Abstract
The 14-3-3 protein family plays an important role in regulating plant growth and development. The genes of the 14-3-3 family have been reported in multiple species. However, little is known about the 14-3-3 gene family in bamboo. In this study, a total of 58 genes belonging to the 14-3-3 family were identified in three representative bamboo species, i.e., Olyra latifolia, Phyllostachys edulis, and Bonia amplexicaulis, whose encoding proteins were grouped into ε and non-ε groups by phylogeny analysis with 14-3-3 proteins from Arabidopsis thaliana and Oryza sativa. The 14-3-3s had diverse gene structures and motif characteristics among the three bamboo species. Collinearity analysis suggested that the genes of the 14-3-3 family in bamboo had undergone a strong purification selection during evolution. Tissue-specific expression analysis showed the expression of Pe14-3-3s varied in different tissues of P. edulis, suggesting that they had functional diversity during growth and development. Co-expression analysis showed that four Pe14-3-3s co-expressed positively with eight ribosomal genes. Yeast two-hybrid (Y2H) assays showed that Pe14-3-3b/d could interact with Pe_ribosome-1/5/6, and qPCR results demonstrated that Pe14-3-3b/d and Pe_ribosome-1/5/6 had similar expression trends with the increase in shoot height, which further confirmed that they would work together to participate in the shoot growth and development of bamboo. Additionally, the transgenic Arabidopsis plants overexpressing Pe14-3-3b had longer roots, a larger stem diameter, an earlier bolting time and a faster growth rate than wild-type Arabidopsis, indicating that Pe14-3-3b acted as a growth promoter. Our results provide comprehensive information on 14-3-3 genes in bamboo and highlight Pe14-3-3b as a potential target for bamboo improvement.
1. Introduction
The 14-3-3 protein family was originally discovered and isolated from bovine brain [1]. In plants, the first 14-3-3 protein was isolated from Arabidopsis thaliana and named GF14 [2]. Subsequently, 14-3-3 proteins were detected in many plant species and named GF or GRF [2,3]. The latest research clearly confirmed that 14-3-3s were widely involved in regulating plant growth and development, such as cell elongation and cell division, seed germination and seed dormancy [4]. For example, in cotton (Gossypium hirsutum), 14-3-3 proteins were involved in cell elongation, which were highly induced during the rapid cell-elongation phase in fiber development [5]. In rice (Oryza sativa), the overexpression of OsGF14c delayed flowering, while its knockout mutants displayed early flowering [6]. In soybean (Glycine soja), GsGF14o overexpression caused deficits in root-hair formation and development [7]. In A. thaliana, the 14-3-3μ T-DNA insertion mutant, 14-3-3μ-1, had shorter roots than the wild type [8], and the overexpression of AtGRF9 favored the allocation of shoot carbon to roots and enhanced proton extrusion in the root-elongation zone and the root-hair zone [9]. In cassava (Manihot esculenta), 14-3-3 proteins participated in carbohydrate metabolism and starch accumulation during cassava root tuberization, and an overexpression of a cassava 14-3-3 gene increased the sugar and starch content of older leaves in A. thaliana [10]. However, the overexpression of Ta14-3-3 in A. thaliana inhibited the growth of primary roots [11]. Additionally, a prominent role of 14-3-3 proteins their involvement in regulating plant immunity against pathogens to influence plant growth and development. For example, a G. hirsutum 14-3-3 was rapidly expressed in response to Verticillium dahliae in a cultivar with enhanced wilt resistance [12], and in O. sativa, OsGF14c positively regulated immunity by modulating the protein homoeostasis of the GRAS protein OsSCL7 [13]. These results indicated the diverse function of 14-3-3 genes.
Furthermore, the interaction between 14-3-3 proteins and other proteins during plant growth and development has been reported in recent years. For instance, the GhGRF3/6/9/15 in cotton interacted with GhFT and GhFD to inhibit flowering, while GhGRF14 interacted with GhFT and GhFD to promote flowering [14]. In apples (Malus domestica), 14-3-3 proteins (MdGF14a, MdGF14d, MdGF14i, and MdGF14j) participated in the development of flowering through interaction with MdTFL1 and MdFT [15]. In rice roots, 14-3-3 proteins (GF14b, GF14c, GF14f and GF14e) interacted with root hair development-related proteins involved in regulating root growth [5]. In Arabidopsis, 14-3-3ε isoforms interacted with four seed storage proteins (At4g27170.1, At4g28520.1, At4g28520.2 and At1g07750) in the process of seed development [16]. A large number of 14-3-3 proteins and ribosomal proteins were found during the development of bud differentiation in Pinus pinaster, and the numbers increased with bud maturation [17]. It was found that 14-3-3 proteins could interact with p90 ribosomal S6 kinase (RSK), negatively regulating its activity to affect growth [18]. In Arabidopsis, most proteins identified in protein synthesis during seed development were ribosomal-related and could interact with the 14-3-3 isoforms χ and ε [16]. Additionally, studies have shown that 14-3-3 proteins could bind ADF4 (one protein of the actin-depolymerizing factor family) in regulating hypocotyl growth [19]. These results indicated that 14-3-3 proteins functioned together with other proteins during plants growth and development.
Moso bamboo (Phyllostachys edulis) is the most widely planted bamboo species in China with a large yield [20]. Moso bamboo is characterized by fast growth, with the ability to grow more than 1.0 m per day during the rapid-growth period and reaching its maximum height within one and a half months [21]. Before bamboo shoots were excavated, the number of nodes of bamboo culm were determined, and the growth of bamboo was mainly demonstrated by the growth of internodes after the excavation [22]. Cell division and cell elongation played an important role in the rapid growth of bamboo shoots [23], which were regulated by a series of factors, including hormones [18], proteins [24], transcriptional factors [25] and so on. Although 14-3-3 proteins play an important role in cell division and cell elongation during plant growth and development as an important regulatory protein [5,26], the role of the 14-3-3 family in bamboo is still unclear. In this study, members of the 14-3-3 gene family were identified in the genomes of Olyra latifolia, P. edulis and Bonia amplexicaulis, and further comprehensive analyses of molecular characteristics and the evolutionary relationship of 14-3-3 genes were conducted. Moreover, the function of Pe14-3-3b was validated by ectopic expression in A. thaliana, and a yeast two-hybrid (Y2H) assay. The results provided a comprehensive understanding of the 14-3-3 gene family in bamboo and revealed that Pe14-3-3b is a candidate gene for the improvement of bamboo breeding.
