1. Introduction
Tuberous sclerosis complex (TSC) is an autosomal dominant inherited genetic disorder characterized by multiple organ lesions, epilepsy, and neuropsychiatric manifestations. Its estimated frequency in the population is about one in 6000 to 10,000, of whom approximately two-thirds are sporadic, with the remainder segregated in families [
1]. TSC lesions are composed of disarranged undifferentiated dysplastic cells. In some lesions, these dysplastic cells do not grow more rapidly than normal cells (hamartia); but in other lesions, they grow into benign tumors (hamartomas) or rarely grow into malignant tumors, such as renal cell carcinoma [
2]. The representative hamartia of TSC is brain cortical tubers, whose disease name is derived from macroscopic pathological findings, described by Bourneville as “tuberous sclerosis of the cerebral convolutions” [
3]. In addition, representative hamartomas are facial angiofibromas (AF) and renal angiomyolipomas (AML).
In practice, entire body organs are involved in patients with TSC, including the brain, skin, retina, lung, liver, kidney, heart, pancreas, intestines, endocrine system, bone, and teeth [
4], and the established clinical diagnostic criteria are widely used [
1]. All hamartias are embryonic lesions, whereas hamartomas appear in an age-dependent fashion. TSC is also associated with epilepsy and various neuropsychiatric symptoms, called tuberous sclerosis-associated neuropsychiatric disorders (TAND) [
5]. TAND includes many psychiatric disorders and symptoms, such as intellectual disability, learning disorders, autism spectrum disorder, attention deficit hyperactivity disorder, higher brain dysfunction, mood disorders, and so on. Despite the diverse lesions and symptoms that can occur with TSC, their combination and severity in individual patients vary widely between families or even within a family. Some patients are diagnosed early in life, because of developmental delay and epilepsy, while others are diagnosed in adulthood with renal AML as the chief complaint and without any neuropsychiatric symptoms. This diversity makes it challenging to understand the overall picture of TSC.
The disease is caused by loss of function mutations of one of two genes,
TSC1 and
TSC2, which encode hamartin and tuberin, respectively [
6,
7]. These proteins form a functional heterodimer. This intracellular complex (TSC complex) is a universal sensor of several intracellular and extracellular signals, e.g., growth factor stimulation, hypoxia, and energy levels [
8]. The most remarkable function of the TSC complex is the GTPase activating protein (GAP) activity for Ras homolog enriched in the brain (Rheb), which converts Rheb-GTP to Rheb-GDP. While Rheb-GTP activates the mechanistic target of rapamycin kinase (MTOR), especially in the mTORC1 complex. In cells lacking proper functioning of the TSC complex, there are increased levels of Rheb-GTP, which leads to activation of mTORC1, causing increased cell size and growth through phosphorylation of eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), ribosomal protein S6 kinase beta-1 (S6K1), and eukaryotic translation initiation factor 2 (eEF-2K) [
8,
9,
10].
TSC1 and
TSC2 gene mutation patterns are also diverse, with no mutational hotspots in relatively large-size genes. Moreover, adding to point mutations, large deletions, splicing abnormalities due to deep intronic mutations, and mosaic mutations for all these mutations are included in a non-negligible proportion. This makes the genetic diagnosis of TSC difficult. To date, several large-scale TSC mutation studies have been completed worldwide. Several aspects of the molecular genetics of TSC have been historically elucidated through these studies. The overall mutation distribution among patients with TSC shows a higher frequency of
TSC2 than
TSC1 [
11]. Pathogenic missense mutations in
TSC1 are rare [
12].
TSC2 mutations produce a more severe phenotype than
TSC1 mutations, particularly in intercultural disability [
12,
13]. Some
TSC2 missense mutations (i.e., p.Arg905Gln and p.Gln1503Pro); however, not all missense mutations are associated with milder disease phenotypes [
14,
15], and
TSC2/
PKD1 contiguous gene deletion is associated with polycystic kidney disease [
16].
After that, TSC research progressed further, with the launch of the tuberous sclerosis registry to increase disease awareness (TOSCA) global patient registry, which published baseline data of 2093 patients in 2017 [
17]. In Japan, we analyzed 57 patients with TSC in 2013 [
18]. In this study, the frequency of patients with a
TSC1 mutation was higher than that reported in previous studies, suggesting that differences in the prevalence of diagnostic brain imaging, due to differences in medical insurance systems, may result in the diagnosis of more patient populations with mild TSC. However, a more detailed analysis was impossible, due to an insufficient number of patients. Since then, we have continued to perform genetic tests of TSC for clinical purposes, receiving patient specimens from hospitals all over Japan. We have also developed a new analysis method that can adapt to various types of mutations using a single platform of next generation sequencing (NGS), named combined long amplicon sequencing (CoLAS) [
19,
20]. In this study, we analyzed the phenotype and genotype details of 283 TSC patients from 260 families, to clarify the overall picture of TSC in Japan.
