Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus
Abstract
1. Introduction
2. Results and Discussion
2.1. Features of Stentor Introns
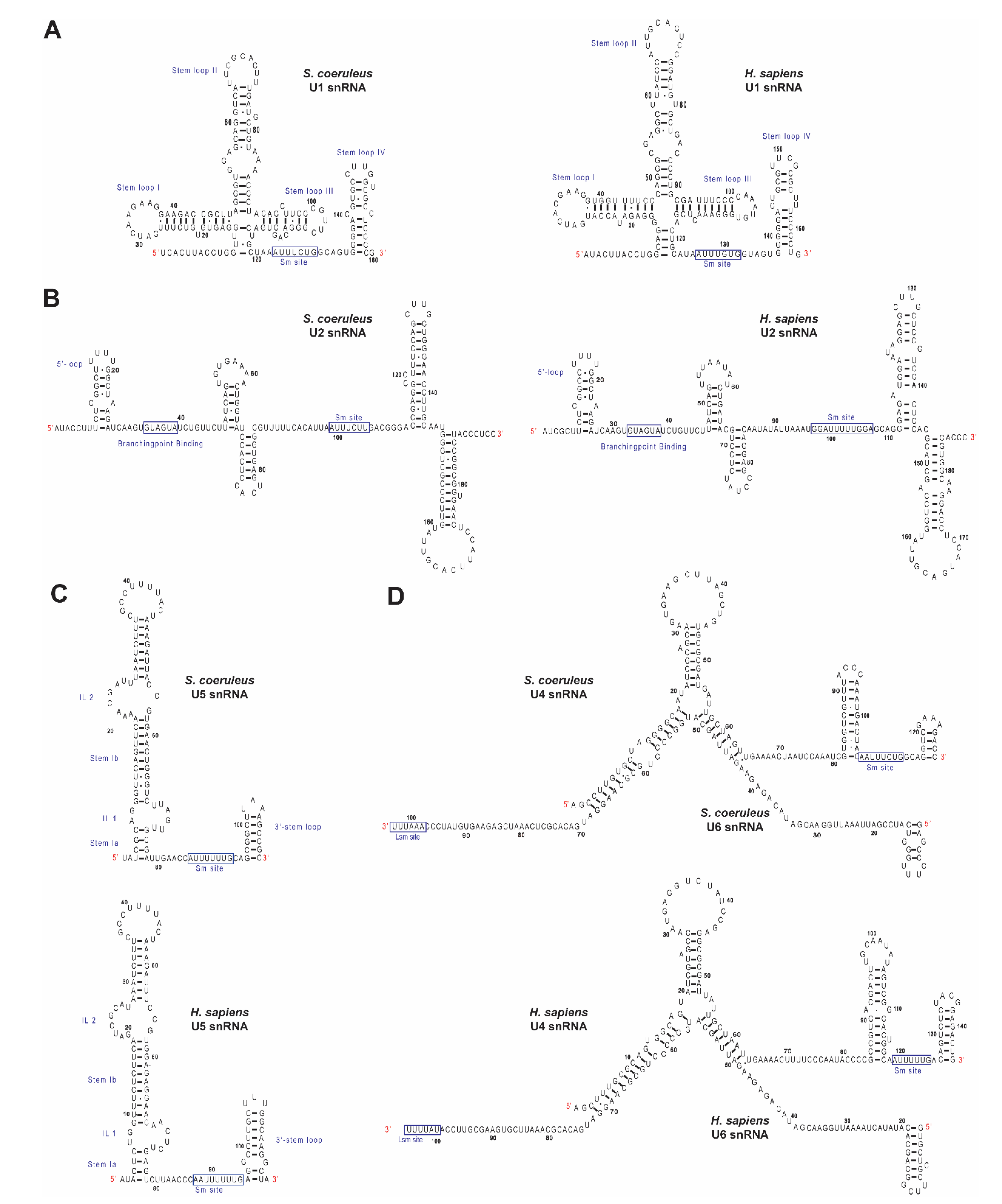
2.2. Identification of Spliceosomal snRNAs in S. coeruleus
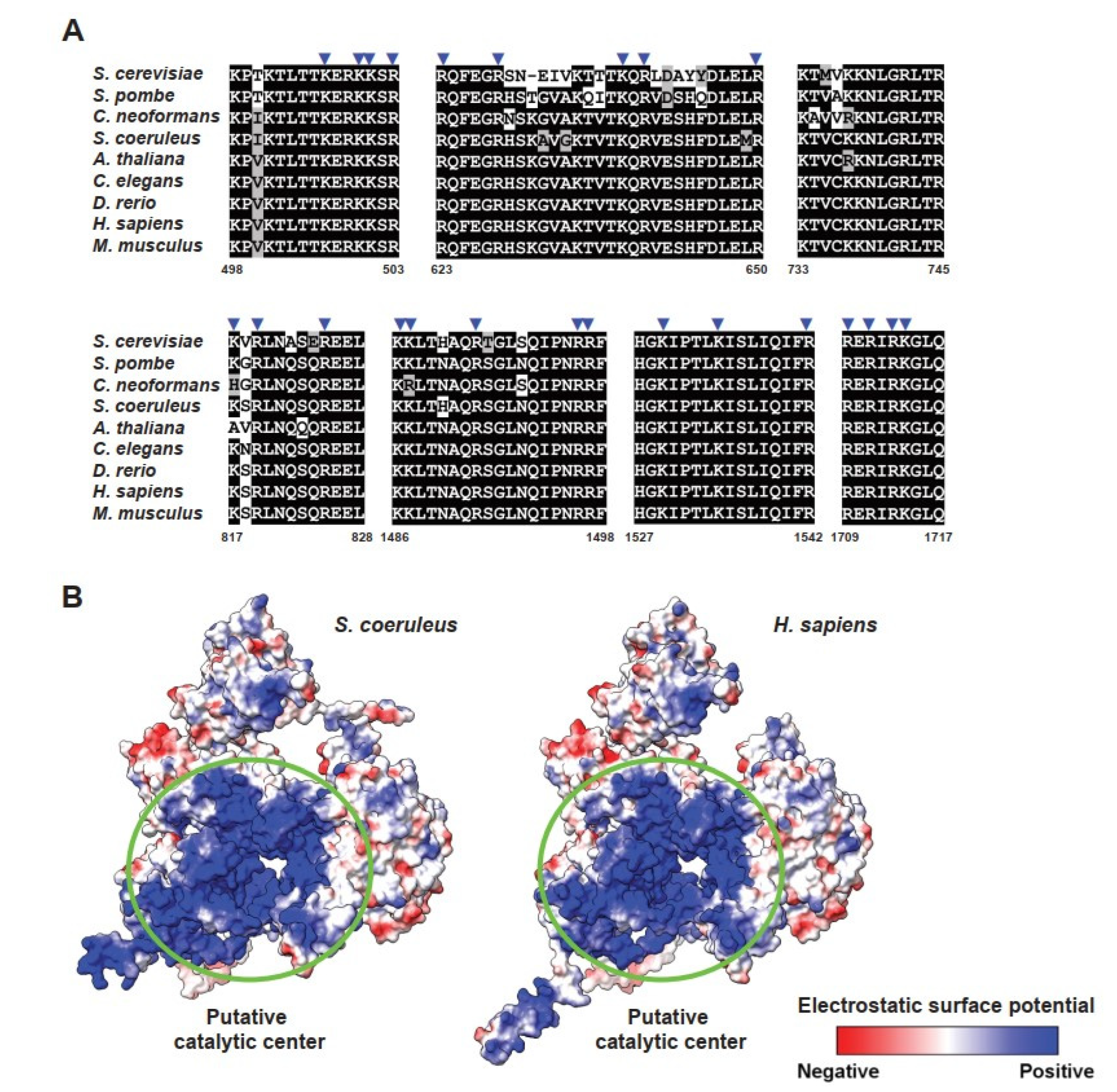
2.3. Identification of Protein Components of Spliceosomal snRNPs in S. coeruleus
2.4. Identification of Stentor Spliceosomal RNA Helicases and Other Non-snRNP Proteins Involving Spliceosome Assembly and Activation
2.5. A Model of RNA–RNA Interaction Network in Stentor Spliceosomal Active Site
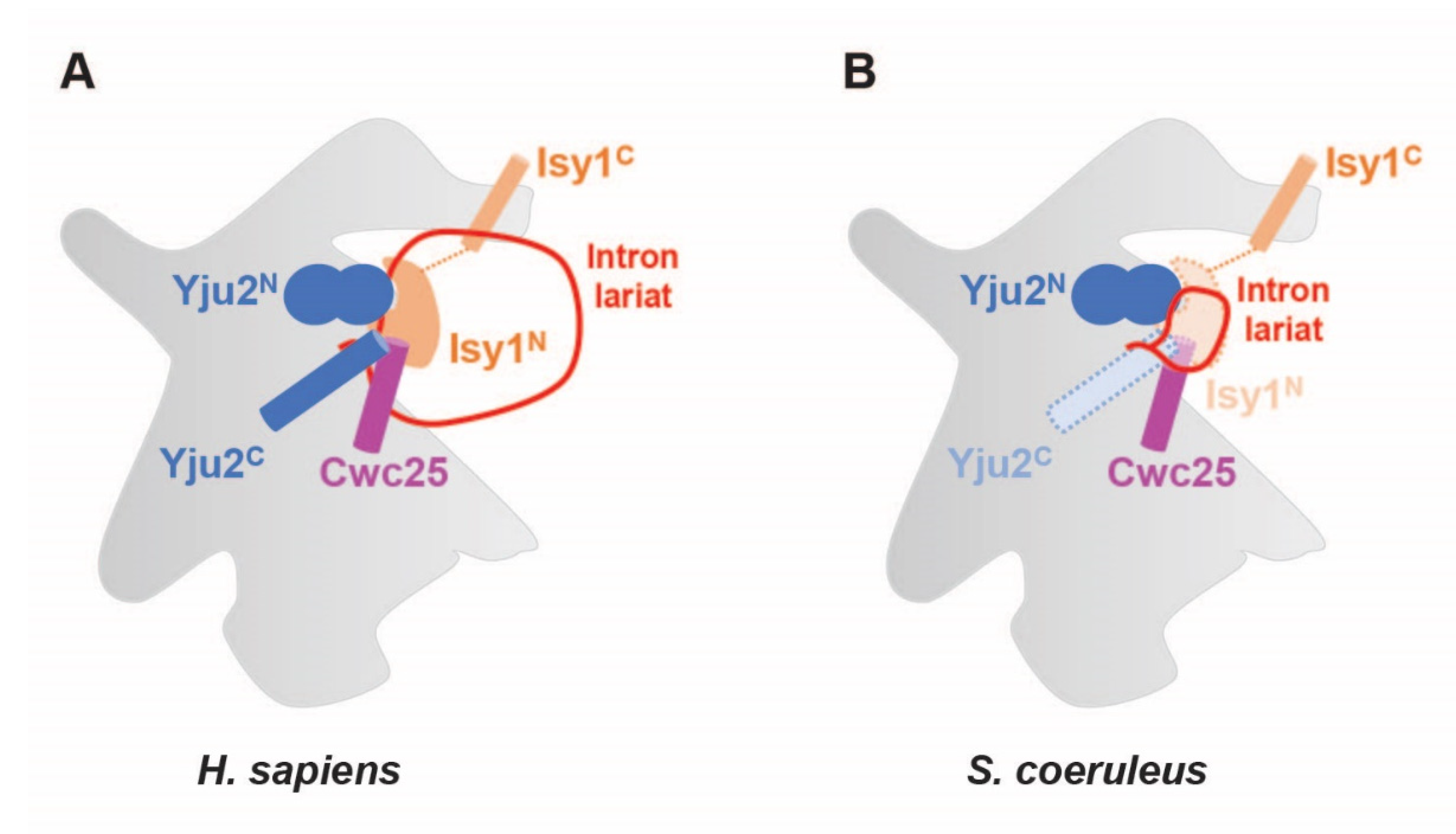
2.6. Regions of Branching Factors Projecting to the Spliceosomal Active Site May Be Unique in Stentor
3. Materials and Methods
3.1. Computational Analyses of Features of Introns of S. coeruleus
3.2. Identification of Spliceosomal Proteins in the S. coeruleus Genome
3.3. Prediction of S. coeruleus U snRNA Candidates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| snRNA | small nuclear RNA |
| snRNP | small nuclear ribonucleoproteins |
| SS | splice site |
| BP-A | branchpoint adenosine |
References
