Integration of TE Induces Cancer Specific Alternative Splicing Events
Abstract
1. Introduction
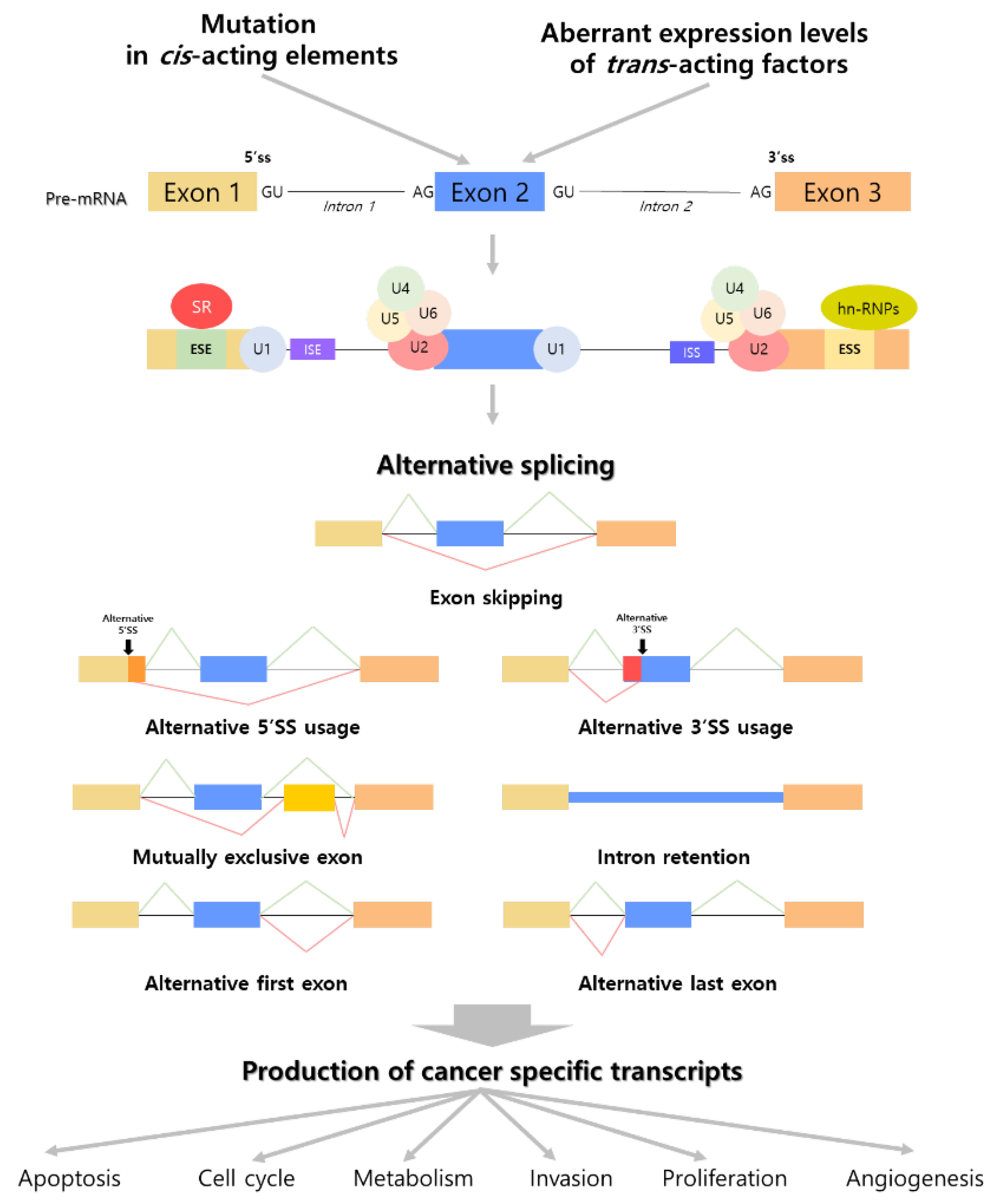
2. The Intimate Connection between Alternative Splicing and Cancer
2.1. Cancer Promoting Mutations in Alternative Splicing Causing Factors
2.2. Cancer-Specific Transcripts Generated from Several Mechanisms of Alternative Splicing
3. Regulation of Cancer Related-Biological Processes by TE Induced Alternative Splicing
3.1. Exonization and Exon Disruption
3.2. Providing Splicing Donor/Acceptor Site and Stop Codon
3.3. Providing Alternative Regulatory Sequences Such as Enhancer, Repressor and Promoter
3.4. Epigenetic Alteration
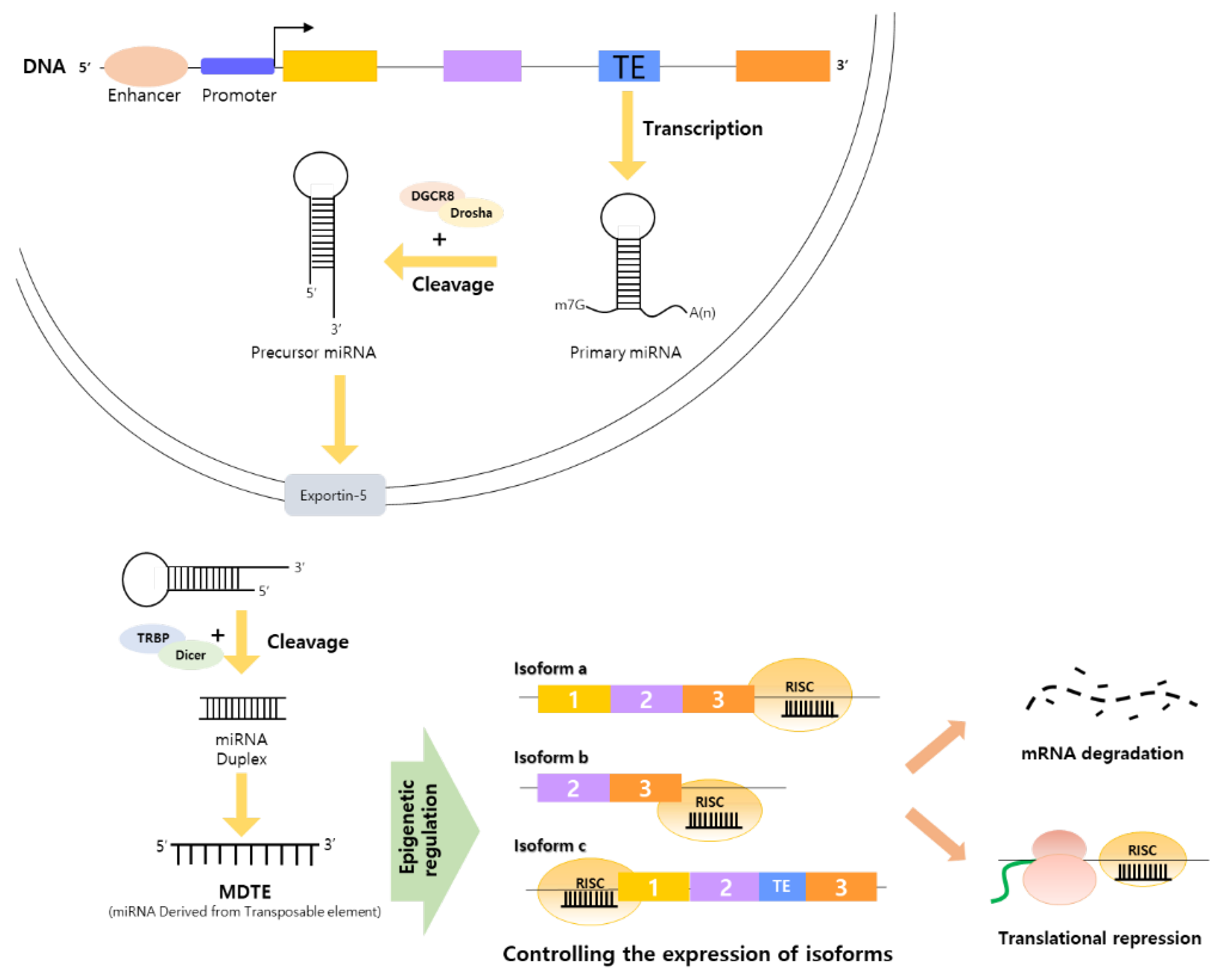
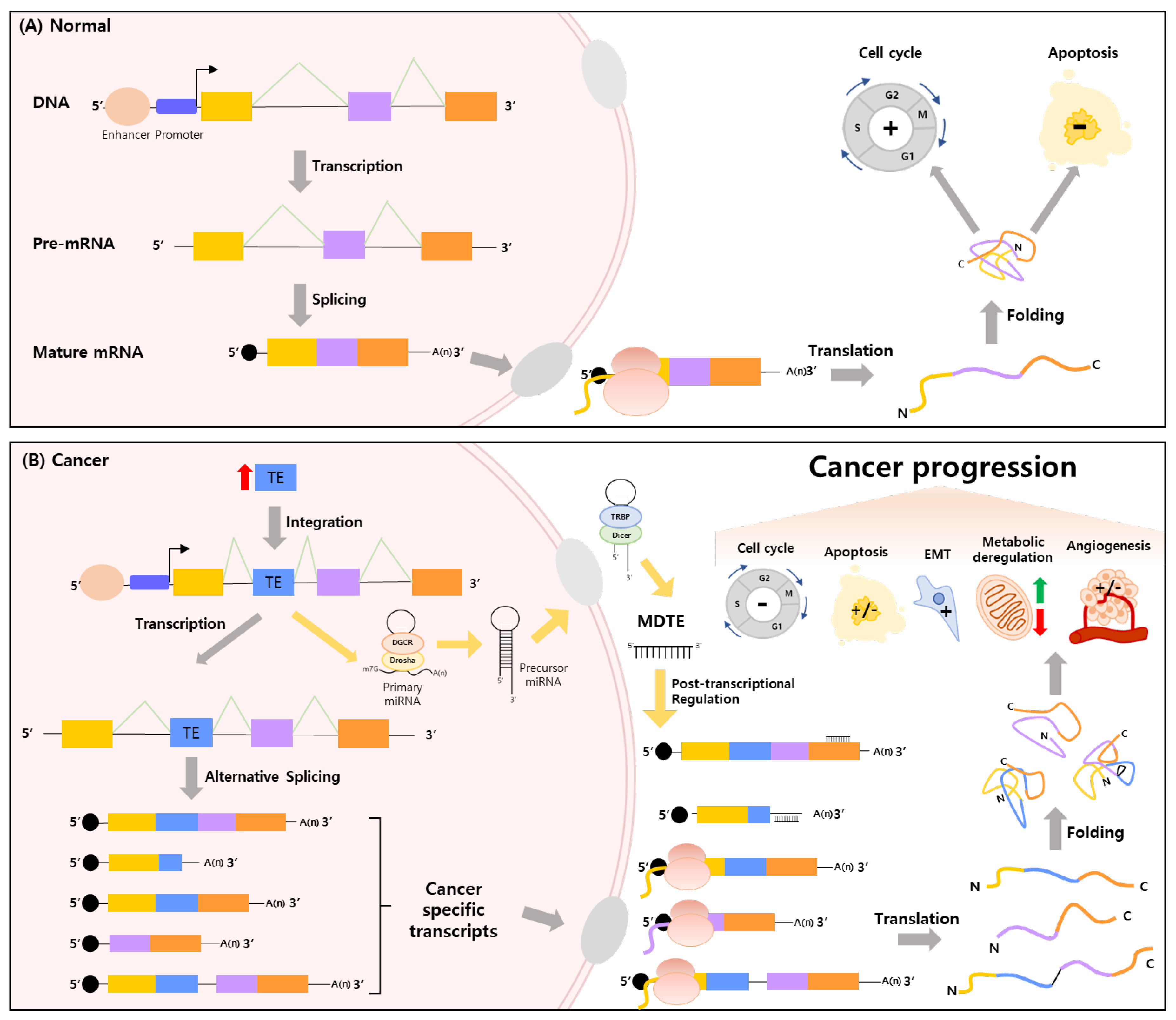
4. MDTEs as Regulatory Elements Linking Alternative Splicing by TE Integration and Cancer Organically
4.1. MDTEs’ Regulatory Processes Related to Cancer Progression and Cancer Therapy
4.2. Cancer Controlling MDTEs in the Top 5 Mortality Cancers
4.2.1. Lung Cancer
4.2.2. Colon Cancer
4.2.3. Liver Cancer
4.2.4. Stomach Cancer
4.2.5. Breast Cancer
| Cancer Type | MiRNA | Subclass | Superfamily | Target Gene | Reference |
|---|---|---|---|---|---|
| Lung cancer | hsa-miR-421 | LINE | L2 | - | [178] |
| hsa-miR-4317 | SINE | MIR | FGF9, CCND2 | [180] | |
| hsa-miR-330-5p | SINE | MIR | RASSF1A | [221] | |
| FAM83A | [185] | ||||
| hsa-miR-374b-5p | LINE | L2 | SRSF7 | [184] | |
| hsa-miR-544a | DNA transposon | hAT-Charlie | FBXW7 | [222] | |
| hsa-miR-1183 | LINE | L2 | PDPK1 | [223] | |
| hsa-miR-181a | LINE | RTE-BovB | GAS7 | [224] | |
| hsa-miR-181b | LINE | RTE-BovB | Bcl-2 | [182] | |
| hsa-miR-340 | DNA transposon | TcMar-Mariner | - | [225] | |
| hsa-miR-340-5p | DNA transposon | TcMar-Mariner | KPNA4 | [226] | |
| hsa-miR-885-3p | SINE | MIR | Aurora A | [183] | |
| hsa-miR-378a-3p | SINE | MIR | CDK4/CDK6 | [227] | |
| hsa-miR-1246 | LTR | ERVL-MaLR | CXCR4 | [181] | |
| hsa-miR-1290 | DNA transposon | TcMar-Tigger | - | [179] | |
| hsa-miR-326 | DNA transposon | hAT-Tip100 | hsa_circ_0003998 | [228] | |
| Sp1 | [229] | ||||
| hsa-miR-608 | LINE | L2 | MIF | [230] | |
| Colon cancer | hsa-miR-585-3p | LTR | ERVL-MaLR | PSME3 | [186] |
| hsa-miR-335-5p | SINE | MIR | RASA1 | [188] | |
| hsa-miR-181a | LINE | RTE-BovB | SRCIN1 | [190] | |
| hsa-miR-708 | LINE | L2 | ZEB1 | [192] | |
| hsa-miR-1273 | SINE | Alu | MAGEA3/6 | [187] | |
| circPIP5K1A | [231] | ||||
| hsa-miR-340-5p | DNA transposon | TcMar-Mariner | CLDN8, IL22 | [189] | |
| hsa-miR-552 | LINE | L1 | PTEN, p53 | [191] | |
| hsa-miR-28-5p | LINE | L2 | HOXB3 | [193] | |
| hsa-miR-374b-5p | LINE | L2 | LRH-1 | [232] | |
| Liver cancer | hsa-miR-23c | SINE | MIR | ERBB2IP | [233] |
| hsa-miR-575 | SINE | MIR | ST7L | [202] | |
| hsa-miR-608 | LINE | L2 | BRD4 | [197] | |
| hsa-miR-326 | DNA transposon | hAT-Tip100 | TWIST1 | [201] | |
| hsa-miR-645 | DNA transposon | hAT-Charlie | SOX30 | [234] | |
| hsa-miR-493-5p | LINE | L2 | VAMP2 | [196] | |
| hsa-miR-224 | DNA transposon | DNA transposon | GNMT | [195] | |
| hsa-miR-342 | SINE | tRNA-RTE | IGF-1R | [199] | |
| MCT1 | [235] | ||||
| hsa-miR-378a | SINE | MIR | VEGFR, PDGFRβ, c-Raf | [236] | |
| hsa-miR-885-5p | SINE | MIR | HK2 | [200] | |
| hsa-miR-421 | LINE | L2 | MAPK14 | [237] | |
| Stomach cancer | hsa-miR-575 | SINE | MIR | PTEN | [238] |
| hsa-miR-581 | DNA transposon | hAT-Charlie | MUC4 | [206] | |
| hsa-miR-552 | LINE | L1 | FOXO1 | [205] | |
| hsa-miR-885-3p | SINE | MIR | CDK4 | [207] | |
| hsa-miR-421 | LINE | L2 | Hsacirc0001546 | [212,239] | |
| hsa-miR-181a | LINE | RTE-BovB | caprin-1 | [240] | |
| hsa-miR-612 | SINE | MIR | IGF2BP1, FOXM1 | [208] | |
| hsa-miR-224-5p | DNA transposon | DNA transposon | circ-LDLRAD3 | [211] | |
| hsa-miR-3200-5p | LTR | ERVL | BCAT1 | [209] | |
| hsa-miR-1269 | LTR | ERVL | RASSF9 | [204] | |
| hsa-miR-585 | LTR | ERVL-MaLR | MAPK1 | [203] | |
| hsa-miR-4317 | SINE | MIR | ZNF322 | [241] | |
| hsa-miR-548c-3p | DNA transposon | TcMar-Mariner | CLIC1 | [210] | |
| Breast cancer | hsa-miR-130a-3p | LINE | RTE-BovB | FOSL1, RAB5B | [242] |
| hsa-miR-582-5p | LINE | CR1 | CMTM8 | [218] | |
| hsa-miR-224-5p | DNA transposon | DNA transposon | CASP9 | [220] | |
| Smad4 | [243] | ||||
| hsa-miR-1246 | LTR | ERVL-MaLR | - | [244] | |
| hsa-miR-326 | DNA transposon | hAT-Tip100 | EGFR, ErbB2, ErbB3, AKT1, AKT2, AKT3 | [214] | |
| hsa-miR-708 | LINE | L2 | - | [245] | |
| hsa-miR-374a-5p | LINE | L2 | ARRB1 | [219] | |
| hsa-miR-335-5p | SINE | MIR | EphA4 | [246] | |
| hsa-miR-181a | LINE | RTE-BovB | AK024094 | [161] | |
| hsa-miR-340-5p | DNA transposon | TcMar-Mariner | LGR5 | [215] | |
| hsa-miR-421 | LINE | L2 | PDCD4 | [213] |
4.3. Cancer Regulatory MDTEs in Other Cancer Types
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Abascal, F.; Tress, M.L.; Valencia, A. Alternative splicing and co-option of TEs: The case of TMPO/LAP2α and ZNF451 in mammals. Bioinformatics 2015, 31, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Hallegger, M.; Llorian, M.; Smith, C.W. Alternative splicing: Global insights. FEBS J. 2010, 277, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, C.; Hartmann, B.; Lundgren, J.; Valcárcel, J. Genome-wide analysis of alternative pre-mRNA splicing. J. Biol. Chem. 2008, 283, 1229–1233. [Google Scholar] [CrossRef]
- Effenberger, K.A.; Perriman, R.J.; Bray, W.M.; Lokey, R.S.; Ares, M., Jr.; Jurica, M.S. A high-throughput splicing assay identifies new classes of inhibitors of human and yeast spliceosomes. J. Biomol. Screen. 2013, 18, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef]
- Matlin, A.J.; Clark, F.; Smith, C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef]
- Faustino, N.A.; Cooper, T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003, 17, 419–437. [Google Scholar] [CrossRef]
- Venables, J.P.; Klinck, R.; Bramard, A.; Inkel, L.; Dufresne-Martin, G.; Koh, C.; Gervais-Bird, J.; Lapointe, E.; Froehlich, U.; Durand, M. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008, 68, 9525–9531. [Google Scholar] [CrossRef]
- Kahles, A.; Lehmann, K.-V.; Toussaint, N.C.; Hüser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Caesar-Johnson, S.J. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 2018, 34, 211–224.e6. [Google Scholar] [CrossRef]
- Kim, E.; Goren, A.; Ast, G. Alternative splicing: Current perspectives. Bioessays 2008, 30, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Herron, P. Mobile DNA II. Heredity 2004, 92, 476. [Google Scholar] [CrossRef][Green Version]
- Wells, J.N.; Feschotte, C. A field guide to eukaryotic TEs. Annu. Rev. Genet. 2020, 54, 539. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.; Satish, L.; Sharma, A.; Vinod, K.K.; Emamverdian, A.; Zhou, M.; Wei, Q. Transposable elements in plants: Recent advancements, tools and prospects. Plant Mol. Biol. Rep. 2022, 1–18. [Google Scholar] [CrossRef]
- Mercan, L.; Bülbül, C.E.; Bilgi, F.; Marakli, S. Determination of plant-specific retrotransposons in chicken. Turk. J. Agric. For. 2022, 46, 67–73. [Google Scholar]
- de Assis, R.; Baba, V.Y.; Cintra, L.A.; Gonçalves, L.S.A.; Rodrigues, R.; Vanzela, A.L.L. Genome relationships and LTR-retrotransposon diversity in three cultivated Capsicum L. (Solanaceae) species. BMC Genom. 2020, 21, 237. [Google Scholar] [CrossRef]
- Liu, G.; Jiang, H.; Sun, W.; Zhang, J.; Chen, D.; Murchie, A.I. The function of twister ribozyme variants in non-LTR retrotransposition in Schistosoma mansoni. Nucleic Acids Res. 2021, 49, 10573–10588. [Google Scholar] [CrossRef]
- Wicker, T.; Stritt, C.; Sotiropoulos, A.G.; Poretti, M.; Pozniak, C.; Walkowiak, S.; Gundlach, H.; Stein, N. TE Populations Shed Light on the Evolutionary History of Wheat and the Complex Co-Evolution of Autonomous and Non-Autonomous Retrotransposons. Adv. Genet. 2022, 3, 2100022. [Google Scholar] [CrossRef]
- Kubota, S.; Ishikawa, T.; Kawata, K.; Hattori, T.; Nishida, T. Retrotransposons manipulating mammalian skeletal development in chondrocytes. Int. J. Mol. Sci. 2020, 21, 1564. [Google Scholar] [CrossRef]
- Maquat, L.E. Short interspersed nuclear element (SINE)-mediated post-transcriptional effects on human and mouse gene expression: SINE-UP for active duty. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190344. [Google Scholar] [CrossRef]
- Maxwell, P.H. Consequences of ongoing retrotransposition in mammalian genomes. Adv. Genom. Genet. 2014, 4, 129–142. [Google Scholar] [CrossRef][Green Version]
- Ullastres, A.; Merenciano, M.; González, J. Regulatory regions in natural TE insertions drive interindividual differences in response to immune challenges in Drosophila. Genome Biol. 2021, 22, 265. [Google Scholar] [CrossRef] [PubMed]