3. Discussion
3.1. Patient Population
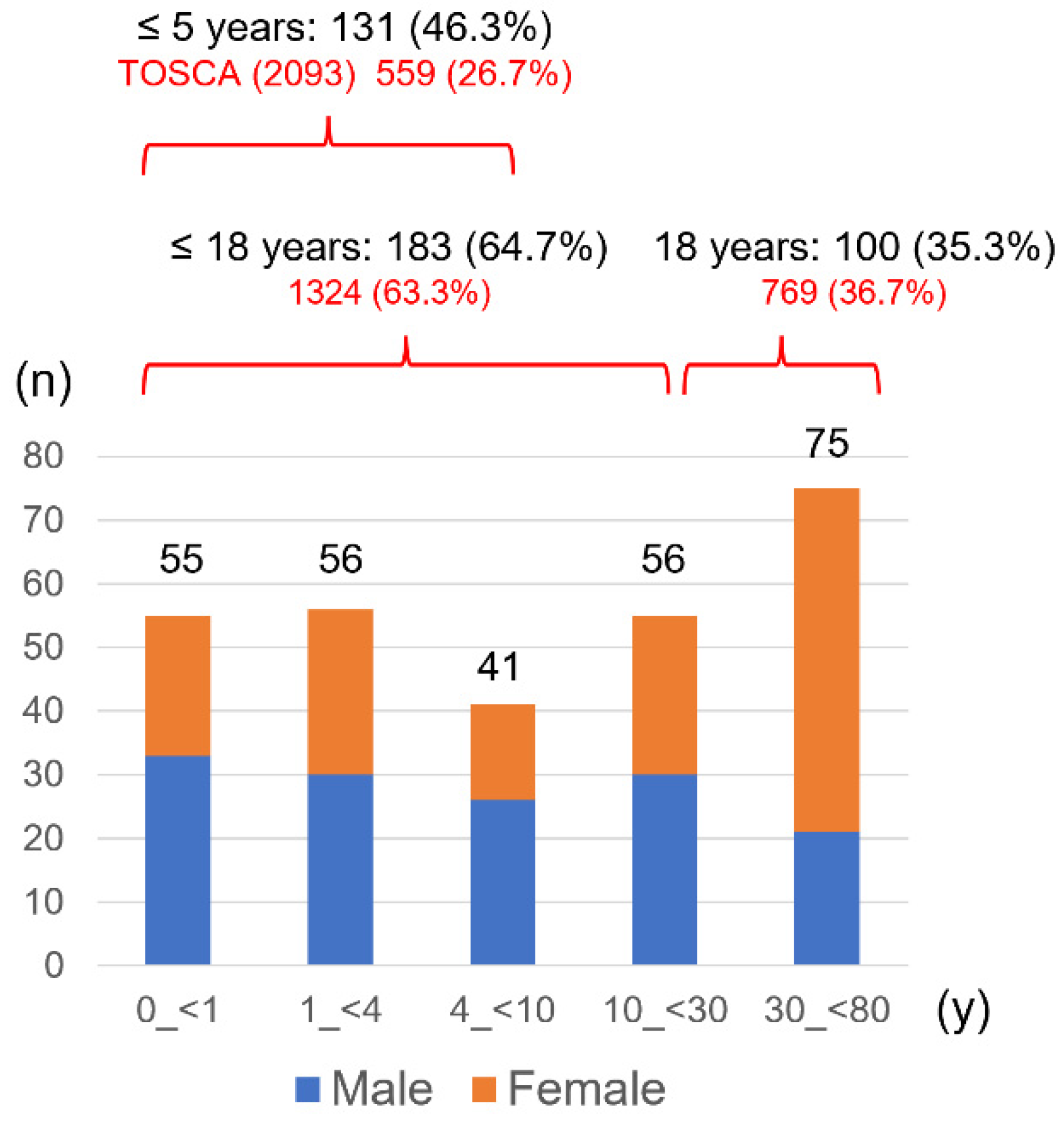
This study obtained clinical symptoms from patients who underwent genetic testing. Notably, there was a selection bias because of the recruited patients who required testing. Compared to the TOSCA baseline data, a higher proportion of infants and toddlers were included in this study, as patients aged 5 years or younger accounted for 46.3% vs. 26.7% for TOSCA (
Figure 1). The need for genetic testing in infants and toddler lies in estimating developmental prognosis and determining heritability when parents wish to have a second child. Genetic testing is also indicated for a neonate with cardiac rhabdomyomas, to provide the necessary treatment, as the diagnosis cannot be confirmed clinically because of no other TSC lesions. This study included six patients with such a situation, and gene mutations were detected in five of them, one
TSC1 and four
TSC2. Cardiac rhabdomyomas were found in at least 50% of TSC and grew during late pregnancy, influenced by maternal hormones, but some showed spontaneous regression after birth. In recent years, the number of diagnoses in the fetal period has increased along with technological advances in fetal ultrasound [
30]. In most cases, it is asymptomatic and does not require treatment, but it is a lesion that can cause perinatal to neonatal death and is one of the emergency states of TSC [
31]. The treatment with mTOR inhibitors (everolimus, sirolimus) is effective for cardiac rhabdomyomas of TSC [
32,
33], and the Japanese Society of Tuberous Sclerosis has published an “Expert opinion consensus for everolimus treatment” (
http://jstsc.kenkyuukai.jp/information/information_detail.asp?id=98767 (in Japanese), last accessed on 27 August 2022).
On the other hand, adult patients in this study include those whose diagnosis could not be confirmed clinically, who wished to have children, and obtained positive results from genetic testing after mutations were identified in their children. Despite these biases, the proportions of patients aged 18 years and younger and older than 18 years were similar to those in TOSCA, indicating that this study included patients of a wide age range and reflected the actual distribution of TSC patients in Japan. In Japan, the genetic testing for TSC was covered by national insurance from April 2022, and a clinical laboratory company started accepting samples. Prior to that, facilities that could perform genetic testing were limited, and our facility had more than 90% of the domestic share. This is thought to be the reason why we were able to recruit patients from a wide range of populations.