- Tartar, V. The Biology of Stentor; Elsevier: Amsterdam, The Netherlands, 1961; ISBN 978-0-08-009343-7. [Google Scholar]
- Tartar, V. Extreme alteration of the nucleocytoplasmic ratio in Stentor coeruleus. J. Protozool. 1963, 10, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.H. Regeneration of proportionate structures in stentor. Biol. Bull. 1901, 2, 311–328. [Google Scholar] [CrossRef]
- Slabodnick, M.M.; Ruby, J.G.; Reiff, S.B.; Swart, E.C.; Gosai, S.; Prabakaran, S.; Witkowska, E.; Larue, G.E.; Fisher, S.; Freeman, R.M.; et al. The macronuclear genome of stentor coeruleus reveals tiny introns in a giant cell. Curr. Biol. 2017, 27, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Sood, P.; Lin, A.; Yan, C.; McGillivary, R.; Diaz, U.; Makushok, T.; Nadkarni, A.V.; Tang, S.K.; Marshall, W.F. Modular, cascade-like transcriptional program of regeneration in stentor. eLife 2022, 11, e80778. [Google Scholar] [CrossRef]
- Grate, L.; Ares, M. Searching yeast intron data at Ares Lab web site. Methods Enzym. 2002, 350, 380–392. [Google Scholar] [CrossRef]
- Hong, X.; Scofield, D.G.; Lynch, M. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006, 23, 2392–2404. [Google Scholar] [CrossRef]
- Hang, J.; Wan, R.; Yan, C.; Shi, Y. Structural basis of pre-MRNA splicing. Science 2015, 349, 1191–1198. [Google Scholar] [CrossRef]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Fica, S.M.; Nagai, K. Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 2017, 24, 791–799. [Google Scholar] [CrossRef]
- Chanarat, S. UBL5/Hub1: An atypical ubiquitin-like protein with a typical role as a stress-responsive regulator. Int. J. Mol. Sci. 2021, 22, 9384. [Google Scholar] [CrossRef] [PubMed]
- Galej, W.P.; Wilkinson, M.E.; Fica, S.M.; Oubridge, C.; Newman, A.J.; Nagai, K. Cryo-EM structure of the spliceosome immediately after branching. Nature 2016, 537, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Beggs, J.D. RNA helicases in splicing. RNA Biol. 2013, 10, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Hahn, D.; Beggs, J.D. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 2012, 24, 431–438. [Google Scholar] [CrossRef] [PubMed]
- StentorDB. Stentor Genome Database Wiki. Available online: http://stentor.ciliate.org/index.php/home/welcome (accessed on 3 March 2022).