- Szatkowska, M.; Krupa, R. Regulation of DNA damage response and homologous recombination repair by miRNA in human cells exposed to ionizing radiation. Cancers 2020, 12, 1838. [Google Scholar] [CrossRef]
- Campo, S.; Sánchez-Sanuy, F.; Camargo-Ramírez, R.; Gómez-Ariza, J.; Baldrich, P.; Campos-Soriano, L.; Soto-Suárez, M.; San Segundo, B. A novel TE-derived miRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 2021, 19, 1798–1811. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Rösch, N.; Hausser, J.; Manoharan, M.; Zavolan, M.; Stoffel, M. MiRNA-194 is a target of transcription factor 1 (Tcf1, HNF1α) in adult liver and controls expression of frizzled-6. Hepatology 2012, 55, 98–107. [Google Scholar] [CrossRef]
- Liz, J.; Esteller, M. lncRNAs and miRNAs with a role in cancer development. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 169–176. [Google Scholar] [CrossRef]
- Grundy, E.E.; Diab, N.; Chiappinelli, K.B. TE regulation and expression in cancer. FEBS J. 2022, 289, 1160–1179. [Google Scholar] [CrossRef]
- Beckmann, P.J.; Largaespada, D.A. Transposon insertion mutagenesis in mice for modeling human cancers: Critical insights gained and new opportunities. Int. J. Mol. Sci. 2020, 21, 1172. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Leoni, G.; Le Pera, L.; Ferrè, F.; Raimondo, D.; Tramontano, A. Coding potential of the products of alternative splicing in human. Genome Biol. 2011, 12, R9. [Google Scholar] [CrossRef]
- Papasaikas, P.; Valcárcel, J. The spliceosome: The ultimate RNA chaperone and sculptor. Trends Biochem. Sci. 2016, 41, 33–45. [Google Scholar] [CrossRef]
- Kastner, B.; Will, C.L.; Stark, H.; Lührmann, R. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb. Perspect. Biol. 2019, 11, a032417. [Google Scholar] [CrossRef] [PubMed]
- Furlanis, E.; Traunmüller, L.; Fucile, G.; Scheiffele, P. Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat. Neurosci. 2019, 22, 1709–1717. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, S.C.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Zhang, J.; Manley, J.L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013, 3, 1228–1237. [Google Scholar] [CrossRef]
- Visconte, V.; Nakashima, M.O.; Rogers, H.J. Mutations in splicing factor genes in myeloid malignancies: Significance and impact on clinical features. Cancers 2019, 11, 1844. [Google Scholar] [CrossRef]
- Anczuków, O.; Akerman, M.; Cléry, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y. SRSF1-regulated alternative splicing in breast cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef]
- Liang, Y.; Tebaldi, T.; Rejeski, K.; Joshi, P.; Stefani, G.; Taylor, A.; Song, Y.; Vasic, R.; Maziarz, J.; Balasubramanian, K. SRSF2 mutations drive oncogenesis by activating a global program of aberrant alternative splicing in hematopoietic cells. Leukemia 2018, 32, 2659–2671. [Google Scholar] [CrossRef]
- Jafari, P.A.; Ayatollahi, H.; Sadeghi, R.; Sheikhi, M.; Asghari, A. Prognostic significance of SRSF2 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: A meta-analysis. Hematology 2018, 23, 778–784. [Google Scholar] [CrossRef]
- Zhang, J.; Lieu, Y.K.; Ali, A.M.; Penson, A.; Reggio, K.S.; Rabadan, R.; Raza, A.; Mukherjee, S.; Manley, J.L. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. USA 2015, 112, E4726–E4734. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Hu, X.; Zhou, Y.; Shi, Y.; Qian, R.; Wan, Y. Characterization of the aberrant splicing of DVL2 induced by cancer-associated SF3B1 mutation. Biochem. Biophys. Res. Commun. 2021, 546, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, Z.; Qian, R.; Liu, G.; Fan, M.; Liang, Z.; Hu, X.; Wan, Y. Cancer-associated mutations in SF3B1 disrupt the interaction between SF3B1 and DDX42. J. Biochem. 2022, 172, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Palangat, M.; Anastasakis, D.G.; Fei, D.L.; Lindblad, K.E.; Bradley, R.; Hourigan, C.S.; Hafner, M.; Larson, D.R. The splicing factor U2AF1 contributes to cancer progression through a noncanonical role in translation regulation. Genes Dev. 2019, 33, 482–497. [Google Scholar] [CrossRef]
- Lin, J.-C.; Lin, C.-Y.; Tarn, W.-Y.; Li, F.-Y. Elevated SRPK1 lessens apoptosis in breast cancer cells through RBM4-regulated splicing events. RNA 2014, 20, 1621–1631. [Google Scholar] [CrossRef]
- Zhang, Y.-P.; Liu, K.-L.; Wang, Y.-X.; Yang, Z.; Han, Z.-W.; Lu, B.-S.; Qi, J.-C.; Yin, Y.-W.; Teng, Z.-H.; Chang, X.-L. Down-regulated RBM5 inhibits bladder cancer cell apoptosis by initiating an miR-432-5p/β-catenin feedback loop. FASEB J. 2019, 33, 10973–10985. [Google Scholar] [CrossRef]
- Dou, X.Q.; Chen, X.J.; Wen, M.X.; Zhang, S.Z.; Zhou, Q.; Zhang, S.Q. Alternative splicing of VEGFA is regulated by RBM10 in endometrial cancer. Kaohsiung J. Med. Sci. 2020, 36, 13–19. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, F.; Zhong, W.; Ling, H.; Wang, J.; Cui, J.; Xie, T.; Wen, S.; Chen, J. RNA-binding protein RBM6 as a tumor suppressor gene represses the growth and progression in laryngocarcinoma. Gene 2019, 697, 26–34. [Google Scholar] [CrossRef]
- French, P.J.; Peeters, J.; Horsman, S.; Duijm, E.; Siccama, I.; Van Den Bent, M.J.; Luider, T.M.; Kros, J.M.; van der Spek, P.; Sillevis Smitt, P.A. Identification of differentially regulated splice variants and novel exons in glial brain tumors using exon expression arrays. Cancer Res. 2007, 67, 5635–5642. [Google Scholar] [CrossRef]
- Milani, L.; Fredriksson, M.; Syvanen, A.-C. Detection of alternatively spliced transcripts in leukemia cell lines by minisequencing on microarrays. Clin. Chem. 2006, 52, 202–211. [Google Scholar] [CrossRef]
- Li, H.-R.; Wang-Rodriguez, J.; Nair, T.M.; Yeakley, J.M.; Kwon, Y.-S.; Bibikova, M.; Zheng, C.; Zhou, L.; Zhang, K.; Downs, T. Two-dimensional transcriptome profiling: Identification of mRNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006, 66, 4079–4088. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Xu, Q.; Lee, C. Evidence that public database records for many cancer-associated genes reflect a splice form found in tumors and lack normal splice forms. Nucleic Acids Res. 2005, 33, 5026–5033. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boise, L.H.; González-García, M.; Postema, C.E.; Ding, L.; Lindsten, T.; Turka, L.A.; Mao, X.; Nuñez, G.; Thompson, C.B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 1993, 74, 597–608. [Google Scholar] [CrossRef]
- Gayle, S.S.; Sahni, J.M.; Webb, B.M.; Weber-Bonk, K.L.; Shively, M.S.; Spina, R.; Bar, E.E.; Summers, M.K.; Keri, R.A. Targeting BCL-xL improves the efficacy of bromodomain and extra-terminal protein inhibitors in triple-negative breast cancer by eliciting the death of senescent cells. J. Biol. Chem. 2019, 294, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Takehara, T.; Liu, X.; Fujimoto, J.; Friedman, S.L.; Takahashi, H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology 2001, 34, 55–61. [Google Scholar] [CrossRef]
- Prinos, P.; Garneau, D.; Lucier, J.-F.; Gendron, D.; Couture, S.; Boivin, M.; Brosseau, J.-P.; Lapointe, E.; Thibault, P.; Durand, M. Alternative splicing of SYK regulates mitosis and cell survival. Nat. Struct. Mol. Biol. 2011, 18, 673–679. [Google Scholar] [CrossRef]
- Du, Z.; Fan, M.; Kim, J.-G.; Eckerle, D.; Lothstein, L.; Wei, L.; Pfeffer, L.M. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J. Biol. Chem. 2009, 284, 27808–27815. [Google Scholar] [CrossRef]
- Schittenhelm, M.M.; Walter, B.; Tsintari, V.; Federmann, B.; Saipi, M.B.; Akmut, F.; Illing, B.; Mau-Holzmann, U.; Fend, F.; Lopez, C.D. Alternative splicing of the tumor suppressor ASPP2 results in a stress-inducible, oncogenic isoform prevalent in acute leukemia. EBioMedicine 2019, 42, 340–351. [Google Scholar] [CrossRef]
- Liu, J.H.; Wei, S.; Lamy, T.; Li, Y.; Epling-Burnette, P.; Djeu, J.Y.; Loughran, T.P., Jr. Blockade of Fas-dependent apoptosis by soluble Fas in LGL leukemia. Blood Am. J. Hematol. 2002, 100, 1449–1453. [Google Scholar] [CrossRef]
- Inaba, H.; Komada, Y.; Li, Q.S.; Zhang, X.L.; Tanaka, S.; Azuma, E.; Yamamoto, H.; Sakurai, M. mRNA expression of variant Fas molecules in acute leukemia cells. Am. J. Hematol. 1999, 62, 150–158. [Google Scholar] [CrossRef]