2. Results
2.1. Identification, Molecular Characteristics and Phylogenetic Analysis of 14-3-3 Family Members in Bamboos
A total of 58 members of the 14-3-3 gene family were identified (Table S1) in O. latifolia, P. edulis and B. amplexicaulis. Among the 14-3-3 genes of P. edulis, 15 encoding protein members with a complete 14-3-3 domain were named Pe14-3-3a to Pe14-3-3o; the remaining 21 members with an incomplete 14-3-3 domain were named as Pe14-3-3like1 to Pe14-3-3like21 (Table S1). Similarly, those eight genes identified in O. latifolia and 14 genes identified in B. amplexicaulis were named Ol14-3-3a to Ol14-3-3h and Ba14-3-3a to Ba14-3-3n, respectively (Table S1). The prediction of proteins encoded by Pe14-3-3s showed that they had different physical and chemical properties, including different protein lengths, ranging from 106 aa (Pe14-3-3like8 and Pe14-3-3like15) to 287 aa (Ba14-3-3l), and different molecular weights (MW), ranging from 11.77 kDa (Pe14-3-3like15) to 45.68 kDa (Pe14-3-3like10). Additionally, the theoretical isoelectric points (pI), subcellular localization and aliphatic index were also predicted to be different (Table S1). To understand the evolutionary relationships among the 14-3-3 proteins, an unrooted phylogenetic tree was constructed using Pe14-3-3a~Pe14-3-3o and those of other four plant species (A. thaliana, B. amplexicaulis, O. latifolia, and O. sativa) (Table S2). The results (Figure 1) showed that all the 14-3-3 proteins were obviously divided into ε and non-ε groups, including 12 and 46 members, respectively.
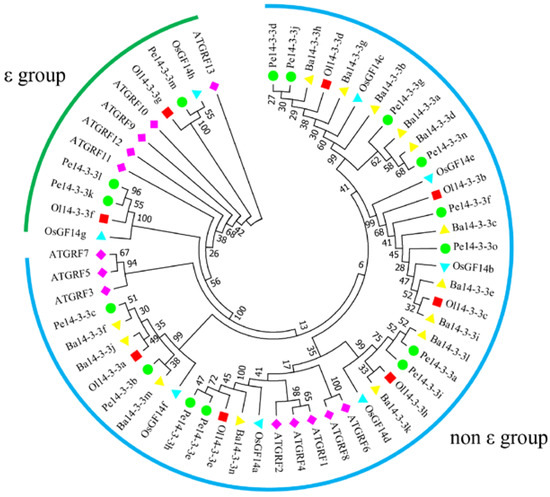
Figure 1.
Phylogenetic tree of 14-3-3s in five plant species (At: A. thaliana; Ba: B. amplexicaulis; Ol: O. latifolia; Os: O. sativa; Pe: P. edulis). The phylogenetic tree was constructed via the maximum-likelihood (ML) method (1000 bootstrap replicates) using MEGA 7.0 software. The different numbers at nodes represent the bootstrap values for homologous genes. Purple prismatic: A. thaliana; yellow triangle: B. amplexicaulis; red square: O. latifolia; blue triangle: O. sativa; green circle: P. edulis; green and blue arcs: ε and non-ε groups.
Furthermore, the gene structure analysis (Figure S1a) showed that Pe14-3-3a~Pe14-3-3o had 4~7 exons with differences in distribution and length, while Pe14-3-3like1~Pe14-3-3like21 had 3~12 exons. None of them had untranslated regions (UTR) structures except Pe14-3-3ike7, Pe14-3-3like8, and Pe14-3-3like12. Motif analysis (Figure S1b) demonstrated that the number of motifs in Pe14-3-3a~Pe14-3-3o ranged from 9 to 11, and their positions were roughly the same: all the members shared motif 1 to motif 6. While the motifs in Pe14-3-3like1~Pe14-3-3like21 ranged from 1 to 9, the location of each motif also varied greatly: Pe14-3-3like1 contained nine motifs, while Pe14-3-3like18 only contained motif 6 (Figure S1b). Differentially, the 14-3-3 members in O. latifolia and B. amplexicaulis showed different gene structures and motif characteristics with those members of P. edulis, whose members had 4~7 exons and 7 to 11 motifs (Figures S2 and S3).
2.2. Scaffold Localization and Collinearity Analysis of 14-3-3 Genes
The genome location analysis revealed that 14-3-3 genes were unevenly distributed on the scaffolds. The identified 14-3-3 genes were located in 8, 20 and 13 scaffolds of O. latifolia, P. edulis, and B. amplexicaulis, respectively. To fully elucidate the genetic evolution in bamboo, three typical circular maps were constructed by comparing 14-3-3 genes of P. edulis and other three plants (O. sativa, O. latifolia and B. amplexicaulis). The intraspecific collinearity results showed that 15 14-3-3 orthologous gene pairs were identified in P. edulis, and interspecific collinearity results revealed that 21, 27, and 40 14-3-3 orthologous gene pairs between P. edulis and O. sativa, O. latifolia and B. amplexicaulis (Figure 2a–c, Table S3) were identified, respectively, suggesting that Pe14-3-3s may have existed before the differentiation of the other three species. To explore the selection pressure of the 14-3-3 genes in these four plants, the Ka (non-synonymous substitution)/Ks (synonymous substitution) ratios of duplicated gene pairs were calculated. All Ka/Ks ratios of 14-3-3 orthologous gene pairs within P. edulis, and between P. edulis and the three other species, were less than 1 (Figure 2d, Table S4), indicating that these 14-3-3 genes had undergone a purifying selection during evolution [27].
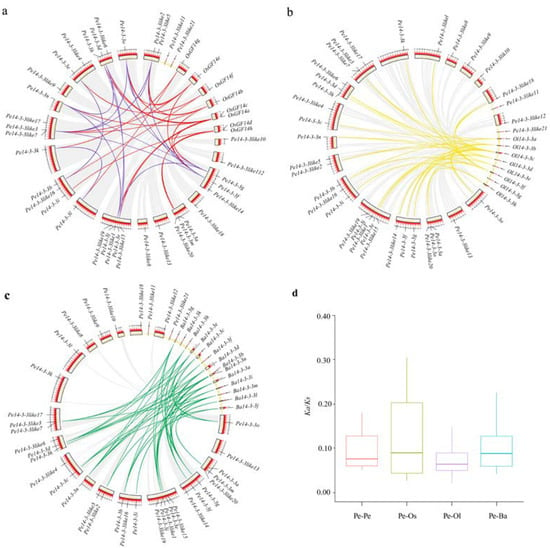
Figure 2.
Collinearity of 14-3-3 genes in P. edulis and four other plants (Ba: B. amplexicaulis; Ol: O. latifolia; Os: O. sativa; Pe. P. edulis). (a) Collinearity among Pe14-3-3s (blue lines) and that between Pe14-3-3s and Os14-3-3s (red lines). (b) Collinearity of Pe14-3-3s and Ol14-3-3s (yellow lines). (c) Collinearity of Pe14-3-3s and Ba14-3-3s (green lines). (d) Boxplot statistics of Ka/Ks values of the 14-3-3 orthologous gene pairs within P. edulis and those between P. edulis and other three species.