- Crooks, G.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- McCoy, M.J.; Fire, A.Z. Intron and gene size expansion during nervous system evolution. BMC Genom. 2020, 21, 360. [Google Scholar] [CrossRef]
- Sakurai, A.; Fujimori, S.; Kochiwa, H.; Kitamura-Abe, S.; Washio, T.; Saito, R.; Carninci, P.; Hayashizaki, Y.; Tomita, M. On biased distribution of introns in various eukaryotes. Gene 2002, 300, 89–95. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef]
- Infernal 1.1: 100-Fold Faster RNA Homology Searches Bioinformatics Oxford Academic. Available online: https://academic.oup.com/bioinformatics/article/29/22/2933/316439?login=false (accessed on 3 March 2022).
- De Maio, A.; Yalamanchili, H.K.; Adamski, C.J.; Gennarino, V.A.; Liu, Z.; Qin, J.; Jung, S.Y.; Richman, R.; Orr, H.; Zoghbi, H.Y. RBM17 interacts with U2SURP and CHERP to regulate expression and splicing of RNA-processing proteins. Cell Rep. 2018, 25, 726–736.e7. [Google Scholar] [CrossRef]
- Chanarat, S.; Sträßer, K. Splicing and beyond: The many faces of the Prp19 complex. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2126–2134. [Google Scholar] [CrossRef]
- Pastor, F.; Shkreta, L.; Chabot, B.; Durantel, D.; Salvetti, A. Interplay between CMGC kinases targeting SR proteins and viral replication: Splicing and beyond. Front. Microbiol. 2021, 12, 658721. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, T.; Hosoya, T.; Shimizu, S.; Sumi, K.; Oshiro, T.; Yoshinaka, Y.; Suzuki, M.; Yamamoto, N.; Herzenberg, L.A.; Herzenberg, L.A.; et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. USA 2006, 103, 11329–11333. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.E.; Frye, M. Noncanonical functions of the serine-arginine-rich splicing factor (SR) family of proteins in development and disease. BioEssays 2021, 43, 2000242. [Google Scholar] [CrossRef]
- Liao, S.E.; Regev, O. Splicing at the phase-separated nuclear speckle interface: A model. Nucleic Acids Res. 2021, 49, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Twyffels, L.; Gueydan, C.; Kruys, V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011, 278, 3246–3255. [Google Scholar] [CrossRef] [PubMed]
- Henry, M.F.; Mandel, D.; Routson, V.; Henry, P.A. The yeast HnRNP-like protein Hrp1/Nab4 accumulates in the cytoplasm after hyperosmotic stress: A novel Fps1-dependent response. MBoC 2003, 14, 3929–3941. [Google Scholar] [CrossRef] [PubMed]
- Loya, T.J.; O’Rourke, T.W.; Reines, D. Yeast Nab3 protein contains a self-assembly domain found in human heterogeneous nuclear Ribonucleoprotein-C (HnRNP-C) that is necessary for transcription termination. J. Biol. Chem. 2013, 288, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- The HnRNP Family: Insights into Their Role in Health and Disease SpringerLink. Available online: https://link.springer.com/article/10.1007/s00439-016-1683-5 (accessed on 20 March 2022).
