Somatic Mutational Landscape in Mexican Patients: CDH1 Mutations and chr20q13.33 Amplifications Are Associated with Diffuse-Type Gastric Adenocarcinoma
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Characteristics of the Patients
2.2. Helicobacter Pylori and EBV Identification
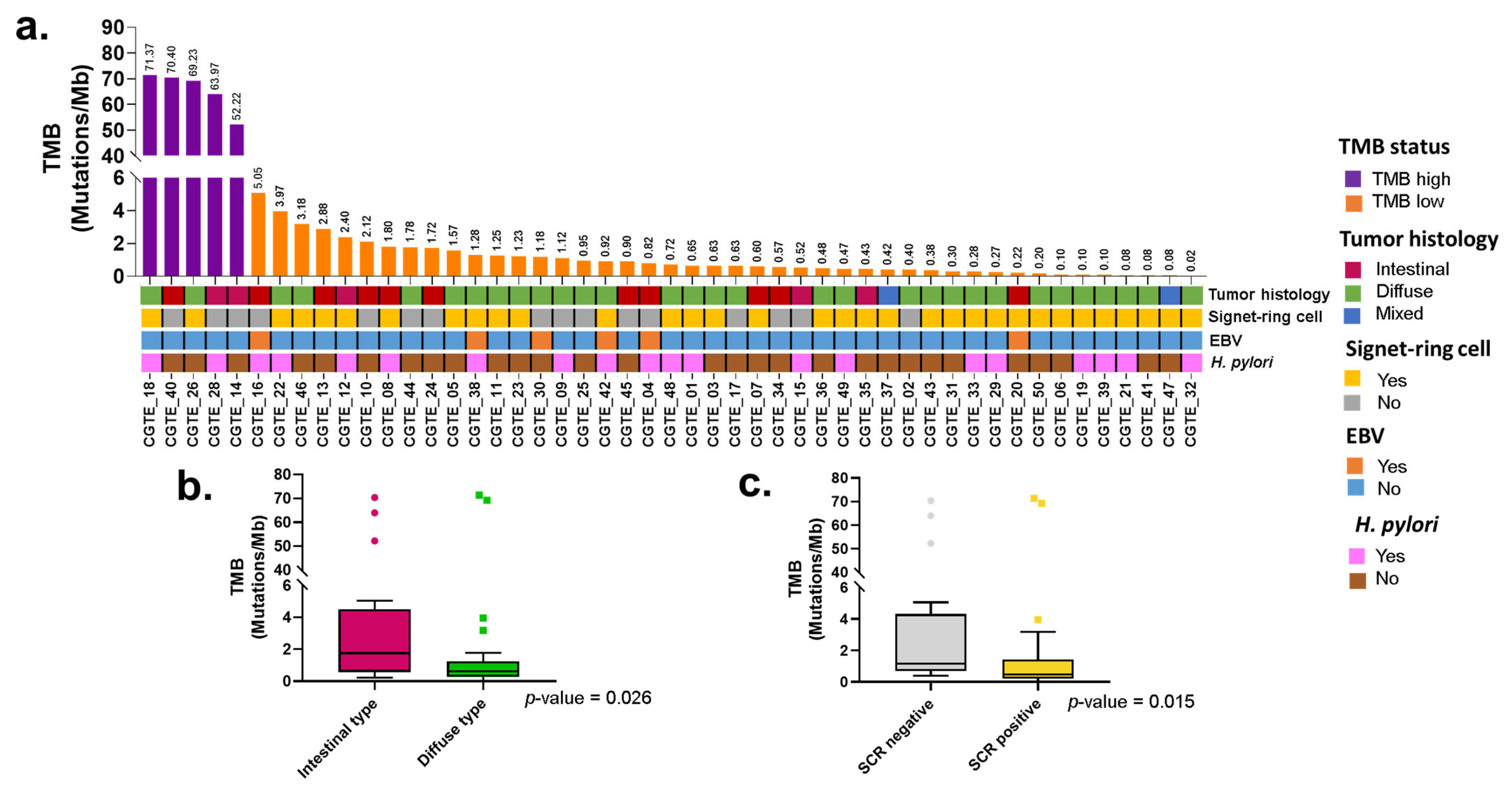
2.3. Tumor Mutational Burden and Clinical Features
2.4. Somatic Variants and Tumor Histology
2.5. Somatic Variants, H. pylori, and EBV
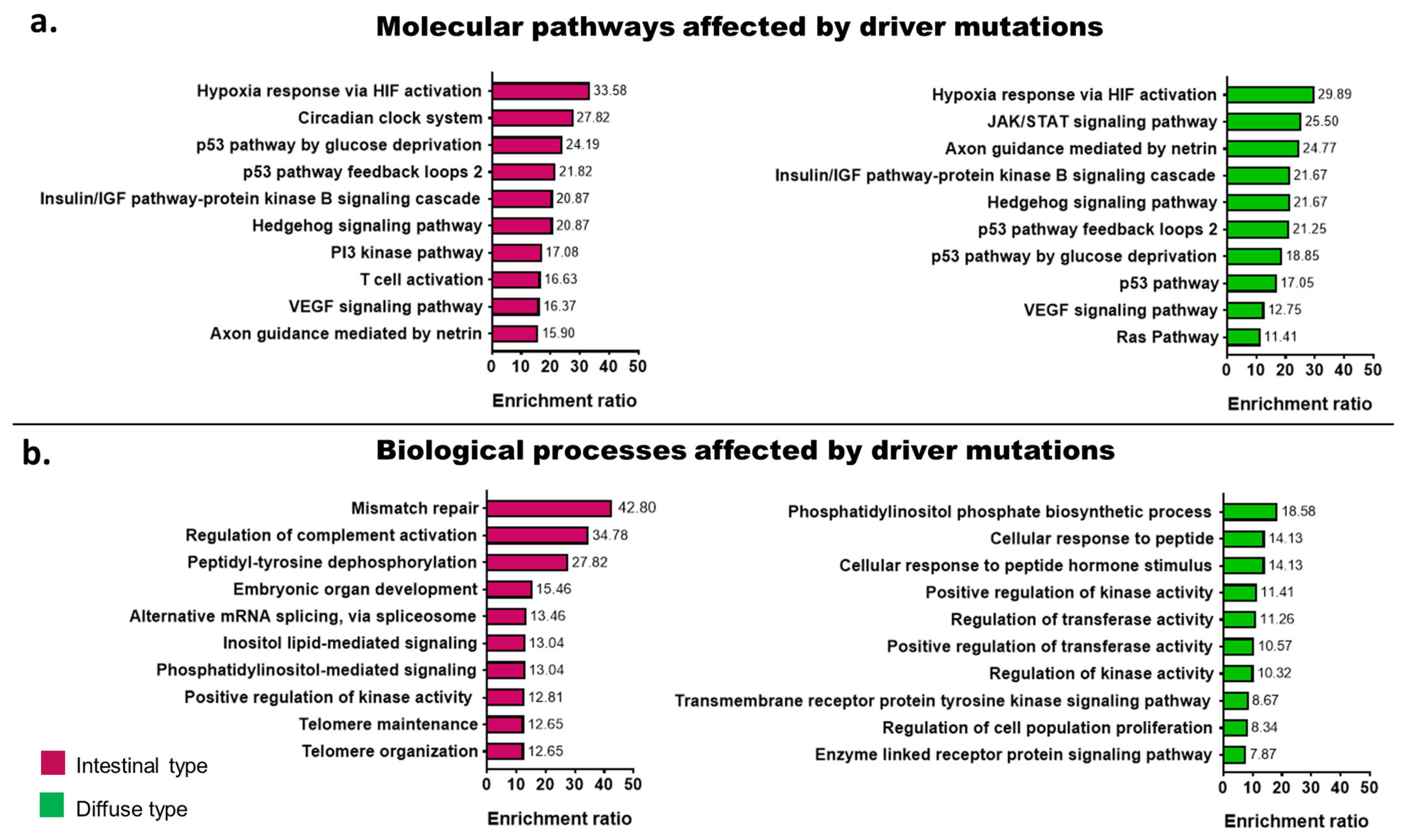
2.6. Signaling Pathways Affected by Somatic Mutations According to Tumor Histology
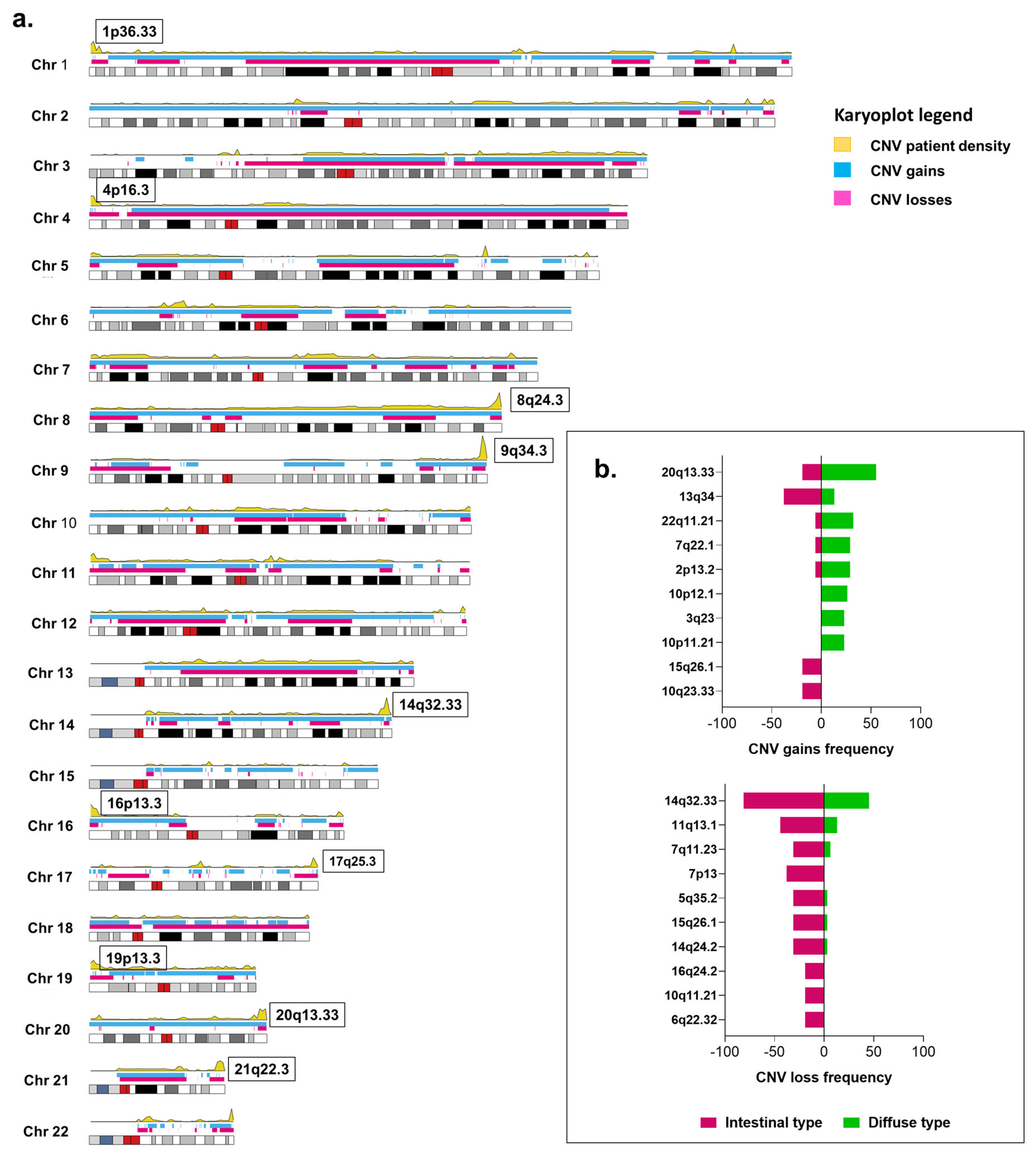
2.7. Copy Number Variations and Tumor Histology
2.8. Copy Number Variations, H. pylori, and EBV
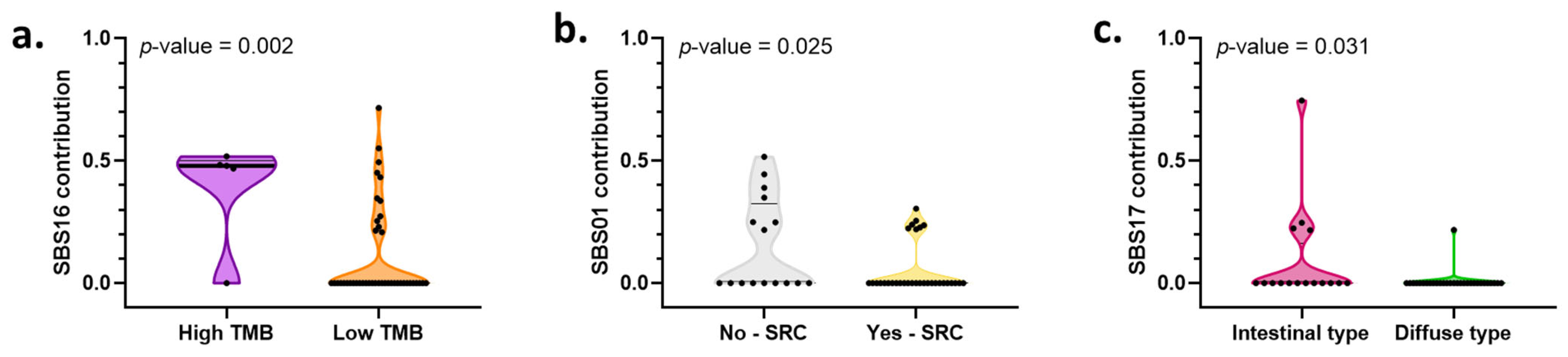
2.9. Mutational Processes and Their Associations with Clinical–Pathological Characteristics
2.10. Comparison of Genes Affected by Somatic Variants and Copy Number Variations between Mexican Patients and Other Cohorts
3. Discussion
4. Materials and Methods
4.1. Population and Sample Collection
4.2. Clinical Data Collection
4.3. DNA Extraction and Quality Control
4.4. PCR
4.5. In Situ Hybridization
4.6. Library Preparation, Hybridization Capture, and WES
4.7. Bioinformatic Analysis Pipeline
4.8. Tumor Mutational Burden
4.9. Enrichment Analysis
4.10. Mutational Signatures
4.11. TCGA and OncoSG Data Acquisition
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asplund, J.; Kauppila, J.H.; Mattsson, F.; Lagergren, J. Survival Trends in Gastric Adenocarcinoma: A Population-Based Study in Sweden. Ann Surg Oncol. 2018, 25, 2693–2702. [Google Scholar] [CrossRef]
- Jim, M.A.; Pinheiro, P.S.; Carreira, H.; Espey, D.K.; Wiggins, C.L.; Weir, H.K. Stomach cancer survival in the United States by race and stage (2001–2009): Findings from the CONCORD-2 study. Cancer 2017, 123, 4994–5013. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cerrato-Izaguirre, D.; Chirino, Y.I.; García-Cuellar, C.M.; Santibáñez-Andrade, M.; Prada, D.; Hernández-Guerrero, A.; Larraga, O.A.; Camacho, J.; Sánchez-Pérez, Y. Mutational landscape of gastric adenocarcinoma in Latin America: A genetic approach for precision medicine. Genes Dis. 2022, 9, 928–940. [Google Scholar] [CrossRef]