2.3. Tissue Expression Patterns of Pe14-3-3s in Moso Bamboo
To reveal the role of Pe14-3-3s, we investigated their spatiotemporal expression patterns based on the published transcriptome data [28], and an expression heatmap was drawn (Figure 3). The results showed that Pe14-3-3s exhibited a tissue-specific expression and were further divided into three groups. In group I, 16 Pe14-3-3s were nearly highly expressed in all the samples of moso bamboo and showed different expression patterns. For example, some members presented a constitutive expression pattern, such as Pe14-3-3g and Pe14-3-3i, suggesting that they played an important role in different tissues during growth and development. Some members, such as Pel4-4-3b/c/a/d/f/like1, had similar patterns both in roots and shoots at different growth stages, and were highly expressed in the top part of the roots, shoots and buds, suggesting that these genes played an important role in different growth stages, especially involved in the development of actively differentiated tissues. Meanwhile, Pe14-3-3k and Pe14-3-3l were highly expressed in specific tissues, including rhizome roots with a 0.5 cm length, the tip of bamboo shoots and the buds, suggesting that they might function in particular tissues. Of 36 genes, 5 Pe14-3-3s belonged to group II, which was only expressed in certain tissues and had a very low expression level, such as Pe14-3-3like7, which was expressed relatively higher in rhizome roots with a 10 cm length and leaf sheath. In group III, the remaining 14 Pe14-3-3s, except Pe14-3-3n, could hardly be detected in all tissues, suggesting that they may be functionally redundant genes.
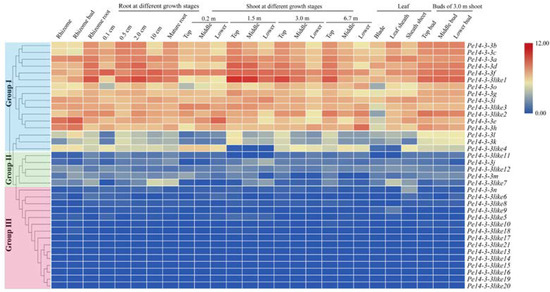
Figure 3.
Expression analysis of Pe14-3-3s in different tissues of moso bamboo. Different color in map represents log2-based fragments per kilobase per million (FPKM) values as shown in bar at right of figure. Pe14-3-3s, divided into three groups on the basis of their expression levels in different tissues, are presented at the left and the names of different tissues are shown on the top.
2.4. Co-expression Analysis of Pe14-3-3s
To further investigate the co-expression genes of Pe14-3-3s and the metabolic pathways they might be involved in, the prediction of co-expression analysis was performed with 16 Pe14-3-3s (group I family members), which were nearly expressed in all detected tissues. A total of 271 genes were found to be co-expressed with these Pe14-3-3s, in which the genes that participated in the ribosome pathway accounted for a higher proportion in biological processes based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (Figure 4a, Table S5). Furthermore, Gene Ontology (GO) enrichment analysis also found that these co-expressed genes participated in the ribosome pathway (Figure 4b, Table S6). Therefore, we speculated that these Pe14-3-3s may be involved in the ribosome pathway in P. edulis. Further correlation analysis showed six of sixteen Pe14-3-3 genes had a high correlation with most Pe_ribosomes in different height shoots of P. edulis (Figure 4c). Similarly, six of sixteen Pe14-3-3s also showed a great correlation with most Pe_ribosomes (Figure 4d) in the root of P. edulis. Finally, there were four Pe14-3-3s that had a strong correlation with eight Pe_ribosomes, both in the shoots and roots of P. edulis (Pearson’s correlation coefficients (PCC) > 0.75, p-value < 0.05)) (Table S7). Furthermore, the intra-group correlation analysis of these twelve genes (four Pe14-3-3s and eight Pe_ribosomes) also showed a strong correlation (Figure S4, Table S8).
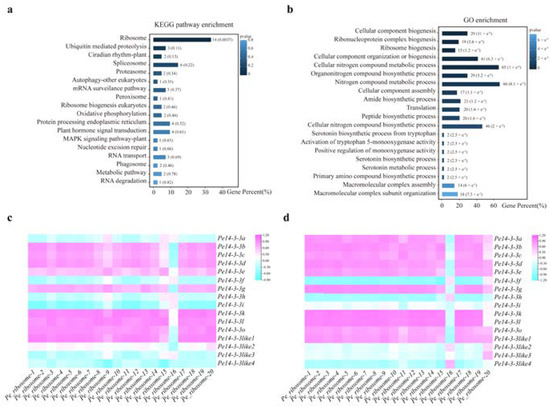
Figure 4.
Function and correlation analysis of the genes co-expressed with Pe14-3-3s involved in the ribosome pathway. (a) KEGG pathway enrichment analysis of the genes co-expressed with 16 Pe14-3-3s. (b) GO enrichment analysis of the genes co-expressed with 16 Pe14-3-3s. Low p-values are shown in dark blue and high p-values are depicted in light blue. (c) The correlation analysis between 16 Pe14-3-3s with genes that participate in the ribosome pathway in the different height shoots of P. edulis. (d) The correlation analysis between 16 Pe14-3-3s with genes participated in the ribosome pathway in the roots of P. edulis with different lengths. The gradual change color from blue to purple indicates negative to positive correlation.
2.5. Co-expression Network Construction and Validation of the Relationship between Pe14-3-3b/d and Pe_ribosomes
Based on the above analysis, a co-expression network was constructed with four Pe14-3-3s and eight Pe_ribosomes with a high correlation (Figure 5a). To further confirm how Pe14-3-3s played a role in the ribosome pathway, we checked the relationship between Pe14-3-3b/d and Pe_ribosome-1/4/5/6 using a targeted Y2H assay. As shown in Figure 5b, positive controls Pe14-3-3b/d and Pe_ribosome-1/5/6 co-transformed into yeast colonies could grow on both synthetic defined (SD)/-Leu/-Trp plates and synthetic defined (SD)/-Ade/-His/-Leu/-Trp/X-α-Gal plates, while the negative controls and the yeast colonies with other co-transformed genes only grew on the synthetic defined (SD)/-Leu/-Trp plates and did not grow on the SD/-Ade/-His/-Leu/-Trp/X-α-Gal plates. These results indicated that Pe14-3-3b/d could interact with Pe_ribosome-1/5/6 instead of Pe_ribosome-4 in yeast. Moreover, the qPCR results showed that Pe14-3-3b/d and Pe_ribosome-1/5/6 had similar up-regulated expression trends with the increase in bamboo shoot height (Figure 5c): this further supported the positive relationship between Pe14-3-3b/d with Pe_ribosome-1/5/6. Therefore, we speculated that Pe14-3-3b/d could interact with Pe_ribosome-1/5/6 and they could work together to participate in the ribosomal metabolic pathway and affect the growth and development of P. edulis.
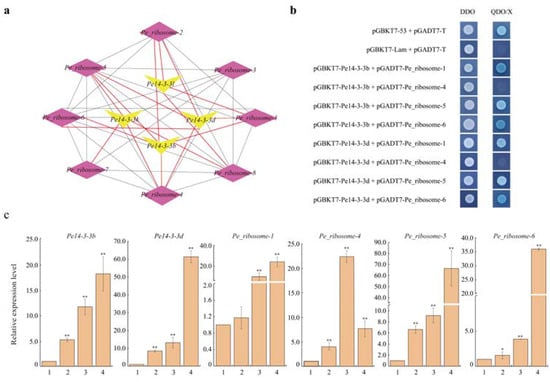
Figure 5.