3.2. Prevalence of Lesions and Symptoms in Patients with TSC
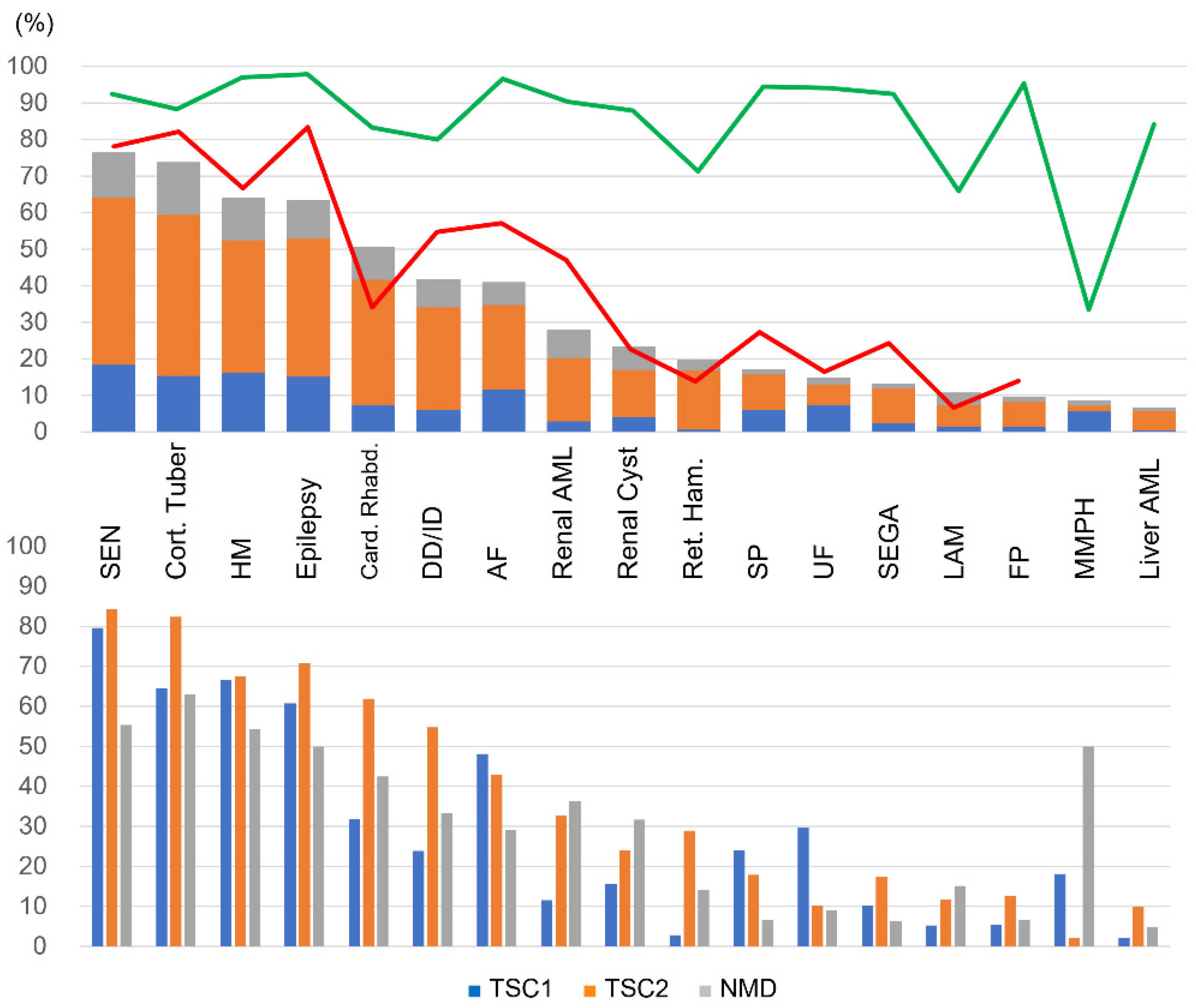
The prevalence of each lesion and symptom in definite TSC patients in this study was roughly similar to that in the TOSCA baseline data (
Figure 2, upper panel). However, the age distribution of patients and the ratio of
TSC1 and
TSC2 were different from that of the TOSCA, the differences of prevalence in some lesions were statistically significant in the overall comparison. A low frequency of AF, SP, and renal AML, and a high frequency of cardiac rhabdomyomas were not observed in patients older than 10 years. A low frequency of DD/ID and cortical tuber disappeared in patients with
TSC2 mutations, and a low frequency of SEGA disappeared in patients with both
TSC1 and
TSC2 mutations. These results reflected the higher percentage of young and TSC1 patients in our study population. On the other hand, the prevalence of epilepsy was consistently low, even after adjusting for age and genes, suggesting that the prevalence of epilepsy in this study is low for different reasons. In the TOSCA epilepsy study, 83.6% (1852/2216) of patients had epilepsy, of which 38.9% presented with infantile spasm (mean age at diagnosis was 0.4 year) and 67.5% with focal seizures (2.7 year) [
34]. This suggests that in the TOSCA study, there are many young patients diagnosed with TSC using epilepsy as the initial symptom. On the other hand, in a previous study by Wataya-Kaneda et al. [
21], which mainly targeted adult patients visiting a dermatologist, the prevalence of epilepsy was 63%, which is almost consistent with the present study. Compared with TOSCA, it seems that more patients were suspected to have TSC from symptoms other than epilepsy in this study, such as cardiac rhabdomyomas and HM in infants, AF in childhood, and renal AML in adulthood. While many patients were diagnosed as clinically definite TSC using brain imaging. In fact, in this study, brain imaging was performed in 100 (87.7%) of 114 patients without epilepsy, more than 95% chose MRI for the imaging test modality, and cortical tuber and/or SEN was detected in 56 patients (
Supplementary Table S1). Indeed, it is known that the adoption rate of CT/MR units in clinics and hospitals in Japan is the world’s best [
35].