- Zenklusen, D.; Vinciguerra, P.; Strahm, Y.; Stutz, F. The yeast HnRNP-like proteins Yra1p and Yra2p participate in MRNA export through interaction with Mex67p. Mol. Cell. Biol. 2001, 21, 4219–4232. [Google Scholar] [CrossRef]
- Sheng, J.; Gan, J.; Soares, A.S.; Salon, J.; Huang, Z. Structural insights of non-canonical U•U pair and Hoogsteen interaction probed with Se atom. Nucleic Acids Res. 2013, 41, 10476–10487. [Google Scholar] [CrossRef]
- Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update Nucleic Acids Research Oxford Academic. Available online: https://academic.oup.com/nar/article/46/W1/W537/5001157 (accessed on 21 March 2022).
- RStudio: Integrated Development for R. 2015. Available online: http://www.rstudio.com (accessed on 4 April 2020).
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice Nucleic Acids Research Oxford Academic. Available online: https://academic.oup.com/nar/article-abstract/22/22/4673/2400290?redirectedFrom=fulltext&login=false (accessed on 27 April 2022).
- pyBoxshade/BS_app.py at master · mdbaron42/pyBoxshade · GitHub. Available online: https://github.com/mdbaron42/pyBox-shade (accessed on 16 October 2021).
- GraphPad Prism 9 User Guide—How to Cite GraphPad Prism. Available online: https://www.graphpad.com/guides/prism/latest/user-guide/citing_graphpad_prism.htm (accessed on 9 August 2022).
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- BLAST: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools Nucleic Acids Research Oxford Academic. Available online: https://academic.oup.com/nar/article/32/suppl_2/W20/1040657?login=false (accessed on 3 March 2022).
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]



| Protein/Complex | S. cerevisiaea | H. sapiensa | S. coeruleusb | BLAST Operation Mode c | |||
|---|---|---|---|---|---|---|---|
| Accession No. | Query Coverage | E-Value | % Identity | ||||
| Sm proteins | SmB | SNRPB | OMJ80606.1 | 48% | 2 × 10−13 | 35.42% | B |
| SmD1 | SNRPD1 | OMJ74037.1 | 75% | 3 × 10−13 | 40.00% | B | |
| SmD2 | SNRPD2 | OMJ84976.1 | 79% | 2 × 10−29 | 55.91% | B | |
| SmD3 | SNRPD3 | OMJ79831.1 | 81% | 2 × 10−30 | 50.00% | B | |
| SmE | SNRPE | OMJ89702.1 | 93% | 3 × 10−21 | 39.77% | B | |
| SmF | SNRPF | OMJ90477.1 | 84% | 2 × 10−20 | 50.68% | B | |
| SmG | SNRPG | OMJ87882.1 | 94% | 1 × 10−14 | 43.84% | B | |
| Lsm proteins | Lsm2 | Lsm2 | MPUH01000040.1 | 94% | 1 × 10−8 | 31.52% | T |
| Lsm3 | Lsm3 | OMJ80606.1 | 75% | 9 × 10−6 | 33.80% | B | |
| Lsm4 | Lsm4 | MPUH01000270.1 | 54% | 2 × 10−16 | 40.79% | T | |
| Lsm5 | Lsm5 | OMJ92196.1 | 69% | 2 × 10−5 | 35.82% | B | |
| Lsm6 | Lsm6 | MPUH01000102.1 | 87% | 1 × 10−13 | 41.43% | T | |
| Lsm7 | Lsm7 | OMJ69462.1 | 68% | 2 × 10−11 | 37.97% | B | |
| Lsm8 | Lsm8 | OMJ73124.1 | 60% | 1 × 10−9 | 34.29% | B | |
| U1 snRNP | Mud1 | SNRPA | OMJ89879.1 | 32% | 5 × 10−28 | 49.46% | B |
| Yhc1 | SNRPC | OMJ82811.1 | 38% | 7 × 10−15 | 48.39% | B | |