- Bonequi, P.; Meneses-González, F.; Correa, P.; Rabkin, C.S.; Camargo, M.C. Risk factors for gastric cancer in Latin America: A meta-analysis. Cancer Causes Control 2013, 24, 217–231. [Google Scholar] [CrossRef]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Chang, E.T.; Gomez, S.L.; Fish, K.; Schupp, C.W.; Parsonnet, J.; DeRouen, M.C.; Keegan, T.H.; Clarke, C.A.; Glaser, S.L. Gastric cancer incidence among Hispanics in California: Patterns by time, nativity, and neighborhood characteristics. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 709–719. [Google Scholar] [CrossRef]
- Corral, J.E.; Delgado Hurtado, J.J.; Domínguez, R.L.; Valdez de Cuéllar, M.; Balmore Cruz, C.; Morgan, D.R. The descriptive epidemiology of gastric cancer in Central America and comparison with United States Hispanic populations. J. Gastrointest. Cancer 2015, 46, 21–28. [Google Scholar] [CrossRef]
- Wang, R.; Song, S.; Harada, K.; Ghazanfari Amlashi, F.; Badgwell, B.; Pizzi, M.P.; Xu, Y.; Zhao, W.; Dong, X.; Jin, J.; et al. Multiplex profiling of peritoneal metastases from gastric adenocarcinoma identified novel targets and molecular subtypes that predict treatment response. Gut 2020, 69, 18–31. [Google Scholar] [CrossRef]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef]
- Shu, Y.; Zhang, W.; Hou, Q.; Zhao, L.; Zhang, S.; Zhou, J.; Song, X.; Zhang, Y.; Jiang, D.; Chen, X.; et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat. Commun. 2018, 9, 2447. [Google Scholar] [CrossRef]
- Tan, P.; Yeoh, K.G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 2015, 149, 1153–1162. [Google Scholar] [CrossRef]
- Ma, J.; Shen, H.; Kapesa, L.; Zeng, S. Lauren classification and individualized chemotherapy in gastric cancer. Oncol. Lett. 2016, 11, 2959–2964. [Google Scholar] [CrossRef]
- Network CGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Liao, P.; Jia, F.; Teer, J.K.; Knepper, T.C.; Zhou, H.H.; He, Y.J.; McLeod, H.L. Geographic variation in molecular subtype for gastric adenocarcinoma. Gut 2019, 68, 1340–1341. [Google Scholar] [CrossRef]
- Schumacher, S.E.; Shim, B.Y.; Corso, G.; Ryu, M.H.; Kang, Y.K.; Roviello, F.; Saksena, G.; Peng, S.; Shivdasani, R.A.; Bass, A.J.; et al. Somatic copy number alterations in gastric adenocarcinomas among Asian and Western patients. PLoS ONE 2017, 12, e0176045. [Google Scholar] [CrossRef]
- Sohn, B.H.; Hwang, J.E.; Jang, H.J.; Lee, H.S.; Oh, S.C.; Shim, J.J.; Lee, K.W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef]