High surveillance testing rates were shown for most lesions, but retinal hamartoma was slightly lower. Regarding pulmonary involvement, LAM had a low testing rate, and MMPH was the most underreported, probably due to the low disease awareness.
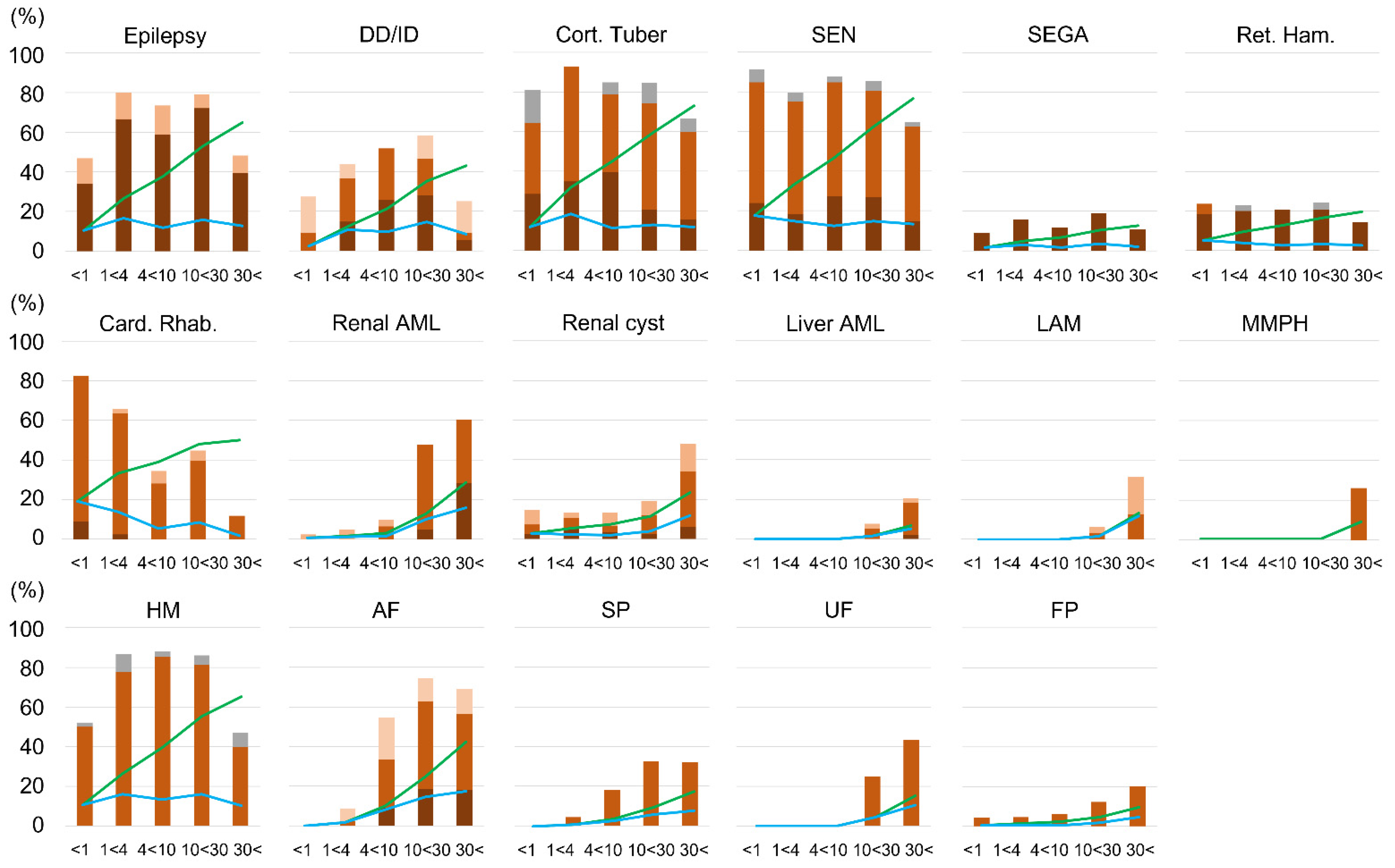
The appearance of lesions in TSC was age-dependent, but lesions of the central nervous system (cortical tuber, SEN, SEGA, retinal hamartoma) and HM were fetal lesions, which were consistently expressed after birth and did not change their frequency with age. As cardiac rhabdomyomas can regress spontaneously, the incidence decreased with age and leveled off after 10 years of age. Other skin and visceral lesions increased in frequency with age. Among these, AF and SP rose sharply after the age of 4. Renal AML and UF then rose after 10 years of age. Liver AML, LAM, and MMPH were even later, rising after 20 years of age, and the occurrence was low. Renal cysts and FP were observed in the neonatal period, and although they remained constant in childhood, the frequency increased from adolescence to adulthood. Moreover, the proportion of patients with severe lesions also increased in lesions that increased in frequency with age (
Figure 3). These results generally agreed with past reports [
4]. However, in this study, the timing of increases in AF and renal AML was not the same. The timing of increases in AF and SP was almost the same, and the increase in renal AML occurred later, similarly to UF rather than AF.
Lesions that increase with age are more likely to develop loss of heterozygosity (LOH). LOH is most frequent in renal AML and less common in central nervous system lesions [
36]. Additionally, some LAMs have been shown to occur as metastatic lesions of the renal AML [
37]. Such differences in onset mechanisms, haploinsufficiency, LOH, and metastasis may determine the timing of onset of each lesion. As with LAM, the possibility of metastatic lesions from renal AML should be considered for liver AML. In fact, of the 17 patients with liver AML, 15 had renal AML, and eight had a score of 3 for severe renal AML (
Supplementary Table S1). Although there are few reports on MMPH, we previously reported that LOH occurs in lesions [
38].
3.3. TSC Mutation Spectrum
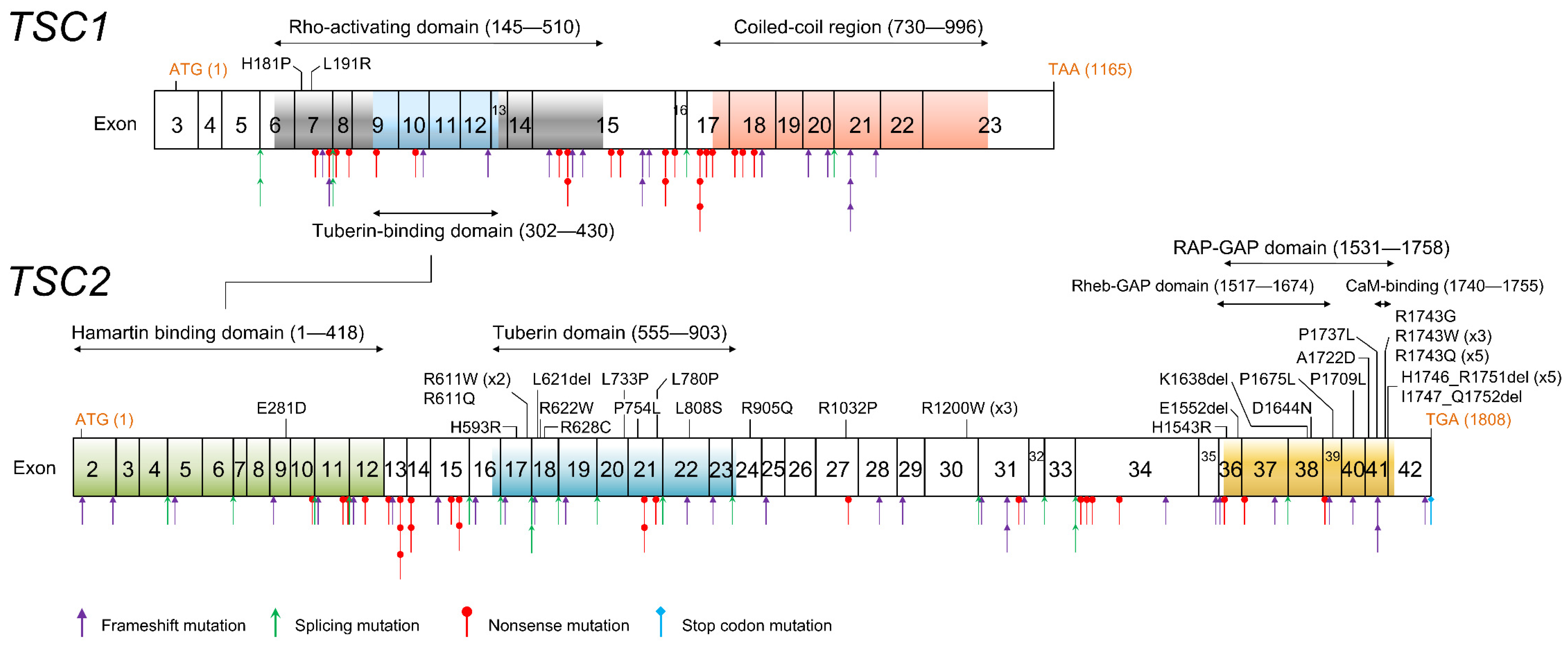
Both
TSC1 and
TSC2 showed various gene mutation patterns. Characteristically, mutations predicted to have protein production were few in
TSC1, but account for about one-third in
TSC2 (
Table 4).