- Kamihira, S.; Yamada, Y.; Tomonaga, M.; Sugahara, K.; Tsuruda, K. Discrepant expression of membrane and soluble isoforms of Fas (CD95/APO-1) in adult T-cell leukaemia: Soluble Fas isoform is an independent risk factor for prognosis. Br. J. Haematol. 1999, 107, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Iwasaki, T.; Okada, T.; Hashimoto, T.; Moon, Y.; Sakaguchi, M.; Fukami, Y.; Nishigori, C.; Oka, M. High expression of Mcl-1L via the MEK-ERK-phospho-STAT 3 (Ser727) pathway protects melanocytes and melanoma from UVB-induced apoptosis. Genes Cells 2016, 21, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Bria, E.; Di Modugno, F.; Sperduti, I.; Iapicca, P.; Visca, P.; Alessandrini, G.; Antoniani, B.; Pilotto, S.; Ludovini, V.; Vannucci, J. Prognostic impact of alternative splicing-derived hMENA isoforms in resected, node-negative, non-small-cell lung cancer. Oncotarget 2014, 5, 11054. [Google Scholar] [CrossRef][Green Version]
- Philippar, U.; Roussos, E.T.; Oser, M.; Yamaguchi, H.; Kim, H.-D.; Giampieri, S.; Wang, Y.; Goswami, S.; Wyckoff, J.B.; Lauffenburger, D.A. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev. Cell 2008, 15, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Oudin, M.J.; Hughes, S.K.; Rohani, N.; Moufarrej, M.N.; Jones, J.G.; Condeelis, J.S.; Lauffenburger, D.A.; Gertler, F.B. Characterization of the expression of the pro-metastatic MenaINV isoform during breast tumor progression. Clin. Exp. Metastasis 2016, 33, 249–261. [Google Scholar] [CrossRef]
- Di Modugno, F.; Iapicca, P.; Boudreau, A.; Mottolese, M.; Terrenato, I.; Perracchio, L.; Carstens, R.P.; Santoni, A.; Bissell, M.J.; Nisticò, P. Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 19280–19285. [Google Scholar] [CrossRef]
- Narla, G.; DiFeo, A.; Yao, S.; Banno, A.; Hod, E.; Reeves, H.L.; Qiao, R.F.; Camacho-Vanegas, O.; Levine, A.; Kirschenbaum, A. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005, 65, 5761–5768. [Google Scholar] [CrossRef]
- Hatami, R.; Sieuwerts, A.M.; Izadmehr, S.; Yao, Z.; Qiao, R.F.; Papa, L.; Look, M.P.; Smid, M.; Ohlssen, J.; Levine, A.C. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci. Transl. Med. 2013, 5, 169ra12. [Google Scholar] [CrossRef]
- Prochazka, L.; Tesarik, R.; Turanek, J. Regulation of alternative splicing of CD44 in cancer. Cell. Signal. 2014, 26, 2234–2239. [Google Scholar] [CrossRef]
- Wielenga, V.J.; Heider, K.-H.; Johan, G.; Offerhaus, A.; Adolf, G.R.; van den Berg, F.M.; Ponta, H.; Herrlich, P.; Pals, S.T. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res. 1993, 53, 4754–4756. [Google Scholar]
- Ni, J.; Cozzi, P.J.; Hao, J.L.; Beretov, J.; Chang, L.; Duan, W.; Shigdar, S.; Delprado, W.J.; Graham, P.H.; Bucci, J. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate 2014, 74, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Jiang, G.; Zhang, X.; Zhao, H.; Wu, J.; Xu, K.; Wang, E. Impact of p120-catenin isoforms 1A and 3A on epithelial mesenchymal transition of lung cancer cells expressing E-cadherin in different subcellular locations. PLoS ONE 2014, 9, e88064. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Hagio, M.; Seya, T.; Ishiwata, T. Fibroblast Growth Factor Receptor 2 IIIc as a Therapeutic Target for Colorectal Cancer CellsFGFR2IIIc in Colorectal Cancer. Mol. Cancer Ther. 2012, 11, 2010–2020. [Google Scholar] [CrossRef] [PubMed]
- Kawase, R.; Ishiwata, T.; Matsuda, Y.; Onda, M.; Kudo, M.; Takeshita, T.; Naito, Z. Expression of fibroblast growth factor receptor 2 IIIc in human uterine cervical intraepithelial neoplasia and cervical cancer. Int. J. Oncol. 2010, 36, 331–340. [Google Scholar]
- Ranieri, D.; Rosato, B.; Nanni, M.; Magenta, A.; Belleudi, F.; Torrisi, M.R. Expression of the FGFR2 mesenchymal splicing variant in epithelial cells drives epithelial-mesenchymal transition. Oncotarget 2016, 7, 5440. [Google Scholar] [CrossRef]
- Ghigna, C.; Giordano, S.; Shen, H.; Benvenuto, F.; Castiglioni, F.; Comoglio, P.M.; Green, M.R.; Riva, S.; Biamonti, G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 2005, 20, 881–890. [Google Scholar] [CrossRef]
- Collesi, C.; Santoro, M.M.; Gaudino, G.; Comoglio, P.M. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol. Cell. Biol. 1996, 16, 5518–5526. [Google Scholar] [CrossRef]
- Ben-Hur, V.; Denichenko, P.; Siegfried, Z.; Maimon, A.; Krainer, A.; Davidson, B.; Karni, R. S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep. 2013, 3, 103–115. [Google Scholar] [CrossRef]
- Mei, H.; Wang, Y.; Fan, J.; Lin, Z. Alternative splicing of S6K1 promotes non-small cell lung cancer survival. Tumor Biol. 2016, 37, 13369–13376. [Google Scholar] [CrossRef]
- Bechara, E.G.; Sebestyén, E.; Bernardis, I.; Eyras, E.; Valcárcel, J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 2013, 52, 720–733. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Kubbutat, M.H.; Jones, S.N.; Vousden, K.H. Regulation of p53 stability by Mdm2. Nature 1997, 387, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Sigalas, I.; Calvert, A.H.; Anderson, J.J.; Neal, D.E.; Lunec, J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: Transforming ability and frequent detection in human cancer. Nat. Med. 1996, 2, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Biselli-Chicote, P.; Oliveira, A.; Pavarino, E.; Goloni-Bertollo, E. VEGF gene alternative splicing: Pro-and anti-angiogenic isoforms in cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 363–370. [Google Scholar] [CrossRef]
- Abou Faycal, C.; Gazzeri, S.; Eymin, B. A VEGF-A/SOX2/SRSF2 network controls VEGFR1 pre-mRNA alternative splicing in lung carcinoma cells. Sci. Rep. 2019, 9, 336. [Google Scholar] [CrossRef]
- Albuquerque, R.J.; Hayashi, T.; Cho, W.G.; Kleinman, M.E.; Dridi, S.; Takeda, A.; Baffi, J.Z.; Yamada, K.; Kaneko, H.; Green, M.G. Alternatively spliced vascular endothelial growth factor receptor-2 is an essential endogenous inhibitor of lymphatic vessel growth. Nat. Med. 2009, 15, 1023–1030. [Google Scholar] [CrossRef]
- Clower, C.V.; Chatterjee, D.; Wang, Z.; Cantley, L.C.; Vander Heiden, M.G.; Krainer, A.R. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 1894–1899. [Google Scholar] [CrossRef]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF (V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef]
- Takeuchi, A.; Yokoyama, S.; Nakamori, M.; Nakamura, M.; Ojima, T.; Yamaguchi, S.; Mitani, Y.; Shively, J.E.; Yamaue, H. Loss of CEACAM1 is associated with poor prognosis and peritoneal dissemination of patients with gastric cancer. Sci. Rep. 2019, 9, 12702. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Z.; Baker, K.; Halvorsen, E.M.; da Cunha, A.P.; Flak, M.B.; Gerber, G.; Huang, Y.-H.; Hosomi, S.; Arthur, J.C. The short isoform of the CEACAM1 receptor in intestinal T cells regulates mucosal immunity and homeostasis via Tfh cell induction. Immunity 2012, 37, 930–946. [Google Scholar] [CrossRef]
- Helfrich, I.; Singer, B.B. Size matters: The functional role of the CEACAM1 isoform signature and its impact for NK cell-mediated killing in melanoma. Cancers 2019, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Comstock, C.E.; Augello, M.A.; Benito, R.P.; Karch, J.; Tran, T.H.; Utama, F.E.; Tindall, E.A.; Wang, Y.; Burd, C.J.; Groh, E.M. Cyclin D1 Splice Variants: Polymorphism, Risk, and Isoform-Specific Regulation in Prostate CancerCyclin D1b Regulation in Prostate Cancer. Clin. Cancer Res. 2009, 15, 5338–5349. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Sen, S.; Wei, C.; Frazier, M.L. Cyclin D1b represses breast cancer cell growth by antagonizing the action of cyclin D1a on estrogen receptor α-mediated transcription. Int. J. Oncol. 2010, 36, 39–48. [Google Scholar] [PubMed]
- Garcia-Perez, J.L.; Widmann, T.J.; Adams, I.R. The impact of TEs on mammalian development. Development 2016, 143, 4101–4114. [Google Scholar] [CrossRef]
- Schrader, L.; Schmitz, J. The impact of TEs in adaptive evolution. Mol. Ecol. 2019, 28, 1537–1549. [Google Scholar] [CrossRef]
- Bernard, A.; Boidot, R.; Végran, F. Alternative Splicing in Cancer and Immune Cells. Cancers 2022, 14, 1726. [Google Scholar] [CrossRef]
- Kaer, K.; Speek, M. Retroelements in human disease. Gene 2013, 518, 231–241. [Google Scholar] [CrossRef]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J., III; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef]
- Krasileva, K.V. The role of TEs and DNA damage repair mechanisms in gene duplications and gene fusions in plant genomes. Curr. Opin. Plant Biol. 2019, 48, 18–25. [Google Scholar] [CrossRef]
- Makałowski, W.; Mitchell, G.A.; Labuda, D. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet. 1994, 10, 188–193. [Google Scholar] [CrossRef]
- Sorek, R.; Ast, G.; Graur, D. Alu-containing exons are alternatively spliced. Genome Res. 2002, 12, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Nekrutenko, A.; Li, W.-H. TEs are found in a large number of human protein-coding genes. Trends Genet. 2001, 17, 619–621. [Google Scholar] [CrossRef]