- Sanjeevaiah, A.; Cheedella, N.; Hester, C.; Porembka, M.R. Gastric Cancer: Recent Molecular Classification Advances, Racial Disparity, and Management Implications. J. Oncol. Pract. 2018, 14, 217–224. [Google Scholar] [CrossRef]
- Holowatyj, A.N.; Ulrich, C.M.; Lewis, M.A. Racial/Ethnic Patterns of Young-Onset Noncardia Gastric Cancer. Cancer Prev. Res. 2019, 12, 771–780. [Google Scholar] [CrossRef]
- Choi, A.H.; Ji, L.; Babcock, B.; Ramos, V.; Kwong, M.L.M.; Morgan, J.W.; Selleck, M.J.; Langridge, W.H.R.; DeLeon, M.; Wall, N.R.; et al. Peritoneal carcinomatosis in gastric cancer: Are Hispanics at higher risk? J. Surg. Oncol. 2020, 122, 1624–1629. [Google Scholar] [CrossRef]
- Merchant, S.J.; Kim, J.; Choi, A.H.; Sun, V.; Chao, J.; Nelson, R. A rising trend in the incidence of advanced gastric cancer in young Hispanic men. Gastric. Cancer 2017, 20, 226–234. [Google Scholar] [CrossRef]
- Wang, S.C.; Yeu, Y.; Hammer, S.T.G.; Xiao, S.; Zhu, M.; Hong, C.; Clemenceau, J.R.; Yoon, L.Y.; Nassour, I.; Shen, J.; et al. Hispanic/Latino Patients with Gastric Adenocarcinoma Have Distinct Molecular Profiles Including a High Rate of Germline. Cancer Res. 2020, 80, 2114–2124. [Google Scholar] [CrossRef]
- Sánchez-Barriga, J.J. Tendencias de mortalidad y años potenciales de vida perdidos por cáncer gástrico en México, 2000–2012. Rev. Gastroenterol. México 2016, 81, 65–73. [Google Scholar] [CrossRef]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernández-López, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuña-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef]
- Romero-Hidalgo, S.; Ochoa-Leyva, A.; Garcíarrubio, A.; Acuña-Alonzo, V.; Antúnez-Argüelles, E.; Balcazar-Quintero, M.; Carnevale, A.; Cornejo-Granados, F.; Fernández-López, J.C. Demographic history and biologically relevant genetic variation of Native Mexicans inferred from whole-genome sequencing. Nat. Commun. 2017, 8, 1005. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignelli, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, L.A.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Erkizan, H.V.; Sukhadia, S.; Natarajan, T.G.; Marino, G.; Notario, V.; Lichy, J.H.; Wadleigh, R.G. Exome sequencing identifies novel somatic variants in African American esophageal squamous cell carcinoma. Sci. Rep. 2021, 11, 14814. [Google Scholar] [CrossRef]
- Guo, Y.A.; Chang, M.M.; Huang, W.; Ooi, W.F.; Xing, M.; Tan, P.; Skanderup, A.J. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 2018, 9, 1520. [Google Scholar] [CrossRef]