Relationship validation of Pe14-3-3s and Pe_ribosomes. (a) The co-expression network of four Pe14-3-3s with eight Pe_ribosomes, which had a strong correlation (PCC > 0.75, p-value < 0.05). The black and red lines indicate the co-expression relationship within Pe_ribosomes and those between Pe_ribosomes and Pe14-3-3s, respectively. (b) Y2H assay between Pe14-3-3b/d and Pe_ribosomes. Positive control, co-transformation with pGBKT7-53 and pGADT7-T; negative control, co-transformation with pGBKT7-Lam and pGADT7-T; DDO: SD/-Leu/-Trp, QDO/X: SD/-Ade/-His/-Leu/-Trp supplemented with X-α-Gal. (c) Expression analysis of Pe14-3-3b/d and four Pe_ribosomes in shoots with different heights (1: 0.5 m, 2: 1.0 m, 3: 4.0 m, 4: 6.0 m; the asterisks indicate the significant difference between the relative expression levels in different height shoots and that in 0.5 m shoots, * p < 0.05 and ** p < 0.01).
2.6. Ectopic Expression of Pe14-3-3b in Arabidopsis
To further elucidate the possible biological roles of Pe14-3-3s, Pe14-3-3b was selected for the recombinant overexpression (OE) vector construction. Pe14-3-3b, driven by 35S, was transformed into Arabidopsis, meditated by Agrobacterium tumefaciens. A total of four hygromycin-resistant T3 plants were generated, of which three homozygous lines (OE-1, OE-2, and OE-3) were selected for further investigation. Under normal conditions, the root length of all transgenic lines was significantly longer than that of the wild type (WT) (Figure 6a–b), especially that of OE-3, wherein the mean root length increased by 58.94% compared with the WT after germination for nine days. Meanwhile, the bolting time of OE-1/2/3 was prior to that of the WT (Figure 6c), and the stem diameter of OE-1/2/3 was significantly larger than that of the WT (Figure 6d). The microscopic observation showed that OE-1/2/3 plants had larger cortical cells than those of the WT (Figure 6e). Additionally, the size of rosette leaves was bigger than those of the WT (Figure S5). Furthermore, qPCR analysis showed that the expression levels of Pe14-3-3b were significantly higher in OE-1/2/3 than that in the WT, especially in OE-3, which was 36.51 times of that in the WT. Similarly, compared with the WT, the expression levels of AtRPL30e, AtRPL4 and AtRPLS4A, which were homologous genes of Pe_ribosome-1/5/6, respectively, showed higher expression levels in OE-1/2/3 than those in the WT (Figure 6f). These results indicated that overexpressing Pe14-3-3b increased the expression of Pe_ribosomes, resulting in a promotion of the growth and development of transgenic Arabidopsis.
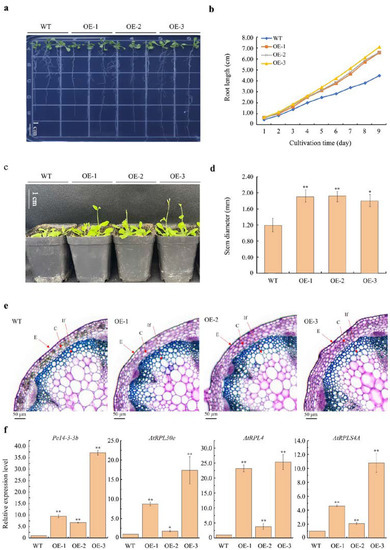
Figure 6.
Ectopic expression of Pe14-3-3b in Arabidopsis. (a) Phenotype of seedlings on vertical plate after germination for 9 days of transgenic lines and WT plants. Bars = 1 cm. (b) Root length changing after germination on 1/2 Murashige and Skoog (MS) medium of transgenic lines and WT plants. The mean length was calculated from 40 individuals. (c) The bolting time was earlier in transgenic lines than that of WT plants (the pictures were taken around 3 weeks after germination, Bars = 1 cm). (d) Stem diameter analysis after germination for 30 days of transgenic lines and WT plants. (e) The microscope observation of apical stem of transgenic lines and WT plants. Bars = 50 μm; E: epidemics; C: cortex; If: interfascicular fibers. (f) Relative expression level of Pe14-3-3b and three ribosomal genes (AtRPL30e, AtRPL4, and AtRPLS4A) in transgenic lines and WT plants (asterisks indicate the significant difference levels (* p < 0.05, ** p < 0.01).
3. Discussion
The 14-3-3 protein family, as a class of important factors for plant growth and development, have been widely studied in many eukaryotes [29]. Although the 14-3-3 gene family have been identified in multiple species, such as O. sativa, barley (Hordeum vulgare), soybean (Glycine max) and Arabidopsis [6,30,31,32], the molecular characteristics and evolution of 14-3-3 proteins in bamboo have not yet been studied. Therefore, the present study was conducted to identify the 14-3-3 gene family in O. latifolia, P. edulis and B. amplexicaulis, which represented diploid, tetraploid and hexaploid bamboo species. Based on the comprehensive analysis of the 14-3-3 gene family in bamboo, the function of Pe14-3-3b was further validated.
3.1. Characteristics Diversity of 14-3-3 Genes in Bamboos
In the present study, 36 Pe14-3-3s were identified in P. edulis, which was more than that identified in O. latifolia (8) and B. amplexicaulis (14) (Table S1). This might be due to the fact that the P. edulis genome had undergone gene duplication events [21]. The physical and chemical characteristics of 14-3-3s in different bamboo species indicated that they were diverse. For example, the amino acid composition of some members of Pe14-3-3s was acidic and some were alkaline, while the amino acid composition of both O. latifolia and B. amplexicaulis was acidic. Likewise, Pe14-3-3s had both hydrophilic and hydrophobic members, but the members of both O. latifolia and B. amplexicaulis were hydrophobic (Table S1). The structural diversity of exons/introns was generally considered to provide further insights into structural, evolutional and functional relationships [33]. In P. edulis, more than 60% of the members contained three or four introns (Figure S1a). Similar results were found in O. latifolia and B. amplexicaulis (Figures S2a and S3a).
In addition, the number of exons and introns in different branches were variable (Figure S1a, S2a), suggesting that evolution may be driving this structural diversity. This was also observed in O. sativa, Medicago truncatula and G. max [32,34,35]. Additionally, the intron lengths and arrangements were different in Pe14-3-3s, and similar results were also found in O. latifolia and B. amplexicaulis, further suggesting that the exon–intron structure could reveal the evolutionary diversity of the 14-3-3 gene family [33,36]. Furthermore, the conserved motifs of 14-3-3s in O. latifolia, P. edulis and B. amplexicaulis were highly variable (Figures S1b, S2b and S3b). The diverse motifs in the 14-3-3 family were the key structures of 14-3-3 proteins that could bind to many ligands [37], which directly affects the interaction of 14-3-3 proteins with other proteins to play a variety of functions.
3.2. Phylogeny and Evolution of 14-3-3 Proteins
Previously, genome duplication studies of 141 sequenced plant genomes demonstrated that, compared with A. thaliana, P. edulis was easier to evolve with O. sativa on shorter branches [28,38], and the relationship between P. edulis and O. sativa, O. latifolia and B. amplexicaulis was closer [39]. In our study, 37 14-3-3s from O. latifolia, P. edulis and B. amplexicaulis were clustered into ε and non-ε groups by phylogenetic analysis (Figure 1), which was consistent with previous reports on Arabidopsis, wheat (Triticum aestivum) and G. soja [7,30,40]. Our study also suggested the same phenomenon that most members of Pe14-3-3s, along with 14-3-3s in O. sativa, O. latifolia and B. amplexicaulis, clustered in close branches, while the evolutionary distance between P. edulis and A. thaliana was relatively far. It was crucial for us to infer the phylogeny and evolutionary direction of the 14-3-3 gene family between P. edulis and other plants (O. sativa, O. latifolia and B. amplexicaulis).