- Singer, S.S.; Männel, D.N.; Hehlgans, T.; Brosius, J.; Schmitz, J. From “junk” to gene: Curriculum vitae of a primate receptor isoform gene. J. Mol. Biol. 2004, 341, 883–886. [Google Scholar] [CrossRef]
- Gotea, V.; Makałowski, W. Do TEs really contribute to proteomes? Trends Genet. 2006, 22, 260–267. [Google Scholar] [CrossRef]
- Zhang, X.H.-F.; Chasin, L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl. Acad. Sci. USA 2006, 103, 13427–13432. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, J.; Lu, J.; Wang, P.; Feng, H.; Zong, Y.; Ou, B.; Zheng, M.; Lu, A. Cadherin-12 enhances proliferation in colorectal cancer cells and increases progression by promoting EMT. Tumor Biol. 2016, 37, 9077–9088. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Y.; Olopade, O.I. MYC and breast cancer. Genes Cancer 2010, 1, 629–640. [Google Scholar] [CrossRef]
- Morse, B.; Rotherg, P.G.; South, V.J.; Spandorfer, J.M.; Astrin, S.M. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 1988, 333, 87–90. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Y.; Tian, N.; Zhao, X.; Du, C.; Han, L.; Zhang, H. Aberrant alternative splicing pattern of ADAR2 downregulates adenosine-to-inosine editing in glioma. Oncol. Rep. 2015, 33, 2845–2852. [Google Scholar] [CrossRef]
- Wang, X.; Ren, X.; Liu, W.; Chen, X.; Wei, J.; Gong, Z.; Yan, Y.; Xu, Z. Role of downregulated ADARB1 in lung squamous cell carcinoma. Mol. Med. Rep. 2020, 21, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Clayton, E.A.; Rishishwar, L.; Huang, T.-C.; Gulati, S.; Ban, D.; McDonald, J.F.; Jordan, I.K. An atlas of TE-derived alternative splicing in cancer. Philos. Trans. R. Soc. B 2020, 375, 20190342. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xu, Y.; Liao, H.; Cai, Y.; Xu, L.; Xiao, D.; Liu, C.; Pu, W.; Zhong, X.; Guo, X. Identification and validation of key genes associated with non-small-cell lung cancer. J. Cell. Physiol. 2019, 234, 22742–22752. [Google Scholar] [CrossRef] [PubMed]
- Dubin, M.J.; Scheid, O.M.; Becker, C. Transposons: A blessing curse. Curr. Opin. Plant Biol. 2018, 42, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Teugels, E.; De Brakeleer, S.; Goelen, G.; Lissens, W.; Sermijn, E.; De Grève, J. De novo Alu element insertions targeted to a sequence common to the BRCA1 and BRCA2 genes. Hum. Mutat. 2005, 26, 284. [Google Scholar] [CrossRef]
- Guilbert, A.; Dhennin-Duthille, I.; Hiani, Y.E.; Haren, N.; Khorsi, H.; Sevestre, H.; Ahidouch, A.; Ouadid-Ahidouch, H. Expression of TRPC6 channels in human epithelial breast cancer cells. BMC Cancer 2008, 8, 125. [Google Scholar] [CrossRef]
- Zhang, Y.; Romanish, M.T.; Mager, D.L. Distributions of TEs reveal hazardous zones in mammalian introns. PLoS Comput. Biol. 2011, 7, e1002046. [Google Scholar] [CrossRef]
- Cajuso, T.; Sulo, P.; Tanskanen, T.; Katainen, R.; Taira, A.; Hänninen, U.A.; Kondelin, J.; Forsström, L.; Välimäki, N.; Aavikko, M. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat. Commun. 2019, 10, 4022. [Google Scholar] [CrossRef]
- Becklin, K.L.; Smeester, B.A.; Moriarity, B.S. Cancer gene discovery utilizing sleeping beauty transposon mutagenesis. In Cancer Driver Genes; Humana Press: New York, NY, USA, 2019; pp. 161–170. [Google Scholar]
- Burns, K.H. TEs in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Lev-Maor, G.; Sorek, R.; Shomron, N.; Ast, G. The birth of an alternatively spliced exon: 3’splice-site selection in Alu exons. Science 2003, 300, 1288–1291. [Google Scholar] [CrossRef]
- Krull, M.; Brosius, J.R.; Schmitz, J.R. Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 2005, 22, 1702–1711. [Google Scholar] [CrossRef] [PubMed]
- Kocher, O.; Pal, R.; Roberts, M.; Cirovic, C.; Gilchrist, A. Targeted disruption of the PDZK1 gene by homologous recombination. Mol. Cell. Biol. 2003, 23, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, R.; Farivar, S.; Mager, D.L. C-GATE-catalogue of genes affected by TEs. Mob. DNA 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Gao, G.; Schneider, S.; Helt, C.; Weiss, C.; O’Reilly, M.A.; Bohmann, D.; Zhao, J. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004, 23, 1133–1143. [Google Scholar] [CrossRef]
- Xie, R.; Medina, R.; Zhang, Y.; Hussain, S.; Colby, J.; Ghule, P.; Sundararajan, S.; Keeler, M.; Liu, L.-J.; Van der Deen, M. The histone gene activator HINFP is a nonredundant cyclin E/CDK2 effector during early embryonic cell cycles. Proc. Natl. Acad. Sci. USA 2009, 106, 12359–12364. [Google Scholar] [CrossRef]
- Lucas, B.A.; Lavi, E.; Shiue, L.; Cho, H.; Katzman, S.; Miyoshi, K.; Siomi, M.C.; Carmel, L.; Ares, M., Jr.; Maquat, L.E. Evidence for convergent evolution of SINE-directed Staufen-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 2018, 115, 968–973. [Google Scholar] [CrossRef]
- Jung, Y.-D.; Huh, J.-W.; Kim, D.-S.; Kim, Y.-J.; Ahn, K.; Ha, H.-S.; Lee, J.-R.; Yi, J.M.; Moon, J.-W.; Kim, T.-O. Quantitative analysis of transcript variants of CHM gene containing LTR12C element in humans. Gene 2011, 489, 1–5. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ji, Z.; Tian, B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of TEs in evolution of the 3′-end of genes. Nucleic Acids Res. 2008, 36, 5581–5590. [Google Scholar] [CrossRef]
- Miki, Y.; Nishisho, I.; Horii, A.; Miyoshi, Y.; Utsunomiya, J.; Kinzler, K.W.; Vogelstein, B.; Nakamura, Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992, 52, 643–645. [Google Scholar]
- Shukla, R.; Upton, K.R.; Muñoz-Lopez, M.; Gerhardt, D.J.; Fisher, M.E.; Nguyen, T.; Brennan, P.M.; Baillie, J.K.; Collino, A.; Ghisletti, S. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 2013, 153, 101–111. [Google Scholar] [CrossRef]
- Savage, A.L.; Schumann, G.G.; Breen, G.; Bubb, V.J.; Al-Chalabi, A.; Quinn, J.P. Retrotransposons in the development and progression of amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Drongitis, D.; Aniello, F.; Fucci, L.; Donizetti, A. Roles of TEs in the different layers of gene expression regulation. Int. J. Mol. Sci. 2019, 20, 5755. [Google Scholar] [CrossRef] [PubMed]
- van de Lagemaat, L.N.; Landry, J.-R.; Mager, D.L.; Medstrand, P. TEs in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003, 19, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Staniunas, R.J.; Barnard, G.F.; Jessup, J.M.; Steele, G.D., Jr.; Chen, L.B. The significance of carbonic anhydrase expression in human colorectal cancer. Gastroenterology 1993, 105, 820–826. [Google Scholar] [CrossRef]
- Kim, H.J.; Moon, S.J.; Kim, S.-H.; Heo, K.; Kim, J.H. DBC1 regulates Wnt/β-catenin-mediated expression of MACC1, a key regulator of cancer progression, in colon cancer. Cell Death Dis. 2018, 9, 831. [Google Scholar] [CrossRef]
- Jun, H.J.; Johnson, H.; Bronson, R.T.; de Feraudy, S.; White, F.; Charest, A. The Oncogenic Lung Cancer Fusion Kinase CD74-ROS Activates a Novel Invasiveness Pathway through E-Syt1 PhosphorylationOncogenic Properties of ROS Fusion Kinases. Cancer Res. 2012, 72, 3764–3774. [Google Scholar] [CrossRef]
- Popowski, M.; Templeton, T.D.; Lee, B.-K.; Rhee, C.; Li, H.; Miner, C.; Dekker, J.D.; Orlanski, S.; Bergman, Y.; Iyer, V.R. Bright/Arid3A acts as a barrier to somatic cell reprogramming through direct regulation of Oct4, Sox2, and Nanog. Stem Cell Rep. 2014, 2, 26–35. [Google Scholar] [CrossRef]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S. TEs drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef]
- Babaian, A.; Mager, D.L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 2016, 7, 24. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014, 63, 635–646. [Google Scholar] [CrossRef]
- Williams, C.S.; Bernard, J.K.; Beckler, M.D.; Almohazey, D.; Washington, M.K.; Smith, J.J.; Frey, M.R. ERBB4 is over-expressed in human colon cancer and enhances cellular transformation. Carcinogenesis 2015, 36, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Mendis, S.R.; Topham, J.T.; Titmuss, E.; Williamson, L.; Pleasance, E.D.; Culibrk, L.; Karasinska, J.; Liu, S.L.; Lee, M.; Aird, J. Comprehensive transcriptome analysis reveals link between epigenetic dysregulation, ERV expression and immunogenicity in metastatic colorectal carcinoma. Am. J. Clin. Oncol. 2019, 37, 3535. [Google Scholar] [CrossRef]
- De Cubas, A.A.; Dunker, W.; Zaninovich, A.; Hongo, R.A.; Bhatia, A.; Panda, A.; Beckermann, K.E.; Bhanot, G.; Ganesan, S.; Karijolich, J. DNA hypomethylation promotes TE expression and activation of immune signaling in renal cell cancer. JCI Insight 2020, 5, e137569. [Google Scholar] [CrossRef] [PubMed]
- Howard, G.; Eiges, R.; Gaudet, F.; Jaenisch, R.; Eden, A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 2008, 27, 404–408. [Google Scholar] [CrossRef]