- Sánchez, Y.; Vaca-Paniagua, F.; Herrera, L.; Oñate, L.; Herrera-Goepfert, R.; Navarro-Martínez, G.; Cerrato, D.; Díaz-Velázquez, C.; Quezada, E.M.; García-Cuellar, C.; et al. Nutritional Indexes as Predictors of Survival and Their Genomic Implications in Gastric Cancer Patients. Nutr. Cancer 2021, 73, 1429–1439. [Google Scholar] [CrossRef]
- Martínez-Galindo, M.G.; Zamarripa-Dorsey, F.; Carmona-Castañeda, A.; Angeles-Labra, A.; Peñavera-Hernández, R.; Ugarte-Briones, C.; Blanco-Vela, C.I. Histopathologic characteristics of gastric adenocarcinoma in Mexican patients: A 10-year experience at the Hospital Juárez de México. Rev. Gastroenterol. México 2015, 80, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Trujillo Rivera, A.; Sampieri, C.L.; Morales Romero, J.; Montero, H.; Acosta Mesa, H.G.; Cruz Ramírez, N.; Novoa Del Toro, E.M.; León Córdoba, K. Risk factors associated with gastric cancer in Mexico: Education, breakfast and chili. Rev. Esp. Enferm. Dig. 2018, 110, 372–379. [Google Scholar] [CrossRef]
- Folprecht, G. Tumor mutational burden as a new biomarker for PD-1 antibody treatment in gastric cancer. Cancer Commun. 2019, 39, 74. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, B.; Kang, S.Y.; Heo, Y.J.; Park, S.H.; Kim, S.T.; Kang, W.K.; Lee, J.; Kim, K.M. Tumor Mutational Burden Determined by Panel Sequencing Predicts Survival After Immunotherapy in Patients With Advanced Gastric Cancer. Front. Oncol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Cai, H.; Jing, C.; Chang, X.; Ding, D.; Han, T.; Yang, J.; Lu, Z.; Hu, X.; Liu, Z.; Wang, J.; et al. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J. Transl. Med. 2019, 17, 189. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, W.; Zhang, X.; Ge, M.; Song, C. Correlation between TMB and MSI in patients with solid tumors. J. Clin. Oncol. 2020, 38, e15169. [Google Scholar] [CrossRef]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Chung, H.C.; Ros, W.; Delord, J.P.; Perets, R.; Italiano, A.; Shapira-Frommer, R.; Manzuk, L.; Piha-Paul, S.A.; Xu, L.; Zeigenfuss, S.; et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Cervical Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2019, 37, 1470–1478. [Google Scholar] [CrossRef]