TSC2 missense and in-frame mutations were concentrated in two functional domains; the tuberin domain and the RAP-GAP domain (
Figure 4). The tuberin domain comprises alpha helices connected by loops, in an arrangement similar to the alpha–alpha superhelix domain. The tuberin domain is thought to stabilize the structure of the tuberin protein by forming a salt bridge between a positively charged amino acid residue on one alpha helix and a negatively charged amino acid residue on another alpha helix [
39]. The TSC complex acts as a GTPase activating protein (GAP) toward a small G-protein GTPase Ras homolog expressed in the brain (RHEB). The RAP-GAP domain of tuberin promotes this reaction and inhibits the RHEB–GTP-dependent stimulation of the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) [
40,
41]. The broad sense of the RAP-GAP domain (amino acids residue 1531–1758) overlaps the core RHEB-GAP domain (1517–1674) and includes a calmodulin (CaM) binding domain (1740–1755) (
Figure 4). All C-terminal
TSC2 missense mutations are contained in this region.
Compared with TOSCA baseline data and previous studies, this study’s proportion of TSC1 patients was significantly higher (
Table 5). This trend was also noted in our previous report [
18], and this study confirmed that more TSC1 patients were diagnosed in Japan. It is unlikely that the regional differences in the frequency of TSC1 patients mean that the barriers to brain imaging diagnosis were low in Japan for national medical insurance, so it is likely that TSC patients with milder phenotypes were more frequently diagnosed. Conversely, there might be more undiagnosed TSC1 patients worldwide. Moreover, the prevalence of epilepsy in TSC2 patients was lower than in the TOSCA study, which indicated that TSC2 patients with milder central nervous system symptoms were diagnosed more frequently in Japan.
3.4. Genotype-Phenotype Correlations
The prevalence of DD/ID was significantly lower in TSC1 than in TSC2 patients (
Table 6). However, this significant difference disappeared in patients aged 20 years or older. In this age group, the prevalence of DD/ID in patients with TSC1 was increased, and the prevalence of patients with TSC2 was decreased. The TSC1 patients with severe neuropsychiatric symptoms continued their treatment, while TSC2 patients without neuropsychiatric symptoms with renal AML as the major chief complaint were increased. In addition, there are significant differences in the prevalence of some lesions between TSC1 and TSC2 patients. Retinal hamartoma was more common in TSC2 patients, and only two of the 28 cases were TSC1 patients. Conversely, more MMPH was found in TSC1 patients, and only one of the seven was a TSC2 patient [
42]. Cardiac rhabdomyomas were also found in TSC1 patients, but were approximately twice as common in TSC2 patients.
UF was more frequent in TSC1 patients and more than three-times as common as in TSC2 patients. This significant difference was maintained even in the group of 10 years and older, and the p-value was at the threshold (0.0579) in the 20 years and older. Likely, the TSC1 significance in UF was not due to the age difference between the groups, which could be a subject for future investigation. Although the severity of UF was not assessed in this study, a previous report showed severe squamous cell carcinoma-like UF in patients with a
TSC1 frameshift mutation [
43].
Comparing the severity of symptoms (
Table 7), DD/ID and renal AML were significantly more severe in TSC2. Regarding epilepsy, no difference in the prevalence was observed between TSC1 and TSC2 patients, but the percentage of patients with residual seizures was significantly lower in TSC1 patients aged ≥10 years. These results suggested that epilepsy in TSC1 patients was more likely to go into remission. Similarly, there was no difference in the prevalence of AF overall. Nevertheless, the proportion of patients with severe AF lesions (score 3 in Table 13) was significantly higher in TSC2 patients aged ≥10 years. Comparing the age group ≥10 years, score 3 DD/ID and renal AML were about ten times higher in TSC2 patients than in TSC1 patients and more than five-times higher in AF (
Table 7).
Previous studies have not shown a clear correlation between the type of TSC2 mutation and disease severity. In this study, comparing TSC2 mutations expected to produce a protein (PP) and mutations predicted to produce truncated or no protein (NP), AF and renal AML were significantly more severe in patients with NP mutations. There were no significant differences in epilepsy and DD/ID, but there were significantly more patients with NP who had six or more SENs. A comparison of TSC1 and TSC2 patients showed a higher number of cortical tubers in TSC2 patients. In contrast, a comparison of TSC2 PP and NP showed no significant differences of cortical tubers. It is suggested that the PP mutation of TSC2 might be a factor that predicts the milder phenotypic consequences in AF and renal AML, but not neuropsychiatric symptoms, and further analysis using more patients will be necessary in the future.
Four patients with
TSC2/
PKD1 contiguous gene deletion syndrome were detected in this study. Of these, only one patient, 42 years old, presented with classic polycystic kidney disease, and it should be noted that young patients do not always develop polycystic kidney disease. None of the four cases were accompanied by DD/ID, and two were a mother and her daughter, indicating that contiguous
TSC2/
PKD1 deletion does not necessarily lead to severe neuropsychiatric symptoms. In the literature, there are reports of familial cases with mild neuropsychiatric symptoms [
44,
45] and cases with severe mental retardation [
46,
47]. Conversely, point mutations of
TSC2 were detected in four out of eight patients with classic polycystic kidney disease (
Supplementary Table S1). This differentiation is important, because
TSC2/
PKD1 deletion carries the risk of polycystic kidney disease 1, PKD1 (OMIM# 173,900), associated with symptoms other than polycystic kidney disease, namely cerebral aneurysms and valvular heart disease; lesions not associated with TSC.
One patient had both TSC1 and TSC2 mutations (family #251), although the severity of this patient could not be said to be higher than other patients. Mutations in both genes were also detected in five patients in the TOSCA study, suggesting that a small number of such patients are likely to exist in reality. Future investigations are required, to determine whether the symptoms of such patients will become severe or remain unchanged. On the other hand, neither our study nor others reported patients with TSC1 or TSC2 germline mutations in both alleles simultaneously. When one cell has TSC mutations in both alleles, it is thought that it will become tumorigenic, and such a zygote is envisioned to be embryonically lethal.