- Ross, J.P.; Rand, K.N.; Molloy, P.L. Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2010, 2, 245–269. [Google Scholar] [CrossRef]
- Watanabe, Y.; Maekawa, M. Methylation of DNA in cancer. Adv. Clin. Chem. 2010, 52, 145–167. [Google Scholar]
- Kulis, M.; Queirós, A.C.; Beekman, R.; Martín-Subero, J.I. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 1161–1174. [Google Scholar] [CrossRef]
- Florl, A.; Steinhoff, C.; Müller, M.; Seifert, H.; Hader, C.; Engers, R.; Ackermann, R.; Schulz, W. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br. J. Cancer 2004, 91, 985–994. [Google Scholar] [CrossRef]
- Santourlidis, S.; Florl, A.; Ackermann, R.; Wirtz, H.C.; Schulz, W.A. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate 1999, 39, 166–174. [Google Scholar] [CrossRef]
- Kim, M.-J.; White-Cross, J.A.; Shen, L.; Issa, J.-P.J.; Rashid, A. Hypomethylation of long interspersed nuclear element-1 in hepatocellular carcinomas. Mod. Pathol. 2009, 22, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.G.; Jares, P.; Rico, D.; Gómez-López, G.; Martínez-Trillos, A.; Villamor, N.; Ecker, S.; González-Pérez, A.; Knowles, D.G.; Monlong, J. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014, 24, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Ecsedi, S.I.; Hernandez-Vargas, H.; Lima, S.C.; Herceg, Z.; Adany, R.; Balazs, M. Transposable hypomethylation is associated with metastatic capacity of primary melanomas. Int. J. Clin. Exp. Pathol. 2013, 6, 2943. [Google Scholar] [PubMed]
- Xiang, S.; Liu, Z.; Zhang, B.; Zhou, J.; Zhu, B.-D.; Ji, J.; Deng, D. Methylation status of individual CpG sites within Alu elements in the human genome and Alu hypomethylation in gastric carcinomas. BMC Cancer 2010, 10, 1–11. [Google Scholar] [CrossRef]
- Turker, M.S.; Bestor, T.H. Formation of methylation patterns in the mammalian genome. Mutat. Res. Rev. Mutat. Res. 1997, 386, 119–130. [Google Scholar] [CrossRef]
- Wang, X.; Fan, J.; Liu, D.; Fu, S.; Ingvarsson, S.; Chen, H. Spreading of Alu methylation to the promoter of the MLH1 gene in gastrointestinal cancer. PLoS ONE 2011, 6, e25913. [Google Scholar] [CrossRef] [PubMed]
- Shirley, K.; Reichard, K.; Grover, N. Small Non-coding RNA, miRNA in Gene Regulation. In Fundamentals of RNA Structure and Function; Springer: Cham, Switzerland, 2022; pp. 167–190. [Google Scholar]
- Ghosh, A.; Platt, R.N., II; Vandewege, M.W.; Tabassum, R.; Hsu, C.-Y.; Isberg, S.R.; Peterson, D.G.; Finger, J.W., Jr.; Kieran, T.J.; Glenn, T.C. Identification and characterization of miRNAs (miRNAs) and their TE origins in the saltwater crocodile, Crocodylus porosus. Anal. Biochem. 2020, 602, 113781. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-N.; Hu, S.; Shang, Y.-P.; Li, L.-Y.; Zhou, H.; Chen, J.-S.; Yang, J.-F.; Li, J.; Huang, Q.; Shen, C.-P. Relevance function of miRNA-708 in the pathogenesis of cancer. Cell. Signal. 2019, 63, 109390. [Google Scholar] [CrossRef]
- Zheng, T.; Li, Y.; Li, W. LncRNA AK024094 aggravates the progression of breast cancer through regulating miRNA-181a. Eur. Rev. Med. Pharmacol. Sci. 2020, 4, 1913–1921. [Google Scholar]
- Fang, C.; Huang, X.; Dai, J.; He, W.; Xu, L.; Sun, F. The circular RNA circFARSA sponges miRNA-330-5p in tumor cells with bladder cancer phenotype. BMC Cancer 2022, 22, 373. [Google Scholar] [CrossRef]
- Lee, H.-E.; Park, S.-J.; Huh, J.-W.; Imai, H.; Kim, H.-S. The enhancer activity of long interspersed nuclear element derived miRNA 625 induced by NF-κB. Sci. Rep. 2021, 11, 3139. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-E.; Huh, J.-W.; Kim, H.-S. Bioinformatics analysis of evolution and human disease related TE-derived miRNAs. Life 2020, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, T.; Amini, M.; Hashemi, Z.S.; Mansoori, B.; Rezaei, S.; Karami, H.; Mosafer, J.; Mokhtarzadeh, A.; Baradaran, B. miRNA-181 serves as a dual-role regulator in the development of human cancers. Free Radic. Biol. Med. 2020, 152, 432–454. [Google Scholar] [CrossRef] [PubMed]
- Makondi, P.T.; Wei, P.-L.; Huang, C.-Y.; Chang, Y.-J. Development of novel predictive miRNA/target gene pathways for colorectal cancer distance metastasis to the liver using a bioinformatic approach. PLoS ONE 2019, 14, e0211968. [Google Scholar] [CrossRef]
- Wu, S.-G.; Chang, T.-H.; Liu, Y.-N.; Shih, J.-Y. MiRNA in lung cancer metastasis. Cancers 2019, 11, 265. [Google Scholar] [CrossRef]
- Liu, B.; Shyr, Y.; Cai, J.; Liu, Q. Interplay between miRNAs and host genes and their role in cancer. Brief. Funct. Genom. 2019, 18, 255–266. [Google Scholar] [CrossRef]
- Cappucci, U.; Torromino, G.; Casale, A.M.; Camon, J.; Capitano, F.; Berloco, M.; Mele, A.; Pimpinelli, S.; Rinaldi, A.; Piacentini, L. Stress-induced strain and brain region-specific activation of LINE-1 transposons in adult mice. Stress 2018, 21, 575–579. [Google Scholar] [CrossRef]
- Curtin, F.; Bernard, C.; Levet, S.; Perron, H.; Porchet, H.; Médina, J.; Malpass, S.; Lloyd, D.; Simpson, R.; RAINBOW-T1D Investigators. A new therapeutic approach for type 1 diabetes: Rationale for GNbAC1, an anti-HERV-W-Env monoclonal antibody. Diabetes Obes. Metab. 2018, 20, 2075–2084. [Google Scholar] [CrossRef]
- Weber, B.; Kimhi, S.; Howard, G.; Eden, A.; Lyko, F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 2010, 29, 5775–5784. [Google Scholar] [CrossRef]
- Houede, N.; Piazza, P.V.; Pourquier, P. LINE-1 as a therapeutic target for castration-resistant prostate cancer. Front. Biosci. 2018, 23, 1292–1309. [Google Scholar]
- Sciamanna, I.; De Luca, C.; Spadafora, C. The reverse transcriptase encoded by LINE-1 retrotransposons in the genesis, progression, and therapy of cancer. Front. Chem. 2016, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Takeshita, F.; Yoshioka, Y.; Hagiwara, K.; Katsuda, T.; Ono, M.; Ochiya, T. Exosomal tumor-suppressive microRNAs as novel cancer therapy: “exocure” is anoTher. choice for cancer treatment. Adv. Drug Deliv. Rev. 2013, 65, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben David, G.; Shlomi, T.; Gil, Z. Transfer of miRNA in Macrophage-Derived Exosomes Induces Drug Resistance in Pancreatic AdenocarcinomaExosomes Induce Gemcitabine Resistance in Pancreatic Cancer. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Li, Y.; Cui, X.; Li, Y.; Zhang, T.; Li, S. Upregulated expression of miR-421 is associated with poor prognosis in non-small-cell lung cancer. Cancer Manag. Res. 2018, 10, 2627. [Google Scholar] [CrossRef]
- Wu, Y.; Wei, J.; Zhang, W.; Xie, M.; Wang, X.; Xu, J. Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. OncoTargets Ther. 2020, 13, 7809. [Google Scholar] [CrossRef]
- He, X.; Chen, S.-Y.; Yang, Z.; Zhang, J.; Wang, W.; Liu, M.-Y.; Niu, Y.; Wei, X.-M.; Li, H.-M.; Hu, W.-N. miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J. Exp. Clin. Cancer Res. 2018, 37, 1–16. [Google Scholar] [CrossRef]
- Xu, X.; Cao, L.; Zhang, Y.; Lian, H.; Sun, Z.; Cui, Y. MiRNA-1246 inhibits cell invasion and epithelial mesenchymal transition process by targeting CXCR4 in lung cancer cells. Cancer Biomark. 2018, 21, 251–260. [Google Scholar] [CrossRef]
- Liu, H.-N.; Qie, P.; Yang, G.; Song, Y.-B. miR-181b inhibits chemoresistance in cisplatin-resistant H446 small cell lung cancer cells by targeting Bcl-2. Arch. Med. Sci. 2018, 14, 745–751. [Google Scholar] [CrossRef]
- Cao, J.; Geng, J.; Chu, X.; Wang, R.; Huang, G.; Chen, L. miRNA-885-3p inhibits docetaxel chemoresistance in lung adenocarcinoma by downregulating Aurora A. Oncol. Rep. 2019, 41, 1218–1230. [Google Scholar] [CrossRef]
- Song, J.; Su, Z.; Shen, Q. Long non-coding RNA MALAT1 regulates proliferation, apoptosis, migration and invasion via miR-374b-5p/SRSF7 axis in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1853–1862. [Google Scholar] [PubMed]
- Wang, Y.; Xu, R.; Zhang, D.; Lu, T.; Yu, W.; Wo, Y.; Liu, A.; Sui, T.; Cui, J.; Qin, Y. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl. Lung Cancer Res. 2019, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, J.; Wu, H.; Li, J. Downregulated miR-585-3p promotes cell growth and proliferation in colon cancer by upregulating PSME3. OncoTargets Ther. 2019, 12, 6525. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Liu, F.; Dong, L.; Yang, H.; He, X.; Li, L.; Zhao, L.; Jin, S.; Li, G. miR-1273g silences MAGEA3/6 to inhibit human colorectal cancer cell growth via activation of AMPK signaling. Cancer Lett. 2018, 435, 1–9. [Google Scholar] [CrossRef]
- Sun, X.; Lin, F.; Sun, W.; Zhu, W.; Fang, D.; Luo, L.; Li, S.; Zhang, W.; Jiang, L. Exosome-transmitted miRNA-335-5p promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Mol. Ther. Nucleic Acids 2021, 24, 164–174. [Google Scholar] [CrossRef]