| Snp1 | SNRNP70 | OMJ72809.1 | 65% | 6 × 10−24 | 33.50% | B | |
| Prp39 | PRPF39 | OMJ71629.1 | 66% | 6 × 10−50 | 27.27% | B | |
| Nam8 | TIA1 | OMJ95152.1 | 14% | 1 × 10−7 | 35.06% | B | |
| Snu71 | - | N/A | N/A | N/A | N/A | B + T | |
| Prp42 | - | N/A | N/A | N/A | N/A | B + T | |
| Snu56 | - | N/A | N/A | N/A | N/A | B + T | |
| U2 snRNP | Prp21 | SF3A1 | OMJ90611.1 | 45% | 8 × 10−40 | 36.92% | B |
| Prp11 | SF3A2 | OMJ89506.1 | 90% | 4 × 10−13 | 25.73% | B | |
| Prp9 | SF3A3 | OMJ96095.1 | 75% | 4 × 10−37 | 27.65% | B | |
| Hsh155 | SF3B1 | OMJ68425.1 | 84% | 0 | 51.95% | B | |
| Cus1 | SF3B2 | OMJ66678.1 | 63% | 2 × 10−40 | 34.05% | B | |
| Rse1 | SF3B3 | OMJ94841.1 | 77% | 6 × 10−50 | 26.73% | B | |
| Hsh49 | SF3B4 | OMJ96224.1 | 90% | 9 × 10−38 | 37.11% | B | |
| Ysf3 | SF3B5 | OMJ86748.1 | 91% | 1 × 10−29 | 53.16% | B | |
| - | SF3B6 | OMJ82020.1 | 84% | 1 × 10−38 | 48.72% | B | |
| Lea1 | SNRPA1 | OMJ66761.1 | 60% | 4 × 10−7 | 31.51% | B | |
| Msl1 | SNRPB2 | OMJ69199.1 | 74% | 3 × 10−16 | 42.35% | B | |
| Rds3 | PHF5A | OMJ89703.1 | 98% | 4 × 10−38 | 53.33% | B | |
| U2 snRNP associated | Prp5 | DDX46 | OMJ88772.1 | 61% | 4 × 10−109 | 38.77% | B |
| Prp43 | DHX15 | OMJ88889.1 | 89% | 0 | 56.81% | B | |
| Cus2 | TATSF1 | MPUH01000067.1 | 29% | 3 × 10−27 | 33.33% | T | |
| Mud2 | U2AF2 | OMJ92895.1 | 68% | 3 × 10−41 | 30.17% | B | |
| - | U2AF1 | OMJ68977.1 | 97% | 2 × 10−70 | 47.66% | B | |
| - | CHERP | OMJ90611.1 | 6% | 2 × 10−7 | 48.33% | B | |
| - | DDX42 | OMJ83405.1 | 46% | 2 × 10−123 | 44.75% | B | |
| - | PUF60 | OMJ96224.1 | 30% | 2 × 10−20 | 29.94% | B | |
| - | U2SURP | N/A | N/A | N/A | N/A | B + T | |
| U5 snRNP | Prp8 | PRPF8 | MPUH01001036.1 | 94% | 0 | 59.68% | T |
| Brr2 | SNRNP200 | OMJ93829.1 | 98% | 0 | 35.16% | B | |
| Snu114 | EFTUD2 | OMJ70706.1 | 99% | 5 × 10−169 | 34.24% | B | |
| Prp6 | PRPF6 | MPUH01000153.1 | 54% | 5 × 10−28 | 34.15% | T | |
| Prp28 | DDX23 | OMJ87031.1 | 62% | 5 × 10−73 | 38.86% | B | |
| Lin1 | CD2BP2 | N/A | N/A | N/A | N/A | B + T | |
| Dib1 | TXNL4A | OMJ67094.1 | 96% | 2 × 10−59 | 61.59% | B | |
| - | SNRNP40 | OMJ86718.1 | 84% | 5 × 10−45 | 32.65% | B | |
| U4/U6 snRNP | Prp31 | PRPF31 | OMJ65893.1 | 76% | 5 × 10-33 | 26.72% | B |
| Prp3 | PRPF3 | OMJ88877.1 | 46% | 7 × 10−21 | 29.91% | B | |
| Prp4 | PRPF4 | OMJ68512.1 | 90% | 3 × 10−38 | 27.25% | B | |
| Snu13 | NHP2L1 | OMJ79450.1 | 100% | 5 × 10−54 | 61.90% | B | |
| - | PPIH | OMJ73551.1 | 94% | 4 × 10−66 | 60.36% | B | |
| - | SART3 | OMJ93468.1 | 18% | 2 × 10−10 | 26.29% | B | |
| U4/U6.U5 tri-snRNP | Snu66 | SART1 | N/A | N/A | N/A | N/A | B + T |
| Sad1 | USP39 | MPUH01000082.1 | 80% | 2 × 10−21 | 24.80% | T | |
| Hub1 | UBL5 | OMJ90719.1 | 98% | 2 × 10−31 | 68.06% | B | |
| Spp381 | - | N/A | N/A | N/A | N/A | B + T | |
| RES complex | Bud13 | BUD13 | OMJ87046.1 | 24% | 1 × 10−11 | 33.12% | B |
| Pml1 | SNIP1 | MPUH01000502.1 | 81% | 3 × 10−12 | 28.74% | T | |
