. Virol. 2015, 89, 4712–4719. [Google Scholar] [CrossRef] [PubMed]
- Kanter, M.R.; Smith, R.E.; Hayward, W.S. Rapid induction of b-cell lymphomas: Insertional activation of c-myb by avian leukosis virus. J. Virol. 1988, 62, 1423–1432. [Google Scholar] [CrossRef]
- Winans, S.; Flynn, A.; Malhotra, S.; Balagopal, V.; Beemon, K.L. Integration of alv into ctdspl and ctdspl2 genes in b-cell lymphomas promotes cell immortalization, migration and survival. Oncotarget 2017, 8, 57302–57315. [Google Scholar] [CrossRef]
- Yang, F.; Xian, R.R.; Li, Y.; Polony, T.S.; Beemon, K.L. Telomerase reverse transcriptase expression elevated by avian leukosis virus integration in b cell lymphomas. Proc. Natl. Acad. Sci. USA 2007, 104, 18952–18957. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.W.; Guo, S.; Shao, W.; Bale, M.J.; Coffin, J.M.; Hughes, S.H.; Wu, X. An analytical pipeline for identifying and mapping the integration sites of hiv and other retroviruses. BMC Genom. 2020, 21, 216. [Google Scholar]
- Uemura, S.; Nagaoka, T.; Yokoyama, M.; Igarashi, M.; Kishi, M. A simple and highly efficient method to identify the integration site of a transgene in the animal genome. Neurosci. Res. 2014, 80, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Mikkers, H.; Kool, J.; van der Weyden, L.; Lund, A.H.; Wilson, C.H.; Rance, R.; Jonkers, J.; van Lohuizen, M.; Berns, A.; et al. A high-throughput splinkerette-pcr method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 2009, 4, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Van Haasteren, J.; Munis, A.M.; Gill, D.R.; Hyde, S.C. Genome-wide integration site detection using cas9 enriched amplification-free long-range sequencing. Nucleic Acids Res. 2021, 49, e16. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Staines, K.; Batra, A.; Mwangi, W.; Maier, H.J.; Van Borm, S.; Young, J.R.; Fife, M.; Butter, C. A versatile panel of reference gene assays for the measurement of chicken mrna by quantitative pcr. PLoS ONE 2016, 11, e0160173. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, S.; Winans, S.; Lam, G.; Justice, J.; Morgan, R.; Beemon, K. Selection for avian leukosis virus integration sites determines the clonal progression of b-cell lymphomas. PLoS Pathog. 2017, 13, e1006708. [Google Scholar] [CrossRef] [PubMed]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of mir-155 and bic rna in human b cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.J.; van den Berg, A. Bic and mir-155 are highly expressed in hodgkin, primary mediastinal and diffuse large b cell lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, A.; Kroesen, B.J.; Kooistra, K.; de Jong, D.; Briggs, J.; Blokzijl, T.; Jacobs, S.; Kluiver, J.; Diepstra, A.; Maggio, E.; et al. High expression of b-cell receptor inducible gene bic in all subtypes of hodgkin lymphoma. Genes Chromosomes Cancer 2003, 37, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Bondada, M.S.; Yao, Y.; Nair, V. Multifunctional mir-155 pathway in avian oncogenic virus-induced neoplastic diseases. Non-Coding RNA 2019, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Schubach, W.; Groudine, M. Alteration of c-myc chromatin structure by avian leukosis virus integration. Nature 1984, 307, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Staller, P.; Eilers, M. Control of cell proliferation by myc. Trends Cell Biol. 1998, 8, 202–206. [Google Scholar] [CrossRef]
- Hartl, M.; Bister, K. Myc analysis in cancer and evolution. Methods Mol. Biol. 2021, 2318, 87–117. [Google Scholar] [PubMed]
- Lu, D.; Wilson, C.; Littlewood, T.D. Methods for determining myc-induced apoptosis. Methods Mol. Biol. 2021, 2318, 209–229. [Google Scholar] [PubMed]
- Zhang, J.; Song, N.; Zang, D.; Yu, J.; Li, J.; Di, W.; Guo, R.; Zhao, W.; Wang, H. C-myc promotes tumor proliferation and anti-apoptosis by repressing p21 in rhabdomyosarcomas. Mol. Med. Rep. 2017, 16, 4089–4094. [Google Scholar] [CrossRef] [PubMed]
- Price, A.M.; Messinger, J.E.; Luftig, M.A. C-myc represses transcription of epstein-barr virus latent membrane protein 1 early after primary b cell infection. J. Virol. 2018, 92, e01178-17. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Jiang, C.; Zhang, Y.; Govande, A.; Trudeau, S.J.; Chen, F.; Fry, C.J.; Puri, R.; Wolinsky, E.; Schineller, M.; et al. Myc controls the epstein-barr virus lytic switch. Mol. Cell 2020, 78, 653–669.e658. [Google Scholar] [CrossRef] [PubMed]
- Nazerian, K.; Witter, R.L. Properties of a chicken lymphoblastoid cell line from marek’s disease tumor. J. Natl. Cancer Inst. 1975, 54, 453–458. [Google Scholar]
- Reddy, E.P.; Reynolds, R.K.; Watson, D.K.; Schultz, R.A.; Lautenberger, J.; Papas, T.S. Nucleotide sequence analysis of the proviral genome of avian myelocytomatosis virus (mc29). Proc. Natl. Acad. Sci. USA 1983, 80, 2500–2504. [Google Scholar] [CrossRef]
- Watson, D.K.; Reddy, E.P.; Duesberg, P.H.; Papas, T.S. Nucleotide sequence analysis of the chicken c-myc gene reveals homologous and unique coding regions by comparison with the transforming gene of avian myelocytomatosis virus mc29, delta gag-myc. Proc. Natl. Acad. Sci. USA 1983, 80, 2146–2150. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).