3.5. Mosaic Mutations
Of the 200 mutations detected in this study, 17 (8.5%) were mosaic mutations (
Table 9). As previously described, this was detected during mutation screening in the proband. In this case, mutations with variant allele frequencies (VAF) of 10% or more were mainly detected. However, in some cases, mosaic mutations with a lower frequency can also be seen (
Figure 5). Estimated VAF is based on NGS variant callers, but in the case of large deletions, VAF could not be estimated, due to the amplification bias of PCR. Moreover, parents were tested and found to have mosaic mutations after the mutation was identified in a proband. In this situation, a single-site test was used, and it was possible to analyze lower-frequency mosaic mutations. Similarly, the mutation was detected in a patient-derived tumor specimen and whether the same mutation was also present in the blood was investigated. Four cases, three with SEGA (Family# SS, 191 and 231) and one with subcutaneous angiofibroma (Family #258), were included in this study.
Regarding SEGA, differences between sporadic SEGA and TSC have been discussed. However, patients with somatic
TSC2 mutations limited to tumors and those with the low-frequency mosaic mutation also detected in blood were reported [
28,
29]. Ultimately, these patients can be interpreted as a spectrum, depending on when the mosaic mutations occur during fetal development. The same may be true for sporadic LAM, which is usually considered to have no mutations other than renal AML and LAM. That is a topic for future investigation.
It is essential to consider the detection limit when analyzing mosaic mutations. Very low-frequency mosaic mutations can be detected by increasing the read depth using NGS, but this is not always true. When preparing the library, the amount of genomic DNA is the first thing to consider. A human cell contains about 6 pg of DNA. Therefore, 60 ng of DNA corresponds to 10
4 cells. If 0.1% of the cells have a mosaic mutation, there will be just 10 copies of the mutated allele in 60 ng of DNA, and 0.01% of mosaics will have a single copy. Therefore, lower frequency mosaics are theoretically undetectable. The next problem is distinguishing between artifacts due to PCR errors during library preparation and real low-frequency mosaic variants. Previously, we examined dual sequencing of NGS and extracted common low-frequency mosaic calls, then removed common artifacts caused by sequence-dependent PCR errors [
19]. This method had a detection sensitivity of 85.2% and a positive predictive value of 96.6% for the experimentally synthesized 10% mosaic, and 69.0% and 95.8% for the 1% mosaic. When the mosaic rate was 10% (VAF 5%) or more, the mutation peak could usually be confirmed by Sanger sequencing (
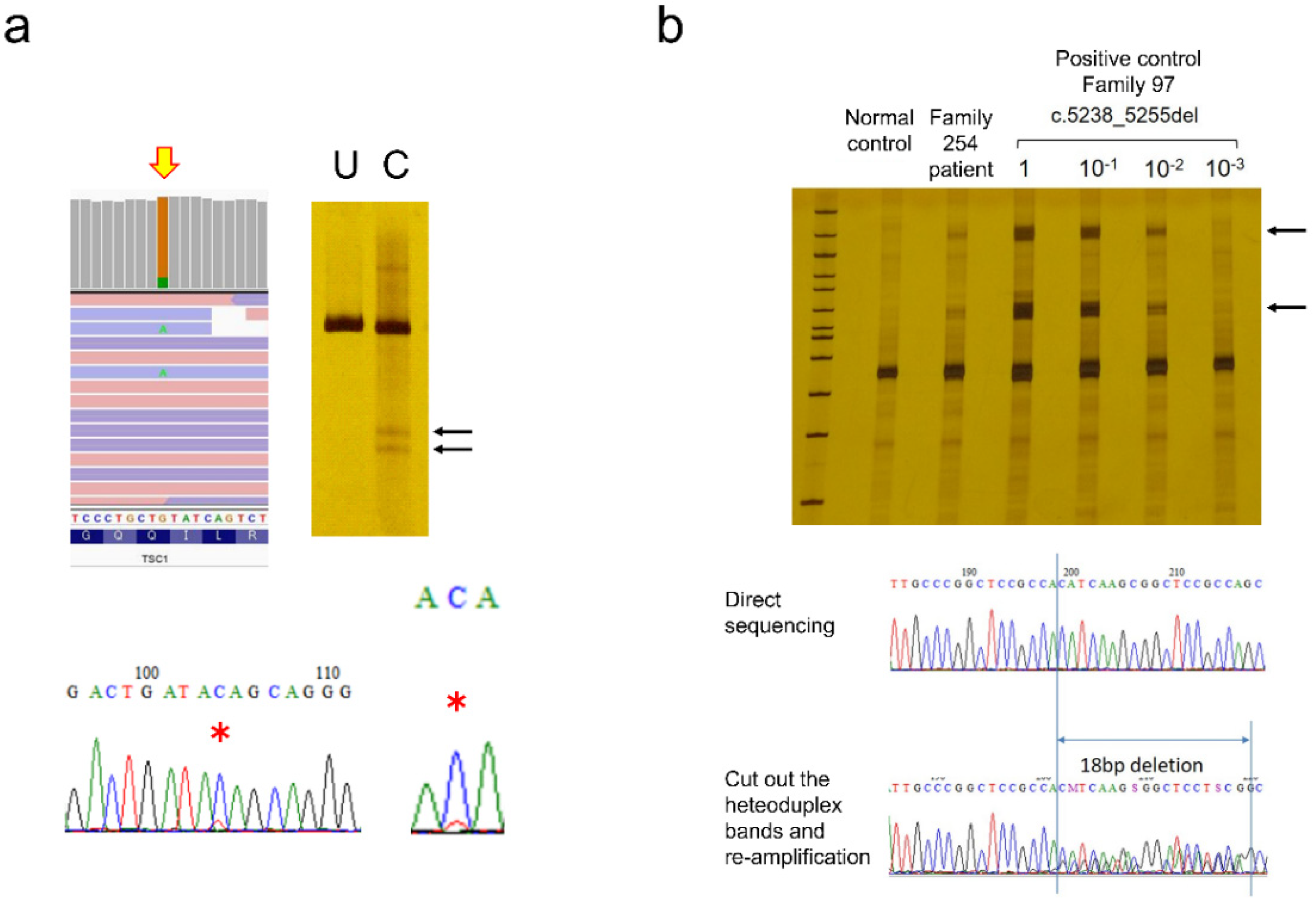
Figure 5a). CEL nuclease-mediated heteroduplex incision with polyacrylamide gel electrophoresis and silver staining (CHIPS) was even more sensitive and could detect less than 1% of mosaic variants (
Figure 5b). If target mutations have been determined by testing a proband or tumor, and if only the wild-type allele can be eliminated by digesting with specific restriction enzymes, mosaic mutations with lower frequencies can be detected. Starting with sufficient amounts of genomic DNA, repeated restriction enzyme digestion, and nested PCR enrichment for mutant alleles allow direct detection [
28,
29]. Empirically, this method is the most reliable for detecting very low-frequency mosaicism, but it cannot be used if the appropriate restriction enzymes are absent. Even if NGS predicts low-frequency mosaic mutations, there is currently no universal and simple confirmation method. In this study, only mosaic mutations that were finally detected by the Sanger method were counted, so the possibility that patients with low-frequency mosaic mutations remained among patients whose mutations could not be detected cannot be denied.