| Ist3 | RBMX2 | OMJ73591.1 | 72% | 6 × 10−34 | 54.21% | B | |
| NTC/Prp19 complex | Syf1 | XAB2 | OMJ71034.1 | 57% | 6 × 10−30 | 23.31% | B |
| Clf1 | CRNKL1 | OMJ94375.1 | 98% | 2 × 10−111 | 33.96% | B | |
| Cef1 | CDC5L | OMJ92975.1 | 36% | 6 × 10−62 | 49.07% | B | |
| Prp19 | PRPF19 | OMJ79960.1 | 61% | 2 × 10−32 | 31.09% | B | |
| Ssa4 | HSPA8 | OMJ95577.1 | 94% | 0 | 73.40% | B | |
| Isy1 | ISY1 | MPUH01001382.1 | 53% | 9 × 10−34 | 44.44% | T | |
| Ecm2/Cwc2 | RBM22 | MPUH01000303.1 | 70% | 6 × 10−28 | 29.46% | T | |
| Syf2 | - | N/A | N/A | N/A | N/A | B + T | |
| Snt309 | SPF27 | N/A | N/A | N/A | N/A | B + T | |
| Ntc20 | - | N/A | N/A | N/A | N/A | B + T | |
| - | PQBP1 | N/A | N/A | N/A | N/A | B + T | |
| - | WBP11 | N/A | N/A | N/A | N/A | B + T | |
| - | CTNNBL1 | N/A | N/A | N/A | N/A | B + T | |
| NTC-Related proteins | Prp46 | PLRG1 | OMJ70863.1 | 73% | 8 × 10−129 | 51.20% | B |
| Prp45 | SNW1 | OMJ67115.1 | 82% | 1 × 10−18 | 28.12% | B | |
| Bud31 | BUD31 | OMJ75480.1 | 100% | 1 × 10−48 | 47.13% | B | |
| - | AQR | OMJ84448.1 | 34% | 3 × 10−26 | 25.33% | B | |
| - | PPIE | OMJ73551.1 | 54% | 3 × 10−68 | 61.18% | B | |
| - | PPIL1 | OMJ93796.1 | 87% | 9 × 10−43 | 46.90% | B | |
| Cwc15 | CWC15 | N/A | N/A | N/A | N/A | B+T | |
| Step 2 proteins | Prp17 | PRP17 | OMJ92236.1 | 73% | 3 × 10−70 | 36.20% | B |
| Prp22 | DHX8 | OMJ81156.1 | 73% | 0 | 50.47% | B | |
| Prp16 | DHX38 | OMJ82483.1 | 75% | 0 | 48.62% | B | |
| Slu7 | SLU7 | N/A | N/A | N/A | N/A | B + T | |
| Prp18 | PRPF18 | N/A | N/A | N/A | N/A | B + T | |
| Cdc40 | CDC40 | N/A | N/A | N/A | N/A | B + T | |
| Recruited at A complex | Prp40 | PRPF40A | OMJ75909.1 | 84% | 2 × 10−24 | 24.30% | B |
| Msl5 | SF1 | OMJ93203.1 | 54% | 2 × 10−26 | 36.64% | B | |
| - | RBM5 | OMJ90804.1 | 27% | 2 × 10−8 | 26.20% | B | |
| - | RBM25 | OMJ73591.1 | 72% | 6 × 10−34 | 54.21% | B | |
| - | RBM10 | N/A | N/A | N/A | N/A | B + T | |
| - | CCAR1 | N/A | N/A | N/A | N/A | B + T | |
| - | SUGP1 | N/A | N/A | N/A | N/A | B + T | |
| - | THRAP3 | N/A | N/A | N/A | N/A | B + T | |
| Recruited at B complex | Prp38 | PRPF38A | OMJ70103.1 | 55% | 1 × 10−49 | 48.84% | B |
| Spp382/Ntr1 | TFIP11 | OMJ89844.1 | 32% | 2 × 10−32 | 35.19% | B | |
| Snu23 | ZMAT2 | OMJ87982.1 | 33% | 1 × 10−10 | 39.71% | B | |
| - | IK | OMJ66867.1 | 30% | 4 × 10−19 | 44.60% | B | |
| - | MFAP1 | OMJ66867.1 | 55% | 1 × 10−28 | 37.75% | B | |
| - | PRPF4B | OMJ69187.1 | 36% | 3 × 10−87 | 41.24% | B | |
| - | SMU1 | OMJ86718.1 | 65% | 1 × 10−28 | 29.32% | B | |
| - | WBP4 | N/A | N/A | N/A | N/A | B + T | |
| Recruited at Bact complex | Cwc22 | CWC22 | OMJ82911.1 | 81% | 1 × 10−58 | 29.73% | B |
| Cwc25 | CWC25 | OMJ67410.1 | 38% | 6 × 10−11 | 34.73% | B | |
| Cwc27 | CWC27 | OMJ68249.1 | 36% | 1 × 10−67 | 58.96% | B | |
| Prp2 | DHX16 | OMJ77262.1 | 75% | 0 | 50.22% | B | |
| Cwc24 | RNF113A | OMJ95226.1 | 45% | 3 × 10−19 | 36.23% | B | |
| Yju2 | YJU2 | OMJ95747.1 | 41% | 1 × 10−21 | 38.79% | B | |