During evolution, plants undergo gene duplication events, among which whole-genome duplication and tandem duplication often promote the expansion of gene families [41,42], resulting in the diversity of gene function [43]. A previous study showed that P. edulis had undergone at least one round of whole-genome duplication, followed by multiple-segment duplication [21], which was considered to be the cause of the expansion of 14-3-3 family members in P. edulis during evolution. The differences of 14-3-3 gene members among three bamboo species might be mainly caused by the occurrence of genome-replication events [44], including series replication, such as fragment replication [45]. In our study, gene duplication and syntenic analyses of 14-3-3s supported this assumption as 15 segmental duplication pairs were found within Pe14-3-3s, and 21 pairs were found between Pe14-3-3s and Os14-3-3s (Figure 2a). Similarly, 27 and 40 pairs were found between Pe14-3-3s and Ol14-3-3s, and Ba14-3-3s, respectively (Figure 2c,d). In addition, the Ka/Ks ratio can be used to indicate the selecting direction of one gene and measure the historical choice of coding sequences [44,45,46,47]. In our study, the Ka/Ks ratios (Figure 2d, Table S4) showed that Pe14-3-3s had undergone a purification selection after duplication with limited functional divergence [27], which provided a better insight into the evolution of the 14-3-3 gene family.
3.3. Multiple Functions of 14-3-3s in Plant Growth and Development
The 14-3-3s are of vital importance during plant growth and development, which mainly manifests in reproductive and vegetative growth [4]. The function in reproductive and vegetative growth and expression patterns of 14-3-3s have been studied in many plant species [48,49,50,51,52], but the possible function of Pe14-3-3s remains unclear. Gene expression patterns are an important manifestation of gene function. The expression patterns of Pe14-3-3s in various organs, and at different plant growth stages in moso bamboo (Figure 3), suggested that they had different functions, which was consistent with previous studies [53,54]. In line with our results shown in Figure 6a,b, the overexpression of Pe14-3-3b in Arabidopsis showed a longer root length than WT plants. Usually, shoot and stem diameter correspond to the number of cortical cells, and the increased number of cortical cells could improve shoot and stem growth [55]. The Pe14-3-3b transgenic Arabidopsis plants showed a larger stem diameter than WT plants (Figure 6d), which was supported by the larger size, instead of the number of cortical cells in transgenic Arabidopsis plants. Therefore, we deduced that Pe14-3-3b may improve shoot growth by increasing the size of cortical cells. The 14-3-3 protein family could also contribute to the development of leaves [51]. Similar results were found in our study (Figure S5): an overexpression of Pe14-3-3b in Arabidopsis led to the formation of bigger rosette leaves than WT plants, suggesting that Pe14-3-3b could accelerate the development of rosette leaves.
Furthermore, 14-3-3s could combine with multiple proteins to perform their functions in plant growth and development. Proteomic analyses revealed that 14-3-3 proteins were ribosomal-related [17]. Our study confirmed that Pe14-3-3b/d could interact with Pe_ribosome-1/5/6, among which Pe_ribosome-1 was the homologue of 40S ribosomal protein (AT5G58420) with an important role in seed germination and seedling transition [56]. The qPCR results showed that, with the increase in shoot height, the expression levels of Pe14-3-3b/d and Pe_ribosome-1/5/6 all showed an up-regulated trend, consistent with previous studies, wherein 14-3-3 proteins could up-regulate the ribosome biogenesis [57]. Meanwhile, the expression level of Pe_ribosome-4 showed a trend of increasing first, and then decreasing with the increase in shoot height, which may be a cause of its inability to interact with 14-3-3 proteins. Moreover, higher expression levels of Pe14-3-3b and Pe_ribosomes homologue genes were found in the transgenic Arabidopsis than those in WT plants (Figure 6f), which further suggested that these Pe_ribosomes work together with Pe14-3-3b to play an important role in the growth and development of moso bamboo. However, how the Pe14-3-3b is involved in ribosome pathways to regulate the development of moso bamboo shoots needs to be further studied.
4. Materials and Methods
4.1. Plant Materials
The moso bamboo (Phyllostachys edulis) samples were taken from the Jiangxi Academy of Forestry; the 15th internode was collected from representative shoots with heights of 0.5 m, 1.0 m, 4.0 m and 6.0 m. After treating with liquid nitrogen, the samples were stored in the refrigerator at −80 °C for further processing.
4.2. Identification, Characteristics, and Phylogenetic Analysis of 14-3-3 Family Members in Bamboos
The 14-3-3 protein sequences of A. thaliana and O. sativa (Table S2) were used as queries against the genomes of O. latifolia, P. edulis and B. amplexicaulis in the database (http://bamboo. bamboogdb.org/, http://www.genobank.org/bamboo (accessed on 14 August 2021)) [58] with an E-value cut-off < 10−10. The HMM profile of the conserved domain of 14-3-3 (PF00244) was downloaded from the PFAM 32.0 database (https://pfam.xfam.org/ (accessed on 14 August 2021)) [59] and used to make sure that all protein sequences had 14-3-3 domain. The integrity of 14-3-3 conserved domain was determined using the online program SMART (http://smart.embl-heidelberg.de/ (accessed on 14 August 2021)) with an E-value < 0.1 [60]. The online ExPasy program (http://www.expasy.org/tools/ (accessed on 15 August 2021)) [61] was used to calculate the molecular characteristics of each 14-3-3 protein. The sequences of 14-3-3 proteins from A. thaliana, B. amplexicaulis, O. latifolia, O. sativa and P. edulis (Table S2) were used to construct an unrooted phylogenetic tree according to the maximum-likelihood (ML) method (1000 bootstrap alignment replicates) by running MEGA 7.0 [62] with ClustalW alignment. Proteins were classified according to the distance of homology with 14-3-3 proteins in A. thaliana [2]. Previously known 14-3-3 protein sequences from A. thaliana (13) and O. sativa (8) used in this study were retrieved from the Phytozome v.12 database (http://phytozome.jgi.doe.gov (accessed on 14 August 2021)) [63]. Gene structure and motif analyses were performed using GSD 2.0 (http://gsds.cbi.pku.edu.cn/ (accessed on 20 August 2021)) [64] and the Multiple EM for Motif Elicitation program v5.4.1 (https://meme-suite.org/meme/ tools/meme/ (accessed on 20 August 2021)) [65], respectively, which were visualized using TBtools [66].
4.3. Scaffold Localization and Collinearity Analysis of 14-3-3 Genes
The scaffold distribution and syntenic relationships of the 14-3-3 genes of P. edulis, O. sativa, O. latifolia and B. amplexicaulis were obtained by MCScanX and visualized using TBtools [66]. The TBtools software was also used to calculate the Ka (non-synonymous substitution)/Ks (synonymous substitution) ratios of paralogous pairs to deduce selective pressure.