- Cheng, B.; Rong, A.; Zhou, Q.; Li, W. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J. Exp. Clin. Cancer Res. 2020, 39, 5. [Google Scholar] [CrossRef]
- Sun, W.; Wang, X.; Li, J.; You, C.; Lu, P.; Feng, H.; Kong, Y.; Zhang, H.; Liu, Y.; Jiao, R. MiRNA-181a promotes angiogenesis in colorectal cancer by targeting SRCIN1 to promote the SRC/VEGF signaling pathway. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Im, J.; Nam, S.K.; Lee, H.S. MiRNA-552 expression in colorectal cancer and its clinicopathological significance. J. Pathol. Transl. Med. 2021, 55, 125–131. [Google Scholar] [CrossRef]
- Sun, S.; Hang, T.; Zhang, B.; Zhu, L.; Wu, Y.; Lv, X.; Huang, Q.; Yao, H. miRNA-708 functions as a tumor suppressor in colorectal cancer by targeting ZEB1 through Akt/mTOR signaling pathway. Am. J. Transl. Res. 2019, 11, 5338. [Google Scholar]
- Cui, M.; Chen, M.; Shen, Z.; Wang, R.; Fang, X.; Song, B. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J. Cell. Biochem. 2019, 120, 6926–6936. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, E.; Janas, D.; Eftekhari, A.; Zare, N. Application of carbon nanotubes in sensing/monitoring of pancreas and liver cancer. Chemosphere 2022, 302, 134826. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xu, H.-F.; Liu, M.-Y.; Xu, Y.-J.; He, J.-C.; Zhou, Y.; Cang, S.-D. Mechanism of exosomal miRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J. Gastroenterol. 2019, 25, 1890. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Fang, X.; Han, M.; Wang, X.; Huang, Q. MiRNA-493-5p promotes apoptosis and suppresses proliferation and invasion in liver cancer cells by targeting VAMP2. Int. J. Mol. Med. 2018, 41, 1740–1748. [Google Scholar] [PubMed]
- He, L.; Meng, D.; Zhang, S.-H.; Zhang, Y.; Deng, Z.; Kong, L.-B. miRNA-608 inhibits human hepatocellular carcinoma cell proliferation via targeting the BET family protein BRD4. Biochem. Biophys. Res. Commun. 2018, 501, 1060–1067. [Google Scholar] [CrossRef]
- Tyagi, K.; Mandal, S.; Roy, A. Recent advancements in therapeutic targeting of the Warburg effect in refractory ovarian cancer: A promise towards disease remission. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188563. [Google Scholar] [CrossRef]
- Liu, W.; Kang, L.; Han, J.; Wang, Y.; Shen, C.; Yan, Z.; Tai, Y.; Zhao, C. miR-342-3p suppresses hepatocellular carcinoma proliferation through inhibition of IGF-1R-mediated Warburg effect. OncoTargets Ther. 2018, 11, 1643. [Google Scholar] [CrossRef]
- Xu, F.; Yan, J.-J.; Gan, Y.; Chang, Y.; Wang, H.-L.; He, X.-X.; Zhao, Q. miR-885-5p negatively regulates warburg effect by silencing hexokinase 2 in liver cancer. Mol. Ther. Nucleic Acids 2019, 18, 308–319. [Google Scholar] [CrossRef]
- Wei, L.Q.; Li, L.; Lu, C.; Liu, J.; Chen, Y.; Wu, H. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J. Cell. Physiol. 2019, 234, 5153–5162. [Google Scholar] [CrossRef]
- Yan, S.; Tang, Z.; Chen, K.; Liu, Y.; Yu, G.; Chen, Q.; Dang, H.; Chen, F.; Ling, J.; Zhu, L. Long non-coding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging miRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res. 2018, 37, 214. [Google Scholar] [CrossRef]
- Hu, L.; Wu, H.; Wan, X.; Liu, L.; He, Y.; Zhu, L.; Liu, S.; Yao, H.; Zhu, Z. MiRNA-585 suppresses tumor proliferation and migration in gastric cancer by directly targeting MAPK1. Biochem. Biophys. Res. Commun. 2018, 499, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-L.; Wang, H.-X.; Shi, C.-X.; Shi, F.-Y.; Zhao, L.-Y.; Zhao, W.; Wang, G.-H. MiRNA-1269 promotes cell proliferation via the AKT signaling pathway by targeting RASSF9 in human gastric cancer. Cancer Cell Int. 2019, 19, 308. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, J.; Yang, W.; Yang, Z.; Zhou, K. MiRNA-552 accelerates the progression of gastric cancer by targeting FOXO1 and regulating PI3K/AKT pathway. J. Oncol. 2021, 2021, 9966744. [Google Scholar] [CrossRef] [PubMed]
- Nabatchian, F.; Naiini, M.R.; Moradi, A.; Tabatabaeian, H.; Hoghoughi, N.; Azadeh, M.; Ghaedi, K. miR-581-related single nucleotide polymorphism, rs2641726, located in MUC4 gene, is associated with gastric cancer incidence. Indian J. Clin. Biochem. 2019, 34, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhou, Z.; Guo, H.; He, Y.; Pang, X.; Zhang, X.; Liu, Y.; Ao, X.; Li, P.; Wang, J. Long non-coding RNA gastric cancer-related lncRNA1 mediates gastric malignancy through miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Xiao, J.; Lin, L.; Luo, D.; Shi, L.; Chen, W.; Fan, H.; Li, Z.; Ma, X.; Ni, P.; Yang, L. Long non-coding RNA TRPM2-AS acts as a miRNA sponge of miR-612 to promote gastric cancer progression and radioresistance. Oncogenesis 2020, 9, 1–15. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, Y.; Yang, P.; Qin, G. Silencing of long non-coding RNA LINC00324 interacts with miRNA-3200-5p to attenuate the tumorigenesis of gastric cancer via regulating BCAT1. Gastroenterol. Res. Pract. 2020, 2020, 4159298. [Google Scholar] [CrossRef]
- Li, B.; Liang, L.; Chen, Y.; Liu, J.; Wang, Z.; Mao, Y.; Zhao, K.; Chen, J. Circ_0008287 promotes immune escape of gastric cancer cells through impairing miRNA-548c-3p-dependent inhibition of CLIC1. Int. Immunopharmacol. 2022, 111, 108918. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, H.; Chen, X. Circ-LDLRAD3 enhances cell growth, migration, and invasion and inhibits apoptosis by regulating MiR-224-5p/NRP2 axis in gastric cancer. Dig. Dis. Sci. 2021, 66, 3862–3871. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, Y.; Wang, L.; Yin, L. Identifying the diagnostic value of miRNA-421 in gastric cancer patients: A meta-analysis. bioRxiv 2018, 468983. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Z.; Shen, J. MiRNA-421-targeted PDCD4 regulates breast cancer cell proliferation. Int. J. Mol. Med. 2019, 43, 267–275. [Google Scholar] [PubMed]
- Ghaemi, Z.; Soltani, B.M.; Mowla, S.J. MiRNA-326 functions as a tumor suppressor in breast cancer by targeting ErbB/PI3K signaling pathway. Front. Oncol. 2019, 9, 653. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Chen, X.; Liu, H.; Yu, K.; Bao, Y.; Chai, J.; Gao, H.; Zou, L. LGR5 acts as a target of miR-340-5p in the suppression of cell progression and drug resistance in breast cancer via Wnt/β-catenin pathway. Gene 2019, 683, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liu, F.; Song, Y. Progress: Targeted therapy, immunotherapy, and new chemotherapy strategies in advanced triple-negative breast cancer. Cancer Manag. Res. 2020, 12, 9375. [Google Scholar] [CrossRef]
- Zeng, X.; Ma, X.; Guo, H.; Wei, L.; Zhang, Y.; Sun, C.; Han, N.; Sun, S.; Zhang, N. MiRNA-582-5p promotes triple-negative breast cancer invasion and metastasis by antagonizing CMTM8. Bioengineered 2021, 12, 10126–10135. [Google Scholar] [CrossRef]
- Son, D.; Kim, Y.; Lim, S.; Kang, H.-G.; Kim, D.-H.; Park, J.W.; Cheong, W.; Kong, H.K.; Han, W.; Park, W.-Y. miR-374a-5p promotes tumor progression by targeting ARRB1 in triple negative breast cancer. Cancer Lett. 2019, 454, 224–233. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X.; Wang, X.; He, M.; Qiao, S. MiRNA-224 promotes tumorigenesis through downregulation of caspase-9 in triple-negative breast cancer. Dis. Markers 2019, 2019, 7378967. [Google Scholar]
- Chen, T.; Yang, Z.; Liu, C.; Wang, L.; Yang, J.; Chen, L.; Li, W. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019, 52, e12548. [Google Scholar] [CrossRef]
- Liu, X.; Ma, J.; Xu, F.; Li, L. TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. Biomed. PharmacoTher. 2018, 99, 9–17. [Google Scholar] [CrossRef]
- Zhou, Y.; Zheng, X.; Xu, B.; Chen, L.; Wang, Q.; Deng, H.; Jiang, J. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 2019, 508, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Ping, W.; Gao, Y.; Fan, X.; Li, W.; Deng, Y.; Fu, X. MiR-181a contributes gefitinib resistance in non-small cell lung cancer cells by targeting GAS7. Biochem. Biophys. Res. Commun. 2018, 495, 2482–2489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Y.; Zhang, W.; Li, J.; Liu, W.; Lu, W. Long non-coding RNA SNHG14 exerts oncogenic functions in non-small cell lung cancer through acting as an miR-340 sponge. Biosci. Rep. 2019, 39, BSR20180941. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, M.; Yang, C. YY1-mediated overexpression of long non-coding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J. Cell. Biochem. 2020, 121, 2258–2267. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, X.; Yang, Y.; Jiao, W. Long non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells and leads to poor prognosis by targeting miR-378a-3p. Thorac. Cancer 2018, 9, 939–949. [Google Scholar] [CrossRef]