- Nemtsova, M.V.; Kalinkin, A.I.; Kuznetsova, E.B.; Bure, I.V.; Alekseeva, E.A.; Bykov, I.I.; Khorobrykh, T.V.; Mikhaylenko, D.S.; Tanas, A.S.; Kutsev, S.I.; et al. Clinical relevance of somatic mutations in main driver genes detected in gastric cancer patients by next-generation DNA sequencing. Sci. Rep. 2020, 10, 504. [Google Scholar] [CrossRef]
- Pan, X.; Ji, X.; Zhang, R.; Zhou, Z.; Zhong, Y.; Peng, W.; Sun, N.; Xu, X.; Xia, L.; Li, P.; et al. Landscape of somatic mutations in gastric cancer assessed using next-generation sequencing analysis. Oncol. Lett. 2018, 16, 4863–4870. [Google Scholar] [CrossRef]
- Zhao, H.; Hu, H.; Chen, B.; Xu, W.; Zhao, J.; Huang, C.; Xing, Y.; Lv, H.; Nie, C.; Wang, J.; et al. Overview on the Role of E-Cadherin in Gastric Cancer: Dysregulation and Clinical Implications. Front. Mol. Biosci. 2021, 8, 689139. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S. CDH1 (E-Cadherin) Mutation and Gastric Cancer: Genetics, Molecular Mechanisms and Guidelines for Management. Cancer Manag. Res. 2019, 11, 10477–10486. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.; Kim, J.H.; Lee, D.H. RHOA in Gastric Cancer: Functional Roles and Therapeutic Potential. Front. Genet. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Lage, L.A.d.P.C.; Barreto, G.C.; Culler, H.F.; Cavalcanti, J.B.; Alves, L.d.O.; Nardinelli, L.; Bendit, I.; Rocha, V.; Pereira, J. Rhoa Mutation Is a Potential Biomarker Associated with Adverse Prognosis and High- Tumor Burden in Patients with Nodal Peripheral Lymphomas with T-Helper Follicular Phenotype (nPTCL-Thf): Data from a Brazilian Retrospective Cohort of Nodal PTCL. Blood 2021, 138, 4482. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, S.; Lim, S.M.; Eom, H.J.; Balch, C.; Lee, J.; Jeong, J.H.; Nam, S.; Kim, Y.H. Rational design of small molecule RHOA inhibitors for gastric cancer. Pharm. J. 2020, 20, 601–612. [Google Scholar] [CrossRef]
- Bui, V.M.; Mettling, C.; Jou, J.; Sun, H.S. Genomic amplification of chromosome 20q13.33 is the early biomarker for the development of sporadic colorectal carcinoma. BMC Med. Genom. 2020, 13, 149. [Google Scholar] [CrossRef]
- Gordon-Weeks, A.; Lim, S.Y.; Yuzhalin, A.; Lucotti, S.; Vermeer, J.A.; Jones, K.; Chen, J.; Muschel, R.J. Tumour-Derived Laminin α5 (LAMA5) Promotes Colorectal Liver Metastasis Growth, Branching Angiogenesis and Notch Pathway Inhibition. Cancers 2019, 11, 630. [Google Scholar] [CrossRef]