4.4. Expression Pattern Analysis of 14-3-3 Genes in Moso Bamboo
The expressions of 14-3-3 genes were analyzed by using the available transcriptome data generated from different tissues of moso bamboo, including rhizomes, rhizome buds, rhizome roots, roots at different growth stages (0.1 cm, 0.5 cm, 2.0 cm, 10 cm), shoots with different heights (0.2 m, 1.5 m, 3.0 m, 6.7 m), leaves and shoot buds (Sequence Read Archive accession number: SRS1847048-SRS1847073) [28]. We retrieved the fragments per kilobase per million (FPKM) values representing the expression levels of Pe14-3-3s, then a heatmap was drawn using the TBtools software [67] to display the expression profiles of Pe14-3-3s.
4.5. Co-expression, KEGG, GO and Correlation Analysis of Pe14-3-3s
The BambooNET (http://bioinformatics.cau.edu.cn/bamboo/index.html/ (accessed on 23 August 2021)) [67] was used to carry out a co-expression prediction analysis of Pe14-3-3s. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses were performed on an online website (https://www.omicshare.com/tools/ (accessed on 24 August 2021)) for genes co-expressed with Pe14-3-3s. Based on the RNA-seq data of moso bamboo in different tissues (roots with different lengths and shoots with different heights), the correlation analysis was conducted by calculating pairwise Pearson’s correlation coefficients (PCC) and p-values on an online website (https://www.omicshare.com/tools/ (accessed on 24 August 2021)) [68] between Pe14-3-3s and Pe_ribosomes: the results were visualized with TBtools [66]. In addition, a co-expression network was constructed using Cytoscape 3.7.1 [69] based on the results of the intra-group correlation analysis of Pe14-3-3s and Pe_ribosomes. PCC > 0.75 and p-values < 0.05 were set in the co-expression relationship analysis.
4.6. RNA Extraction and qPCR Analysis
The total RNA of the samples from moso bamboo was extracted using RNA Plus (Jianshi, Shanghai, China) following the instructions, and was reverse transcribed into cDNA with a PrimeScript™ RT Reagent Kit (TaKaRa, Kyoto, Japan) according to the manufacturer’s instructions. The qPCR experiments were performed using SYBR Green chemistry (Roche, Mannheim, Germany) on a qTOWER 2.2 system (Analytik, Jena, Germany) according to the manufacturer’s directions. PeTIP41 was used as a reference gene for moso bamboo [70] to calculate the relative expression of the selected genes using the 2−∆∆CT method [71]. The specific primers were designed by the Primer 5.0 software (Jin Wang, Soochow, China) (Table S9).
4.7. Y2H Assay
Firstly, the full-length coding sequences of Pe14-3-3b/d and Pe_ribosome-1/4/5/6 were obtained using specific primers (Table S9) and cloned into pGBKT7 and pGADT7 vectors to form recombinant plasmids pGBKT7-Pe14-3-3s and pGADT7-Pe_ribosomes, respectively. According to the Yeast Protocols Handbook (Clontech, Mountain View, CA, USA), these combination constructs, including the positive controls of pGBKT7-53 and pGADT7-T, negative controls of pGBKT7-lam and pGADT7-T7 and the experimental groups of pGBKT7-Pe14-3-3s and pGADT7-Pe_ribosomes, were co-transformed into the yeast strain Y2HGold (Saccharomyces cerevisiae) and then plated on an SD solid-selection medium, including an SD/-Leu/-Trp medium and an SD/-Leu/-Trp/-His/-Ade/X-α-Gal medium, which were incubated at 30 °C until the appearance of colonies. Photographs were taken to record the growth of yeast colonies.
4.8. Transformation and Validation of Transgenic Arabidopsis Plants
The obtained CDS sequence of Pe14-3-3b was cloned into the overexpression (OE) vector Super1300 to form recombinant expression vector Super1300-Pe14-3-3b. The Super1300-Pe14-3-3b vector was introduced into the Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation using the floral dipping method [72]. Positive T1 transgenic plants were identified by PCR analysis and selected on a 1/2 MS solid medium with hygromycin (50 mg/mL); homozygous T3 seeds were used for subsequent experiments.
The seeds of wild-type (WT) Arabidopsis and transgenic Arabidopsis were surface-sterilized and seeded on a 1/2 MS solid medium. After vernalization, kept at 4 °C for two days, the seeds were transferred to an artificial climate chamber (temperature 23 °C, humidity 60–80%, light duration 16 h). About seven days later, the seedlings were transplanted into the nutrient soil (humus: vermiculite: 7:3) for further cultivation and subsequent experiment. For the measurement of root length, the seeds of transgenic lines and the wild type were transformed into vertical plates after germination for three days. The leaf phenotype was observed before bolting (around three weeks after germination). The stem diameter of all transgenic lines and WT plants were determined after germination for 30 days. Three technical and biological replicates were performed for all experimental data. The cDNA of transgenic lines and WT plants were used as templates for Pe14-3-3b expression analysis using AtActin2 as an internal reference [73]. The primer sequences are listed in Table S9.
4.9. Statistical Analysis
The statistical analyses were performed using IBM SPSS Statistics 22.0 (Armonk, NY, USA), and the mean and standard deviation of three biological replicates were presented. Significant differences were indicated at * p < 0.05, ** p < 0.01. For the measurement of the root length, 40 seedlings were statistically analyzed. For the measurement of the stem diameter, 30 seedlings were statistically analyzed.
5. Conclusions
In this study, we identified 8, 36 and 14 genes encoding 14-3-3 proteins from O. latifolia, P. edulis and B. amplexicaulis, respectively. The analyses of gene structures and conserved motifs indicated that 14-3-3s were diverse. Differentially expressed patterns of Pe14-3-3s in different tissues of P. edulis supported their diversity. A co-expression network of Pe14-3-3s revealed that most Pe14-3-3s were ribosomal-related. Pe14-3-3b/d could interact with Pe_ribosome-1/5/6 in yeast two-hybrid assays, and they had similar expression patterns in shoots with different heights, which suggested that Pe14-3-3s worked together with Pe_ribosomes in the growth of P. edulis. Moreover, the overexpression of Pe14-3-3b resulted in the promotion of Arabidopsis growth, which further supported that Pe14-3-3b might play an important role in the rapid growth of P. edulis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911221/s1.