- Yu, W.; Jiang, H.; Zhang, H.; Li, J. Hsa_circ_0003998 promotes cell proliferation and invasion by targeting miR-326 in non-small cell lung cancer. OncoTargets Ther. 2018, 11, 5569. [Google Scholar] [CrossRef]
- Wang, R.; Xu, J.; Xu, J.; Zhu, W.; Qiu, T.; Li, J.; Zhang, M.; Wang, Q.; Xu, T.; Guo, R. MiR-326/Sp1/KLF3: A novel regulatory axis in lung cancer progression. Cell Prolif. 2019, 52, e12551. [Google Scholar] [CrossRef]
- Yu, H.; Wang, X.; Han, X.; Cao, B. MiR-608 exerts tumor suppressive function in lung adenocarcinoma by directly targeting MIF. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4908–4916. [Google Scholar]
- Zhang, Q.; Zhang, C.; Ma, J.-X.; Ren, H.; Sun, Y.; Xu, J.-Z. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J. Gastroenterol. 2019, 25, 5300. [Google Scholar] [CrossRef]
- Qu, R.; Hao, S.; Jin, X.; Shi, G.; Yu, Q.; Tong, X.; Guo, D. MiRNA-374b reduces the proliferation and invasion of colon cancer cells by regulation of LRH-1/Wnt signaling. Gene 2018, 642, 354–361. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Wang, L.; Yin, G.; Li, W.; Xian, Y.; Yang, W.; Liu, Q. miR-23c suppresses tumor growth of human hepatocellular carcinoma by attenuating ERBB2IP. Biomed. PharmacoTher. 2018, 107, 424–432. [Google Scholar] [CrossRef]
- Tao, J.; Liu, Z.; Wang, Y.; Wang, L.; Yin, G.; Yang, W.; Tu, K.; Liu, Q. MiRNA-645 represses hepatocellular carcinoma progression by inhibiting SOX30-mediated p53 transcriptional activation. Int. J. Biol. Macromol. 2019, 121, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Komoll, R.-M.; Hu, Q.; Olarewaju, O.; von Döhlen, L.; Yuan, Q.; Xie, Y.; Tsay, H.-C.; Daon, J.; Qin, R.; Manns, M.P. MiRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J. Hepatol. 2021, 74, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, J.; Pan, T.; Ai, S.; Tang, L.; Wang, F. miR-378a enhances the sensitivity of liver cancer to sorafenib by targeting VEGFR, PDGFRβ and c-Raf. Mol. Med. Rep. 2018, 17, 4581–4588. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Feng, X.; Hao, X.; Wang, P.; Zhang, Y.; Zheng, X.; Li, L.; Ren, S.; Zhang, M.; Xu, M. CircSETD3 (Hsa_circ_0000567) acts as a sponge for miRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. 2019, 38, 98. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Xu, F.; Zhang, P.; Wang, P.; Wei, Y.-N.; Wu, C.; Cheng, S.-J. MiRNA-575 regulates development of gastric cancer by targeting PTEN. Biomed. PharmacoTher. 2019, 113, 108716. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, H.; Liu, L.; Zhu, K.; Yu, W.; Guo, J. Hsa_circ_0001546 acts as a miRNA-421 sponge to inhibit the chemoresistance of gastric cancer cells via ATM/Chk2/p53-dependent pathway. Biochem. Biophys. Res. Commun. 2020, 521, 303–309. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, Y.; Sun, D.; Wang, S.; Ding, K.; Liu, M.; Zhang, Y.; Miao, Y.; Liu, H.; Zhou, F. MiRNA-181a functions as an oncogene in gastric cancer by targeting caprin-1. Front. Pharmacol. 2019, 9, 1565. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, M.; Miao, J.; Wang, X.; Huang, C. miRNA-4317 suppresses human gastric cancer cell proliferation by targeting ZNF322. Cell Biol. Int. 2018, 42, 923–930. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, Z.; Xie, J.; Wang, P.; Zhu, J.; Li, Y.; Wang, Y. MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochem. Biophys. Res. Commun. 2018, 506, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.-Y.; Li, M.-X.; Pan, W.-L.; Chen, Y.; Li, M.-M.; Pang, J.-X.; Zheng, L.; Chen, J.-X.; Duan, W.-J. In situ detection of plasma exosomal miRNA-1246 for breast cancer diagnostics by a Au nanoflare probe. ACS Appl. Mater. Interfaces 2018, 10, 39478–39486. [Google Scholar] [CrossRef] [PubMed]
- Ramchandani, D.; Lee, S.K.; Yomtoubian, S.; Han, M.S.; Tung, C.-H.; Mittal, V. Nanoparticle Delivery of miR-708 Mimetic Impairs Breast Cancer MetastasismiR-708 Mimetic in TNBC Therapy. Mol. Cancer Ther. 2019, 18, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liu, Y.; Jiang, A.; Li, R.; Yin, M.; Wang, Y. MiRNA-335 suppresses the proliferation, migration, and invasion of breast cancer cells by targeting EphA4. Mol. Cell. Biochem. 2018, 439, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hu, J.; Ma, J.; Qi, X.; Zhou, H.; Miao, X.; Zheng, W.; Jia, L. MiR-193a-3p and miR-224 mediate renal cell carcinoma progression by targeting alpha-2, 3-sialyltransferase IV and the phosphatidylinositol 3 kinase/Akt pathway. Mol. Carcinog. 2018, 57, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, W.; Jian, Q.; Xue, K.; Huang, M.; Chi, S.; Li, C.; Li, C. MiRNA-340 inhibits squamous cell carcinoma cell proliferation, migration and invasion by downregulating RhoA. J. Dermatol. Sci. 2018, 92, 197–206. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, G.; Zheng, W.; Xue, Q.; Wei, D.; Zheng, Y.; Yuan, J. MiR-454-3p and miR-374b-5p suppress migration and invasion of bladder cancer cells through targetting ZEB2. Biosci. Rep. 2018, 38, BSR20181436. [Google Scholar] [CrossRef]
- Liu, M.; Chen, Y.; Huang, B.; Mao, S.; Cai, K.; Wang, L.; Yao, X. Tumor-suppressing effects of miRNA-612 in bladder cancer cells by targeting malic enzyme 1 expression. Int. J. Oncol. 2018, 52, 1923–1933. [Google Scholar]
- Li, L.; Ma, L. Upregulation of miR-582-5p regulates cell proliferation and apoptosis by targeting AKT3 in human endometrial carcinoma. Saudi J. Biol. Sci. 2018, 25, 965–970. [Google Scholar] [CrossRef]
- Li, N.; Wang, C.; Zhang, P.; You, S. Emodin inhibits pancreatic cancer EMT and invasion by up-regulating miRNA-1271. Mol. Med. Rep. 2018, 18, 3366–3374. [Google Scholar]
- Xu, X.; Zheng, S. MiR-887-3p negatively regulates STARD13 and promotes pancreatic cancer progression. Cancer Manag. Res. 2020, 12, 6137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Han, T.; Li, B.; Ma, Q.; Yang, P.; Li, H. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J. Ovarian Res. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.M.; Wang, W.W.; Qi, R.; Leng, T.G.; Zhang, X.L. MiRNA-224 inhibition prevents progression of cervical carcinoma by targeting PTX3. J. Cell. Biochem. 2018, 119, 10278–10290. [Google Scholar] [CrossRef] [PubMed]
- Li, G.C.; Cao, X.Y.; Li, Y.N.; Qiu, Y.Y.; Li, Y.N.; Liu, X.J.; Sun, X.X. MiRNA-374b inhibits cervical cancer cell proliferation and induces apoptosis through the p38/ERK signaling pathway by binding to JAM-2. J. Cell. Physiol. 2018, 233, 7379–7390. [Google Scholar] [CrossRef]
- Nakashima, H.; Yoshida, R.; Hirosue, A.; Kawahara, K.; Sakata, J.; Arita, H.; Yamamoto, T.; Toya, R.; Murakami, R.; Hiraki, A. Circulating miRNA-1290 as a potential biomarker for response to chemoradiotherapy and prognosis of patients with advanced oral squamous cell carcinoma: A single-center retrospective study. Tumor Biol. 2019, 41, 1010428319826853. [Google Scholar] [CrossRef]
- Lin, S.-S.; Peng, C.-Y.; Liao, Y.-W.; Chou, M.-Y.; Hsieh, P.-L.; Yu, C.-C. miR-1246 targets CCNG2 to enhance cancer stemness and chemoresistance in oral carcinomas. Cancers 2018, 10, 272. [Google Scholar] [CrossRef]
- Zhu, G.; Zhou, L.; Liu, H.; Shan, Y.; Zhang, X. MiRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1α pathway. Cell. Physiol. Biochem. 2018, 48, 1735–1746. [Google Scholar] [CrossRef]

| Cancer Related Biological Process | Type of Alternative Splicing | Gene | Protein | Isoform | Regulatory Process | Ref |
|---|---|---|---|---|---|---|
| Apoptosis | Alternative 5′ SS usage | BCL2L1 | Bcl-2-Like Protein 1 | BCl-XL | Through the alternative use of two competing 5′ SSs in exon 2, produced BCL-XL which has an antiapoptotic effect and functions as a dominant regulator. | [53,54,55] |
| Exon skipping | SYK | Spleen Associated Tyrosine Kinase | SYK(S) | Switching SYK(L) to SYK(S) generated by exon 9 skipping induces apoptosis in ovarian cancer. | [56] | |
| Intron retention | STAT2 | Signal Transducer and Activator of Transcription 2 | STAT2 + I19 | STAT2 + I19, splice variant containing intron 19 which has a stop codon before the Src homology 2 domain, leads to disruption of STAT dimerization and suppresses IFN-induced apoptosis in IFN-resistant cells. | [57] | |
| Exon skipping | ASPP2 | Apoptosis-Stimulating of P53 Protein 2 | ASPP2K | ASPP2K, which has a truncated C-terminal domain losing the p53 binding regions by exon skipping, possesses dominant-negative activity, impairing the induction of p53 dependent apoptosis and promoting cancer aggressiveness. | [58] | |