Regarding the phenotype of mosaic cases, among 13 patients with low-frequency mosaic mutations in blood DNA, only one had DD/ID (Fisher’s
p = 0.0494 to definite TSC in this study), another one had epilepsy (Fisher’s
p = 6.966×10
−5), and four cases were familial (
Supplementary Table S1). Neuropsychiatric symptoms tended to be milder, and the condition was accompanied by the chief complaint of organ lesions. This result was reasonable because, in the case of mosaic mutations, only cells with mutations produce symptoms.
3.6. Parental Mutation
Single-point mutation tests identified parental mutations in 13 of 49 families. Of the 13 families, only three had been diagnosed with TSC before genetic testing. This fact reveals that a significant proportion (approximately 20%) of the parents of patients not previously thought to have TSC then showed it. Therefore, genetic testing can efficiently diagnose phenotypically mild patients in families.
3.7. Analytical Method: Advantage of CoLAS
Mutation screening in this study was performed using the CHIPS-based method [
18] in 188 patients, and using the newly developed CoLAS method [
19] in the remaining 95 patients. Various advantages of the CoLAS method were observed.
3.7.1. Mutation Detection Rate
Table 11 shows the overall mutation detection rate utilizing CHIPS and CoLAS after removing patients with large chromosomal-level deletions. In definite TSC patients, the mutation detection rate of CoLAS reached 87.5% and was significantly higher than the CHIPS-based screening method. In our previous study, all 138 variants in the
TSC1/
2 gene detected by CHIPS and confirmed by Sanger sequencing were also detectable by CoLAS [
19]. CoLAS also enabled the detection of mutations that CHIPS was unable to detect. In fact, of the 200 mutations detected in this study, 27 were mutations that could only be confirmed by CoLAS, including mosaic mutations, intragenic deletions, and splicing abnormalities due to deep intronic mutations (
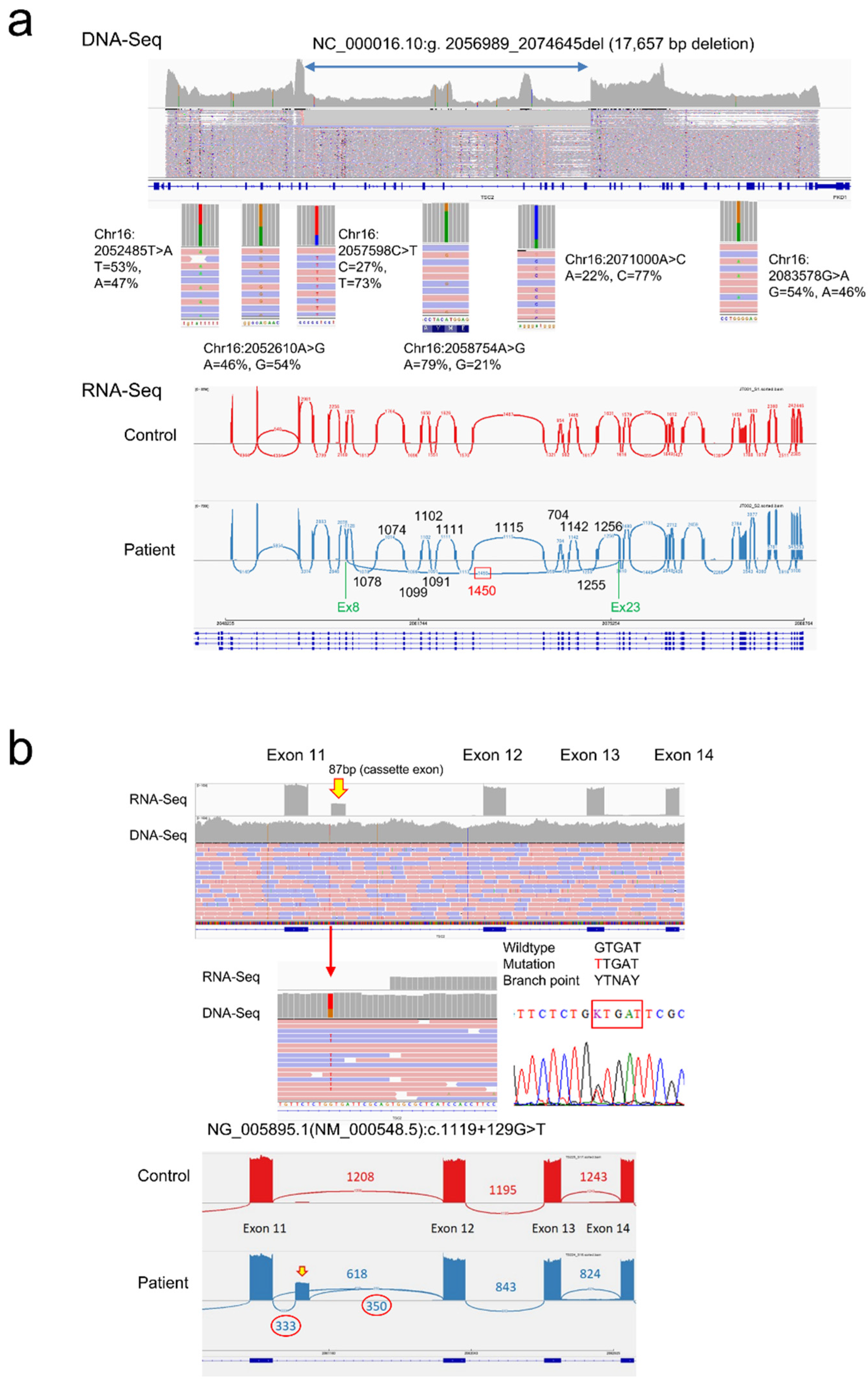
Supplementary Table S1). ColAS has the advantage of direct determination of breakpoint sequences for the intragenic deletions, and the presence of heterozygous SNPs can demonstrate mosaic deletions (
Figure 6a).