Author Contributions
Conceptualization, D.G. and Z.G.; methodology, C.Z. and K.Y.; formal analysis, writing—original draft preparation, D.G.; software, Y.L.; validation, X.X. and Z.L.; supervision, Z.G.; project administration, Z.G.; funding acquisition, Z.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (Grant No. 2021YFD2200502). The funding agency was not involved in the design of the study, collection, analysis, interpretation of data and writing the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moore, B.W.; Perez, V.J. Specific acidic proteins of the nervous system. Physio.l Biochem. Asp. Neverous Integr. 1967, 343–359. [Google Scholar]
- Ferl, R.J.; Lu, G.; Bowen, B.W. Evolutionary implications of the family of 14–3-3 brain protein homologs in Arabidopsis thaliana. Genetica 1994, 92, 129–138. [Google Scholar] [CrossRef]
- De Vetten, N.C.; Ferl, R.J. Two genes encoding GF14 (14–3-3) proteins in Zea mays. Structure, expression, and potential regulation by the G-box binding complex. Plant Physiol 1994, 106, 1593–1604. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Yu, H.; Peng, J.; Hu, Z.; Chen, L. The role of 14–3-3 proteins in plant growth and response to abiotic stress. Plant Cell Rep. 2022, 41, 833–852. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, Y.; Li, Y.; Shao, S.; Li, B.; Shi, H.; Li, X. Interactome analysis of the six cotton 14–3-3s that are preferentially expressed in fibres and involved in cell elongation. J. Exp. Bot. 2010, 61, 3331–3344. [Google Scholar] [CrossRef]
- Purwestri, Y.A.; Ogaki, Y.; Tamaki, S.; Tsuji, H.; Shimamoto, K. The 14–3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 2009, 50, 429–438. [Google Scholar] [CrossRef]
- Sun, X.; Luo, X.; Sun, M.; Chen, C.; Ding, X.; Wang, X.; Yang, S.; Yu, Q.; Jia, B.; Ji, W.; et al. A Glycine soja 14–3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 99–118. [Google Scholar] [CrossRef]
- Mayfield, J.D.; Paul, A.L.; Ferl, R.J. The 14–3-3 proteins of Arabidopsis regulate root growth and chloroplast development as components of the photosensory system. J. Exp. Bot. 2012, 63, 3061–3070. [Google Scholar] [CrossRef]
- He, Y.; Wu, J.; Lv, B.; Li, J.; Gao, Z.; Xu, W.; Baluška, F.; Shi, W.; Shaw, P.; Zhang, J. Involvement of 14–3-3 protein GRF9 in root growth and response under polyethylene glycol-induced water stress. J. Exp. Bot. 2015, 66, 2271–2281. [Google Scholar] [CrossRef]
- Wang, X.; Chang, L.; Tong, Z.; Wang, D.; Yin, Q.; Wang, D.; Jin, X.; Yang, Q.; Wang, L.; Sun, Y.; et al. Proteomics profiling reveals carbohydrate metabolic enzymes and 14–3-3 proteins play important roles for starch accumulation during cassava root tuberization. Sci. Rep. 2016, 6, 19643. [Google Scholar] [CrossRef]
- Li, J.; Song, S.; Zhao, Y.; Guo, W.; Guo, G.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. Wheat 14–3-3 protein conferring growth retardation in Arabidopsis. J. Integr. Agr. 2013, 12, 209–217. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Robatzek, S. 14–3-3 proteins in plant-pathogen interactions. Mol. Plant Microbe Interact. 2015, 28, 511–518. [Google Scholar] [CrossRef]
- Lu, L.; Diao, Z.; Yang, D.; Wang, X.; Zheng, X.; Xiang, X.; Xiao, Y.; Chen, Z.; Wang, W.; Wu, Y.; et al. The 14–3-3 protein GF14c positively regulates immunity by modulating the protein homoeostasis of the GRAS protein OsSCL7 in rice. Plant Cell Env. 2022, 45, 1065–1081. [Google Scholar] [CrossRef]
- Sang, N.; Liu, H.; Ma, B.; Huang, X.; Zhuo, L.; Sun, Y. Roles of the 14–3-3 gene family in cotton flowering. BMC Plant Bio. 2021, 21, 162. [Google Scholar] [CrossRef]
- Zuo, X.; Wang, S.; Xiang, W.; Yang, H.; Tahir, M.M.; Zheng, S.; An, N.; Han, M.; Zhao, C.; Zhang, D. Genome-wide identification of the 14–3-3 gene family and its participation in floral transition by interacting with TFL1/FT in apple. BMC Genom. 2021, 22, 41. [Google Scholar] [CrossRef]
- Swatek, K.N.; Graham, K.; Agrawal, G.K.; Thelen, J.J. The 14–3-3 isoforms chi and epsilon differentially bind client proteins from developing Arabidopsis seed. J. Proteome Res. 2011, 10, 4076–4087. [Google Scholar] [CrossRef]
- Valledor, L.; Guerrero, S.; García-Campa, L.; Meijón, M. Proteometabolomic characterization of apical bud maturation in Pinus pinaster. Tree Physiol. 2021, 41, 508–521. [Google Scholar] [CrossRef]
- Cavet, M.E.; Lehoux, S.; Berk, B.C. 14–3-3beta is a p90 ribosomal S6 kinase (RSK) isoform 1-binding protein that negatively regulates RSK kinase activity. J. Biol. Chem. 2003, 278, 18376–18383. [Google Scholar] [CrossRef]
- Yao, H.; Li, X.; Peng, L.; Hua, X.; Zhang, Q.; Li, K.; Huang, Y.; Ji, H.; Wu, X.; Chen, Y.; et al. Binding of 14–3-3κ to ADF4 is involved in the regulation of hypocotyl growth and response to osmotic stress in Arabidopsis. Plant Sci. 2022, 320, 111261. [Google Scholar] [CrossRef]
- Cui, K.; He, C.; Zhang, J.; Duan, A.; Zeng, Y. Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo. J. Proteome Res. 2012, 11, 2492–2507. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.Y.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461.e4612. [Google Scholar] [CrossRef]
- Xiong, W.; Ding, Z.; Li, Y. Intercalary meristem and internodal elongation of bamboo plants. Scie. Silv. Sin. 1980, 2, 81–91. [Google Scholar]
- Wei, Q.; Guo, L.; Jiao, C.; Fei, Z.; Chen, M.; Cao, J.; Ding, Y.; Yuan, Q. Characterization of the developmental dynamics of the elongation of a bamboo internode during the fast growth stage. Tree Physiol. 2019, 39, 1201–1214. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Li, S.; Jing, Y.; Gu, L.; Zou, S.; Zhang, J.; Liu, B. Genome-wide identification, evolution and expression analysis of the aspartic protease gene family during rapid growth of moso bamboo (Phyllostachys edulis) shoots. BMC Genom. 2021, 22, 45. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, H.; Gao, Y.; Wang, Y.; Wu, M.; Xiang, Y. Genome-wide identification of growth-regulating factors in moso bamboo (Phyllostachys edulis): in silico and experimental analyses. PeerJ 2019, 7, e7510. [Google Scholar] [CrossRef]
- Grønlund, A.L.; Dickinson, J.R.; Kille, P.; Harwood, J.L.; Herbert, R.J.; Francis, D.; Rogers, H.J. Plant WEE1 kinase interacts with a 14–3-3 protein, GF14ω but a mutation of WEE1 at S485 alters their spatial interaction. Open Plant Sci. J. 2009, 3, 40–48. [Google Scholar] [CrossRef][Green Version]