- Diao, B.; Yang, P. Comprehensive Analysis of the Expression and Prognosis for Laminin Genes in Ovarian Cancer. Pathol. Oncol. Res. 2021, 27, 1609855. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Yang, S.; Yang, Y.; Zhang, Q.; Zhang, W.; Zhang, W.; Xiao, H.; Gao, H. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med. Genom. 2012, 5, 14. [Google Scholar] [CrossRef]
- Xia, S.; Tang, Q.; Wang, X.; Zhang, L.; Jia, L.; Wu, D.; Xu, P.; Zhang, X.; Tang, G.; Yang, T.; et al. Overexpression of PSMA7 predicts poor prognosis in patients with gastric cancer. Oncol. Lett. 2019, 18, 5341–5349. [Google Scholar] [CrossRef]
- Yao, Y.; Ni, Y.; Zhang, J.; Wang, H.; Shao, S. The role of Notch signaling in gastric carcinoma: Molecular pathogenesis and novel therapeutic targets. Oncotarget 2017, 8, 53839–53853. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Precision medicine for human cancers with Notch signaling dysregulation (Review). Int. J. Mol. Med. 2020, 45, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Cheng, Z.; Wang, Y.H.; Guo, Z.H.; Zhang, S.Q.; Hu, J.K.; Zhou, Z.G. Role of Notch signaling pathway in gastric cancer: A meta-analysis of the literature. World J. Gastroenterol. 2014, 20, 9191–9199. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Yu, J.; Gan, J.; Song, N.; Shi, L.; Liu, J.; Zhang, Z.; Du, J. Notch1/2/3/4 are prognostic biomarker and correlated with immune infiltrates in gastric cancer. Aging 2020, 12, 2595–2609. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Ulloa, E.; Ramos-Cruz, A.C.; Prada, D.; Avilés-Salas, A.; Chávez-Blanco, A.D.; Herrera, L.A.; Lizano, M.; Contreras-Paredes, A. Loss of nuclear NOTCH1, but not its negative regulator NUMB, is an independent predictor of cervical malignancy. Oncotarget 2018, 9, 27. [Google Scholar] [CrossRef]
- Van Beek, E.J.A.H.; Hernandez, J.M.; Goldman, D.A.; Davis, J.L.; McLoughlin, K.C.; Ripley, R.T.; Kim, T.S.; Tang, L.H.; Hechtman, J.F.; Zheng, J.; et al. Correction to: Rates of TP53 Mutation are Significantly Elevated in African American Patients with Gastric Cancer. Ann. Surg. Oncol. 2020, 27, 963. [Google Scholar] [CrossRef]
- Jia, F.; Teer, J.K.; Knepper, T.C.; Lee, J.K.; Zhou, H.H.; He, Y.J.; McLeod, H.L. Discordance of Somatic Mutations Between Asian and Caucasian Patient Populations with Gastric Cancer. Mol. Diagn. Ther. 2017, 21, 179–185. [Google Scholar] [CrossRef]
- Conomos, M.P.; Laurie, C.A.; Stilp, A.M.; Gogarten, S.M.; McHugh, C.P.; Nelson, S.C.; Sofer, T.; Fernández-Rhodes, L.; Justice, A.E.; Graff, M.; et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2016, 98, 165–184. [Google Scholar] [CrossRef]