| Spp2 | GPKOW | OMJ70620.1 | 65% | 6 × 10−9 | 22.15% | B | |
| - | CCDC12 | OMJ70827.1 | 48% | 3 × 10−6 | 37.04% | B | |
| - | GPATCH1 | OMJ90352.1 | 7% | 5 × 10−13 | 49.30% | B | |
| - | PPIL2 | OMJ93796.1 | 29% | 7 × 10−37 | 45.75% | B | |
| - | PRCC | N/A | N/A | N/A | N/A | B + T | |
| - | ZNF830 | N/A | N/A | N/A | N/A | B + T | |
| Recruited at C complex | - | CACTIN | OMJ67936.1 | 27% | 2 × 10−57 | 40.82% | B |
| - | CDK10 | OMJ71086.1 | 81% | 2 × 10−85 | 44.22% | B | |
| - | DDX41 | OMJ89820.1 | 73% | 5 × 10−117 | 39.47% | B | |
| - | DHX35 | OMJ77262.1 | 91% | 0 | 46.75% | B | |
| - | FAM32A | OMJ85432.1 | 44% | 6 × 10−6 | 44.00% | B | |
| - | NOSIP | OMJ75149.1 | 100% | 1 × 10−20 | 24.36% | B | |
| - | PPIG | OMJ74576.1 | 22% | 3 × 10−66 | 63.10% | B | |
| - | PPIL3 | OMJ93796.1 | 96% | 9 × 10−49 | 51.92% | B | |
| - | PPWD1 | OMJ93796.1 | 88% | 2 × 10−136 | 40.24% | B | |
| - | SDE2 | OMJ76446.1 | 21% | 2 × 10−14 | 34.95% | B | |
| - | WDR83 | OMJ82560.1 | 87% | 4 × 10−52 | 34.29% | B | |
| - | CXorf56 | N/A | N/A | N/A | N/A | B + T | |
| - | C9orf78 | N/A | N/A | N/A | N/A | B + T | |
| - | DGCR14 | N/A | N/A | N/A | N/A | B + T | |
| - | FAM50A | N/A | N/A | N/A | N/A | B + T | |
| - | FRA10AC1 | N/A | N/A | N/A | N/A | B + T | |
| - | LENG1 | N/A | N/A | N/A | N/A | B + T | |
| hnRNPs | - | HNRNPA1 | OMJ89514.1 | 48% | 4 × 10−40 | 41.80% | B |
| - | HNRNPAB | OMJ89514.1 | 49% | 4 × 10−38 | 46.07% | B | |
| - | HNRNPC | OMJ93791.1 | 23% | 2 × 10−5 | 30.99% | B | |
| SR proteins | - | SRSF1-12 | N/A | N/A | N/A | N/A | B + T |
| - | SREK1 | N/A | N/A | N/A | N/A | B + T | |
| - | SFSWAP | N/A | N/A | N/A | N/A | B + T | |
| - | TRA2A | N/A | N/A | N/A | N/A | B + T | |
| - | TRA2B | N/A | N/A | N/A | N/A | B + T | |
| SR related | - | SRRM1 | N/A | N/A | N/A | N/A | B + T |
| - | SRRM2 | N/A | N/A | N/A | N/A | B + T | |
| Others | Pus1 | PUS1 | OMJ75529.1 | 52% | 2 × 10−15 | 35.94% | B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuadthaisong, J.; Phetruen, T.; Techawisutthinan, C.; Chanarat, S. Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus. Int. J. Mol. Sci. 2022, 23, 10973. https://doi.org/10.3390/ijms231810973
Nuadthaisong J, Phetruen T, Techawisutthinan C, Chanarat S. Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus. International Journal of Molecular Sciences. 2022; 23(18):10973. https://doi.org/10.3390/ijms231810973
Chicago/Turabian StyleNuadthaisong, Jirayu, Tanaporn Phetruen, Chanakan Techawisutthinan, and Sittinan Chanarat. 2022. "Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus" International Journal of Molecular Sciences 23, no. 18: 10973. https://doi.org/10.3390/ijms231810973
APA StyleNuadthaisong, J., Phetruen, T., Techawisutthinan, C., & Chanarat, S. (2022). Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus. International Journal of Molecular Sciences, 23(18), 10973. https://doi.org/10.3390/ijms231810973