3.7.2. Consequence of Splicing Mutations
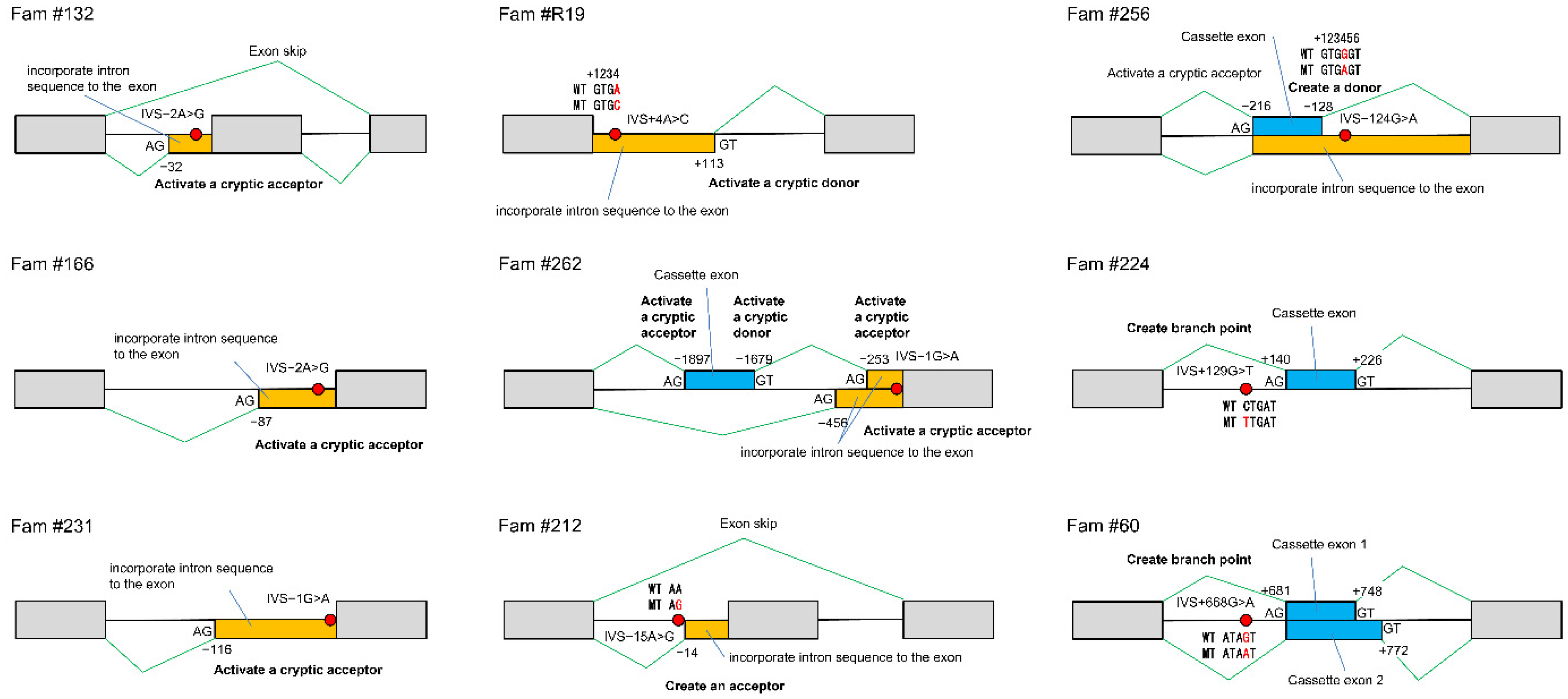
Until now, it has been challenging to determine the consequences of splicing mutations, but the development of the CoLAS method makes it possible to measure abnormal splicing events directly and semi-quantitatively (
Figure 6b). The effects of intronic mutations affecting mRNA splicing are diverse, including activation of cryptic splice acceptor or donor sites, creating a new acceptor, donor or branch point site, and exon skip. A single DNA mutation can cause these splicing abnormalities singly or in various combinations, making it difficult to predict without demonstrating experimentally (
Table 12,
Figure 7).
3.8. Limitation of This Study
Patients in this study were recruited for genetic testing and were not a natural cohort. However, 225 out of 283 patients (79.5%) were clinically diagnosed with definite TSC, and the overall age distribution was the same as the TOSCA baseline data. Although this study included more infants than TOSCA, it is considered to reflect the actual distribution of TSC patients in Japan to some extent.
Epilepsy was less frequent in this study than in TOSCA, even after adjusting for age and causative genes. The prevalence of epilepsy in our study is consistent with previous studies targeting older Japanese patients [
21], and it is likely that the diagnosis rate of TSC patients without epilepsy is actually higher in Japan. This study did not investigate in detail the types and severity of epilepsy. Although this study suggests that TSC1 patients may have a higher remission rate than TSC2 patients, it remains a possibility that a more detailed analysis could prove TSC2 patients have more severe epilepsy. For example, in the TOSCA study, the rate of infantile spasms was higher in TSC2 patients than in TSC1 patients (47.3% vs. 23%) [
34].
This study did not address TAND symptoms. Patients were recruited from hospitals throughout Japan for the purpose of genetic testing, so it was difficult to implement standardized scales and tests for developmental disorders and psychiatric symptoms. DD/ID was used instead, as an index of psychiatric symptoms. Therefore, it is unknown what kind of TAND symptoms patients without intellectual disabilities have, and whether there are any differences between TSC1 and TSC2 patients.
5. Conclusions
Lesions of tuberous sclerosis are divided into three types: those that appear in the fetal period and remain unchanged after birth, those that appear from childhood to adulthood, and those that appear later, mainly in adulthood. The first is hamartia, represented by cortical tubers and HM, which is thought to be due to haploinsufficiency; the next is AF and renal AML, and these hamartomas have LOH; and the last is represented by LAM, which has LOH, and some metastatic lesions from renal AMLs are included. Retinal hamartomas were generally TSC2-specific lesions. MMPH seemed to be TSC1 dominant lesion, but this requires further investigations. The prevalence of UF and cardiac rhabdomyoma was higher in TSC1 and TSC2 patients, respectively. TSC2 patients showed a more severe phenotype from high DD/ID frequency, tough AF and renal AML lesions, and non-remission of epilepsy. TSC2 PP mutations may attenuate AF and renal AML compared with NP mutations. CoLAS had an advantage in detecting broad TSC mutations. Approximately 20% of parents of the proband showed mild TSC that might have been missed. The patient demographics presented in this study had a high frequency of patients with TSC1 and a low prevalence of epilepsy. The reason why many patients with mild neuropsychiatric symptoms were diagnosed seems to be the high prevalence of brain imaging in Japan, suggesting the possibility that mild TSC patients might not be correctly diagnosed worldwide.