- González Burchard, E.; Borrell, L.N.; Choudhry, S.; Naqvi, M.; Tsai, H.-J.; Rodriguez-Santana, J.R.; Chapela, R.; Rogers, S.D.; Mei, R.; Rodriguez-Cintron, W.; et al. Latino Populations: A Unique Opportunity for the Study of Race, Genetics, and Social Environment in Epidemiological Research. Am. J. Public Health 2005, 95, 2161–2168. [Google Scholar] [CrossRef]
- Peña-Llopis, S.; Brugarolas, J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat. Protoc. 2013, 8, 2240–2255. [Google Scholar] [CrossRef]
- Chang, K.L.; Chen, Y.-Y.; Shibata, D.; Weiss, L.M. Description of an In Situ Hybridization Methodology for Detection of Epstein-Barr Virus RNA in Paraffin-Embedded Tissues, with a Survey of Normal and Neoplastic Tissues. Diagn. Mol. Pathol. 1992, 1, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Benjamin, D.; Sato, T.; Cibulskis, K.; Getz, G.; Stewart, C.; Lichtenstein, L. Calling Somatic SNVs and Indels with Mutect2. bioRxiv 2019, 861054. [Google Scholar] [CrossRef]
- Saunders, C.T.; Wong, W.S.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef]
- Tamborero, D.; Rubio-Perez, C.; Deu-Pons, J.; Schroeder, M.P.; Vivancos, A.; Rovira, A.; Tusquets, I.; Albanell, J.; Rodon, J.; Tabernero, J.; et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018, 10, 25. [Google Scholar] [CrossRef]
- Muiños, F.; Martínez-Jiménez, F.; Pich, O.; Gonzalez-Perez, A.; Lopez-Bigas, N. In silico saturation mutagenesis of cancer genes. Nature 2021, 596, 428–432. [Google Scholar] [CrossRef]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, R.; McGranahan, N.; Herrero, J.; Taylor, B.S.; Swanton, C. DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Schultz, N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]


| Variable | Mean | SD |
|---|---|---|
| Age (years) | 54.94 | 15.49 |
| BMI (Kg/m2) | 23.60 | 5.18 |
| Follow-up (months) | 12.53 | 14.17 |
| Variable | n | % |
| Sex | ||
| Female | 21 | 42.00 |
| Clinical stage | ||
| I–II | 3 | 6.00 |
| III−IV | 47 | 94.00 |
| Tumor location | ||
| Antrum and pylorus | 11 | 22.00 |
| Cardia | 0 | 0.00 |
| Fundus and body | 18 | 36.00 |
| Two or more locations | 21 | 42.00 |
| Endoscopic impression 1 | ||
| I | 1 | 2.00 |
| II | 3 | 6.00 |
| III | 15 | 30.00 |
| IV | 22 | 44.00 |
| V | 6 | 12.00 |