- Hurst, L.D. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Z.; Wang, L.; Wang, J.; Wang, S.; Fei, B.; Chen, C.; Shi, C.; Liu, X.; Zhang, H.; et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience 2018, 7, giy115. [Google Scholar] [CrossRef]
- Aitken, A.; Collinge, D.B.; Van Heusden, B.P.; Isobe, T.; Roseboom, P.H.; Rosenfeld, G.; Soll, J. 14–3-3 proteins: A highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 1992, 17, 498–501. [Google Scholar] [CrossRef]
- Wu, K.; Rooney, M.F.; Ferl, R.J. The Arabidopsis 14–3-3 multigene family. Plant Physiol. 1997, 114, 1421–1431. [Google Scholar] [CrossRef]
- Schoonheim, P.J.; Sinnige, M.P.; Casaretto, J.A.; Bunney, T.D.; Quatrano, R.S.; De Boer, R.S. 14–3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 2007, 49, 289–301. [Google Scholar] [CrossRef]
- Wang, Y.; Ling, L.; Jiang, Z.; Tan, W.; Liu, Z.; Wu, L.; Zhao, Y.; Xia, S.; Ma, J.; Wang, G.; et al. Genome-wide identification and expression analysis of the 14–3-3 gene family in soybean (Glycine max). PeerJ 2019, 7, e7950. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H.Z. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Chen, F.; Li, Q.; Sun, L.X.; He, Z. The rice 14–3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 2006, 13, 53–63. [Google Scholar] [CrossRef]
- Qin, C.; Cheng, L.; Shen, J.; Zhang, Y.; Cao, H.; Lu, D.; Shen, C. Genome-wide identification and expression analysis of the 14–3-3 family genes in Medicago truncatula. Front. Plant. Sci. 2016, 7, 320. [Google Scholar] [CrossRef]
- DeLille, J.M.; Sehnke, P.C.; Ferl, R.J. The Arabidopsis 14–3-3 family of signaling regulators. Plant Physiol. 2001, 126, 35–38. [Google Scholar] [CrossRef]
- Yang, X.; Lee, W.H.; Sobott, F.; Papagrigoriou, E.; Robinson, C.V.; Grossmann, J.G.; Sundström, M.; Doyle, D.A.; Elkins, J.M. Structural basis for protein-protein interactions in the 14–3-3 protein family. Proc. Natl. Acad. Sci. USA 2006, 103, 17237–17242. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, K.; Li, G.; Li, Y.; Gao, Z. Identification and Expression Analyses of invertase genes in moso bamboo reveal their potential drought stress functions. Front. Genet. 2021, 12, 696300. [Google Scholar] [CrossRef]
- Shao, W.; Chen, W.; Zhu, X.; Zhou, X.; Jin, Y.; Zhan, C.; Liu, G.; Liu, X.; Ma, D.; Qiao, Y. Genome-wide identification and characterization of wheat 14–3-3 genes unravels the role of TaGRF6-A in salt Stress tolerance by binding MYB transcription factor. Int. J. Mol. Sci. 2021, 22, 1904. [Google Scholar] [CrossRef]
- Doyle, J.J.; Flagel, L.E.; Paterson, A.H.; Rapp, R.A.; Soltis, D.E.; Soltis, P.S.; Wendel, J.F. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 2008, 42, 443–461. [Google Scholar] [CrossRef]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Wu, S.; Yan, H.; Zhang, A.; Huang, L.; Yin, G.; Lee, S. Identification and characterization of the 14–3-3 gene family in switchgrass. Gene Mol. Res. 2016, 15, 10–4238. [Google Scholar] [CrossRef]
- Li, B.; Xiao, G.; Luo, K.; Wang, Z.; Mao, B.; Lin, X.; Guo, X. Overexpression of PvGF14c from Phyllostachys violascens delays flowering time in transgenic Arabidopsis. Front. Plant. Sci. 2018, 9, 105. [Google Scholar] [CrossRef]
- Dou, Y.; Liu, X.; Yin, Y.; Han, S.; Lu, Y.; Liu, Y.; Hao, D. Affinity chromatography revealed insights into unique functionality of two 14–3-3 protein species in developing maize kernels. J. Proteom. 2015, 114, 274–286. [Google Scholar] [CrossRef]
- Mayfield, J.D.; Folta, K.M.; Paul, A.L.; Ferl, R.J. The 14–3-3 Proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiol. 2007, 145, 1692–1702. [Google Scholar] [CrossRef]
- Lee, J.H.; Kwak, G.; Lim, Y.P.; Oh, M.H. 14–3-3 proteins contribute to leaf and root development via brassinosteroid insensitive 1 in Arabidopsis thaliana. Genes Genom. 2020, 42, 347–354. [Google Scholar] [CrossRef]
- Adams, E.; Diaz, C.; Hong, J.P.; Shin, R. 14–3-3 proteins participate in light signaling through association with PHYTOCHROME INTERACTING FACTORs. Int. J. Mol. Sci. 2014, 15, 22801–22814. [Google Scholar] [CrossRef]
- Daugherty, C.J.; Rooney, M.F.; Miller, P.W.; Ferl, R.J. Molecular organization and tissue-specific expression of an Arabidopsis 14–3-3 gene. Plant Cell 1996, 8, 1239–1248. [Google Scholar] [CrossRef]
- Xu, W.; Jia, L.; Shi, W.; Baluska, F.; Kronzucker, H.J.; Liang, J.; Zhang, J. The Tomato 14–3-3 protein TFT4 modulates H+ efflux, basipetal auxin transport, and the PKS5-J3 pathway in the root growth response to alkaline stress. Plant Physiol. 2013, 163, 1817–1828. [Google Scholar] [CrossRef]
- Guo, X.; Zhao, J.; Wang, R.; Zhang, H.; Xing, B.; Naeem, M.; Yao, T.; Li, R.; Xu, R.; Zhang, Z.; et al. Effects of graphene oxide on tomato growth in different stages. Plant Physiol Biochem. 2021, 162, 447–455. [Google Scholar] [CrossRef]
- Luo, T.; Zhang, Y.; Zhang, C.; Nelson, M.N.; Yuan, J.; Guo, L.; Xu, Z. Genome-wide association mapping unravels the genetic control of seed vigor under low-temperature conditions in rapeseed (Brassica napus L.). Plants 2021, 10, 426. [Google Scholar] [CrossRef]
- Trembley, M.A.; Berrus, H.L.; Whicher, J.R.; Humphrey-Dixon, E.L. The yeast 14–3-3 proteins BMH1 and BMH2 differentially regulate rapamycin-mediated transcription. Biosci. Rep. 2014, 34, e00099. [Google Scholar] [CrossRef]
- Guo, Z.H.; Ma, P.F.; Yang, G.Q.; Hu, J.Y.; Liu, Y.L.; Xia, E.H.; Zhong, M.C.; Zhao, L.; Sun, G.L.; Xu, Y.X.; et al. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos. Mol. Plant 2019, 12, 1353–1365. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, H.; Xu, W.; You, Q.; Yan, H.; Gao, Z.; Su, Z. Co-expression gene network analysis and functional module identification in bamboo growth and development. Front. Genet. 2018, 9, 574. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation coefficients: Appropriate use and interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PloS One 2013, 8, e56573. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Alazem, M.; He, M.H.; Moffett, P.; Lin, N.S. Abscisic acid induces resistance against bamboo mosaic virus through argonaute2 and 3. Plant Physiol. 2017, 174, 339–355. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).