| Exon skipping | FAS | Fas Cell Surface Death Receptor | sFAS | An alternatively spliced isoform, soluble Fas (sFAS), generated by the skipping of exon 6 that encodes the transmembrane domain, cannot localize to the plasma membrane. As a result, upregulated sFAS inhibits the extrinsic pathway of apoptosis in various cancer types. | [59,60,61] | |
| Exon inclusion | MCL1 | Myeloid Leukemia Cell Differentiation Protein Mcl-1 | MCL1-L | Melanocytes upregulate MCL-1L, a splicing variant of MCL1 by exon 2 inclusion, in response to UVB radiation to protect themselves against apoptosis, whereas melanoma cells elevating MCL1-L expression without UV exposure are resistant to apoptosis. | [62] | |
| Invasion (EMT) | Exon skipping | ENAH (MENA) | ENAH Actin Regulator | MENA v6 | ENAH (known as Mena), controls actin nucleation as well as cell morphology and motility. Expression of the exon skipped splicing isoform, Mena11a, has been correlated with epithelial markers and decreased invasion. Inversely, increased expression of MenaINV by exon inclusion has been associated with mesenchymal markers and increased invasion and metastasis. | [63,64,65,66] |
| Exon inclusion | MENA INV | |||||
| Alternative 5′ SS usage | KLF6 | Kruppel Like Factor 6 | KLF6-SV1 | KLF6-SV1 uses an alternative 5′SS, causing frameshift, and produces a protein isoform that contains 21 novel amino acids but lacking all three of the zinc finger domains. Upregulated expression of KLF6-SV1 increases cell survival, migration, and invasion in various cancer cells. | [67,68] | |
| Exon inclusion | CD44 | CD44 Antigen | CD44v8-10 | Expression of CD44v8-10, an alternative isoform including the variable portion of exon 8 to 10, induces a higher metastatic potential of cancer cells than the standard form of CD44 in breast cancer cell lines. | [69] | |
| CD44v6 | CD44 variant including variable exon 6 (CD44v6) has been identified that promotes the development of metastasis by involving epithelial-mesenchymal transition in cancers. | [70,71] | ||||
| Alternative 5′ SS usage | CTNND1 | p120-catenin | p120-1A | p120-catenin (p120ctn) isoforms produced by alternative 5′SS usage, p120-1A, and -3A, induced the EMT of tumor cells. Especially, in non-small cell lung cancer (NSCLC), both p120-1A and - 3A inhibited EMT and decreased cell invasiveness in cells with membrane E-cadherin. In cells with cytoplasmic E-cadherin, p120-1A stimulated EMT and cell invasiveness, while p120-3A prevented EMT and decreased cell invasiveness. | [72] | |
| p120-3A | ||||||
| Mutually exclusive exon | FGFR | Fibroblast Growth Factor Receptor | FGFR lllc | Increased level of FGFR-IIIc by mutually exclusive exon9 has been detected in a variety of tumors and correlated with tumor progression, such as increased grading and invasiveness, by promoting cancer cells to acquire mesenchymal characteristics. | [73,74,75] | |
| Exon skipping | RON | Macrophage Stimulating 1 Receptor | ΔRON | The skipping of Exon 11(ΔRON) brought about the deletion of an extra cellular domain that affects the proteolytic maturation of protein and increases cancer invasiveness through sustaining constitutively active status. | [76,77] | |
| Proliferation | Exon inclusion | RPS6KB1 | Ribosomal Protein S6 Kinase B1 | RPS6KB1-2 | RPS6KB1-2 made by inclusion of three cassette exons 6a, 6b, and 6c, caused the shorter isoform to lack a portion of the kinase domain. RPS6KB1-2 has contributed to cell proliferation and tumor growth via mTORC1 and 4E-BP1 phosphorylation. | [78,79] |
| Exon inclusion | NUMB | NUMB Endocytic Adaptor Protein | NUMB-PRR(L) | In lung cancer cells, RBM10 mutations identified that disrupt splicing regulation of NUMB (Exon 9 inclusion) which is a key target of RBM5, 6, and 10 in the control of cell proliferation, to correlate with cell growth. | [80] | |
| Exon inclusion | SYK | Spleen Associated Tyrosine Kinase | SYK(L) | SYK(L), which includes exon 9 compared to the shorter isoform (SYK(S)), stimulates cell survival and tumor malignancy in many cancers by driving expression of epidermal growth factor. | [56] | |
| Exon skipping | MDM2 | E3 Ubiquitin-Protein Ligase Mdm2 | MDM2-A | Normal type of MDM2 could bind to p53 and facilitate proteasomal degradation of p53 as an ubiquitin ligase. Four of the splice isoforms (MDM2-A, -B, -C, and -D) by exon skipping in human cancers lack part of the p53-binding domain. Spliced isoforms could not bind to p53, enhancing degradation of p53 and cell proliferation. | [81,82,83] | |
| MDM2-B | ||||||
| MDM2-C | ||||||
| MDM2-D | ||||||
| Angiogenesis | Alternative 3′ SS usage | VEGF | Vascular Endothelial Growth Factor | VEGFxxx | VEGFxxx isoforms, produced by alternative 3′SS usage, were overexpressed in many cancers, and resulted in proangiogenic effects. | [84] |
| Alternative 3′ SS usage | VEGF | Vascular Endothelial Growth Factor | VEGF 165 | sVEGFR1-113, a truncated version of VEGFR1, lacks its transmembrane and tyrosine kinase domains due to intron 13 retention.VEGF165 is a pro-angiogenic factor made by 3′ SS usage. One study identified that sVEGFR1-113 is considered to be a natural antagonist of VEGFA and upregulated under the mechanism associating the VEGF165/SOX2/SRSF2 network in anti-angiogenic therapied squamous lung carcinoma cells. | [85,86] | |
| Intron retention | VEGFR | Vascular Endothelial Growth Factor Receptor 1 | sVEGFR1-113 | |||
| Dysregulated metabolism | Mutually exclusive exon | PKM | Pyruvate Kinase M2 | PKM2 | PKM2 had mutually exclusive exons containing Exon 10 not Exon 9 and is ubiquitously expressed in tumors. Substituting PKM2 with PKM1 in the tumor decreases lactate production and increases oxidative phosphorylation. Therefore, tumor growth is repressed. | [87,88] |
| Immune response | Exon skipping | BRAF | B-Raf Proto-Oncogene, Serine/Threonine Kinase | BRAF(V600E) | BRAF(V600E) transcripts by exon skipping (exon 4–8) brought about in-frame deletion of the N-terminal RAS-binding domain, resulting in melanoma cell resistance which is insensitive to inhibitors such as drug (PLX4032). | [89] |
| Exon skipping | CEACAM1 | CEA Cell Adhesion Molecule 1 | CEACAM1(S) | The short isoform of CEACAM1, CEACAM1 (S) upregulated in many cancer types. This variant enlarged secretory IgA production by B cells and was associated with poor prognosis and peritoneal dissemination in gastric cancer. | [90,91,92] | |
| Cell cycle | Intron retention | CCND1 | CyclinD1 | CyclinD1b | The formation of the cyclin D1b variant was linked with intron 4 retention, also concerned with cell cycle progression and proliferation in various cancers, competing with the same target, CDK4 with Cyclin D1a. | [93,94] |
| Cancer Type | MiRNA | Subclass | Superfamily | Target Gene | Reference |
|---|---|---|---|---|---|
| Renal cancer | hsa-miR-224-3p | DNA transposon | DNA transposon | ST3GalIV | [247] |
| Skin cancer | hsa-miR-340-5p | DNA transposon | TcMar-Mariner | RhoA | [248] |
| Bladder cancer | hsa-miR-374b-5p | LINE | L2 | ZEB2 | [249] |
| hsa-miR-330-5p | SINE | MIR | circFARSA | [162] | |
| hsa-miR-612 | SINE | MIR | ME1 | [250] | |
| Cervical cancer | hsa-miR-224-5p | DNA transposon | DNA transposon | PTX3 | [255] |
| hsa-miR-374b-5p | LINE | L2 | JAM2 | [256] | |
| Endometrial cancer | hsa-miR-582-5p | LINE | CR1 | AKT3 | [251] |
| Oral cancer | hsa-miR-1290 | DNA transposon | TcMar-Tigger | - | [257] |
| hsa-miR-1246 | LTR | ERVL-MaLR | CCNG2 | [258] | |
| Pancreatic cancer | hsa-miR-1271 | LINE | L2 | E-cadherin, ZEB1, TWIST1 | [252] |
| hsa-miR-224-5p | DNA transposon | DNA transposon | TXNIP | [259] | |
| hsa-miR-887-3p | LINE | L2 | STARD13 | [253] | |
| Ovarian cancer | hsa-miR-552 | LINE | L1 | PTEN | [254] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, W.R.; Park, E.G.; Lee, Y.J.; Bae, W.H.; Lee, D.H.; Kim, H.-S. Integration of TE Induces Cancer Specific Alternative Splicing Events. Int. J. Mol. Sci. 2022, 23, 10918. https://doi.org/10.3390/ijms231810918
Kim WR, Park EG, Lee YJ, Bae WH, Lee DH, Kim H-S. Integration of TE Induces Cancer Specific Alternative Splicing Events. International Journal of Molecular Sciences. 2022; 23(18):10918. https://doi.org/10.3390/ijms231810918
Chicago/Turabian StyleKim, Woo Ryung, Eun Gyung Park, Yun Ju Lee, Woo Hyeon Bae, Du Hyeong Lee, and Heui-Soo Kim. 2022. "Integration of TE Induces Cancer Specific Alternative Splicing Events" International Journal of Molecular Sciences 23, no. 18: 10918. https://doi.org/10.3390/ijms231810918
APA StyleKim, W. R., Park, E. G., Lee, Y. J., Bae, W. H., Lee, D. H., & Kim, H.-S. (2022). Integration of TE Induces Cancer Specific Alternative Splicing Events. International Journal of Molecular Sciences, 23(18), 10918. https://doi.org/10.3390/ijms231810918