| Not reported | 3 | 6.00 |
| Tumor histology 2 | ||
| Intestinal | 16 | 32.00 |
| Diffuse | 31 | 62.00 |
| Mixed | 3 | 6.00 |
| Tumor grade | ||
| Well-differentiated | 0 | 0.00 |
| Moderately differentiated | 8 | 16.00 |
| Poorly differentiated | 42 | 84.00 |
| Signet-ring cell | ||
| Positive | 34 | 68.00 |
| H. pylori | ||
| Positive | 21 | 42.00 |
| EBV | ||
| Positive | 6 | 12.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerrato-Izaguirre, D.; Chirino, Y.I.; Prada, D.; Quezada-Maldonado, E.M.; Herrera, L.A.; Hernández-Guerrero, A.; Alonso-Larraga, J.O.; Herrera-Goepfert, R.; Oñate-Ocaña, L.F.; Cantú-de-León, D.; et al. Somatic Mutational Landscape in Mexican Patients: CDH1 Mutations and chr20q13.33 Amplifications Are Associated with Diffuse-Type Gastric Adenocarcinoma. Int. J. Mol. Sci. 2022, 23, 11116. https://doi.org/10.3390/ijms231911116
Cerrato-Izaguirre D, Chirino YI, Prada D, Quezada-Maldonado EM, Herrera LA, Hernández-Guerrero A, Alonso-Larraga JO, Herrera-Goepfert R, Oñate-Ocaña LF, Cantú-de-León D, et al. Somatic Mutational Landscape in Mexican Patients: CDH1 Mutations and chr20q13.33 Amplifications Are Associated with Diffuse-Type Gastric Adenocarcinoma. International Journal of Molecular Sciences. 2022; 23(19):11116. https://doi.org/10.3390/ijms231911116
Chicago/Turabian StyleCerrato-Izaguirre, Dennis, Yolanda I. Chirino, Diddier Prada, Ericka Marel Quezada-Maldonado, Luis A Herrera, Angélica Hernández-Guerrero, Juan Octavio Alonso-Larraga, Roberto Herrera-Goepfert, Luis F. Oñate-Ocaña, David Cantú-de-León, and et al. 2022. "Somatic Mutational Landscape in Mexican Patients: CDH1 Mutations and chr20q13.33 Amplifications Are Associated with Diffuse-Type Gastric Adenocarcinoma" International Journal of Molecular Sciences 23, no. 19: 11116. https://doi.org/10.3390/ijms231911116
APA StyleCerrato-Izaguirre, D., Chirino, Y. I., Prada, D., Quezada-Maldonado, E. M., Herrera, L. A., Hernández-Guerrero, A., Alonso-Larraga, J. O., Herrera-Goepfert, R., Oñate-Ocaña, L. F., Cantú-de-León, D., Meneses-García, A., Basurto-Lozada, P., Robles-Espinoza, C. D., Camacho, J., García-Cuellar, C. M., & Sánchez-Pérez, Y. (2022). Somatic Mutational Landscape in Mexican Patients: CDH1 Mutations and chr20q13.33 Amplifications Are Associated with Diffuse-Type Gastric Adenocarcinoma. International Journal of Molecular Sciences, 23(19), 11116. https://doi.org/10.3390/ijms231911116






