The Structure of Stable Cellulolytic Consortia Isolated from Natural Lignocellulosic Substrates
Abstract
1. Introduction
2. Results
2.1. Taxonomic Analysis of Cellulolytic Consortia by Amplicon Sequencing
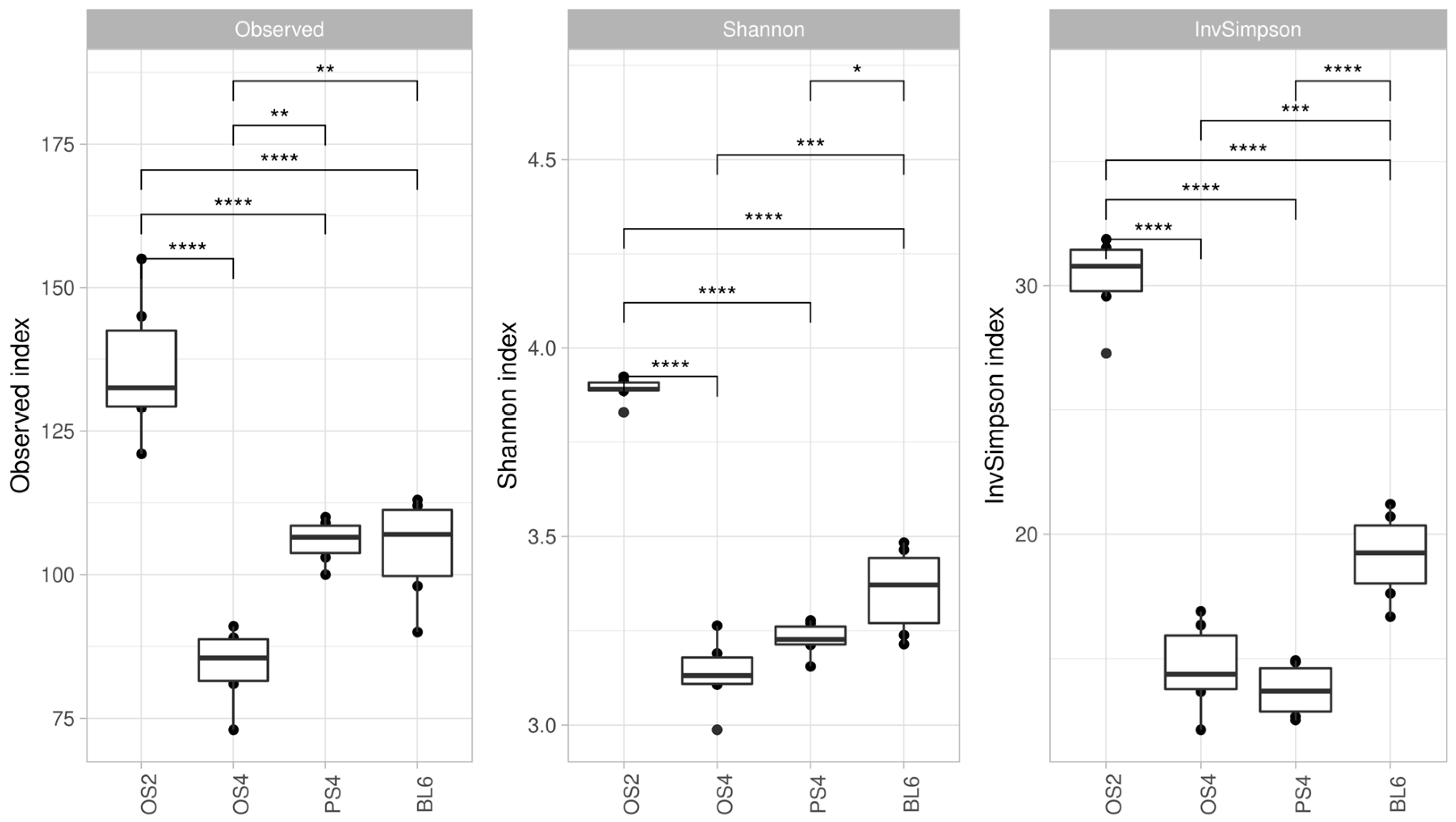
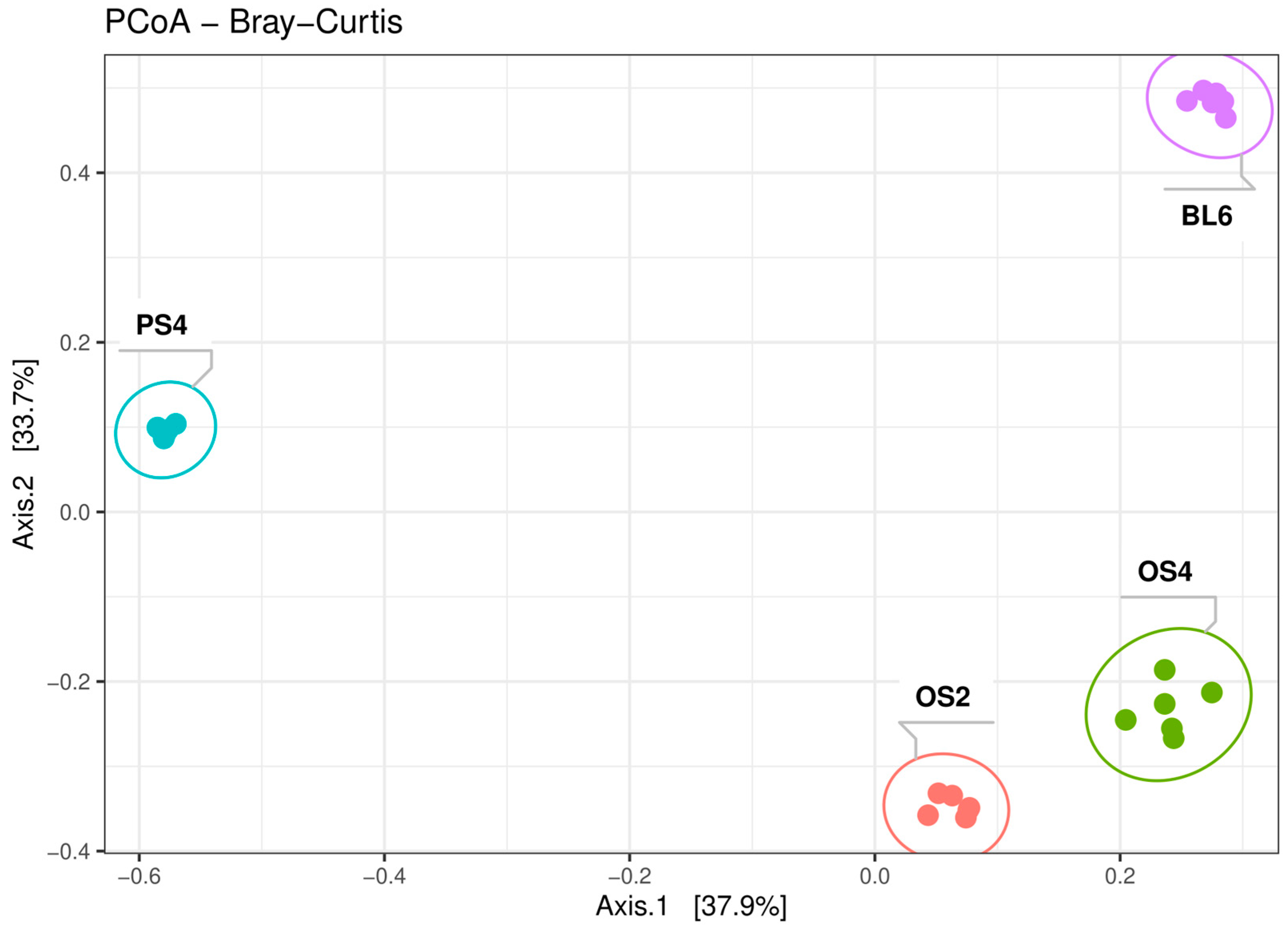
2.1.1. Alpha and Beta-Diversity of Consortia
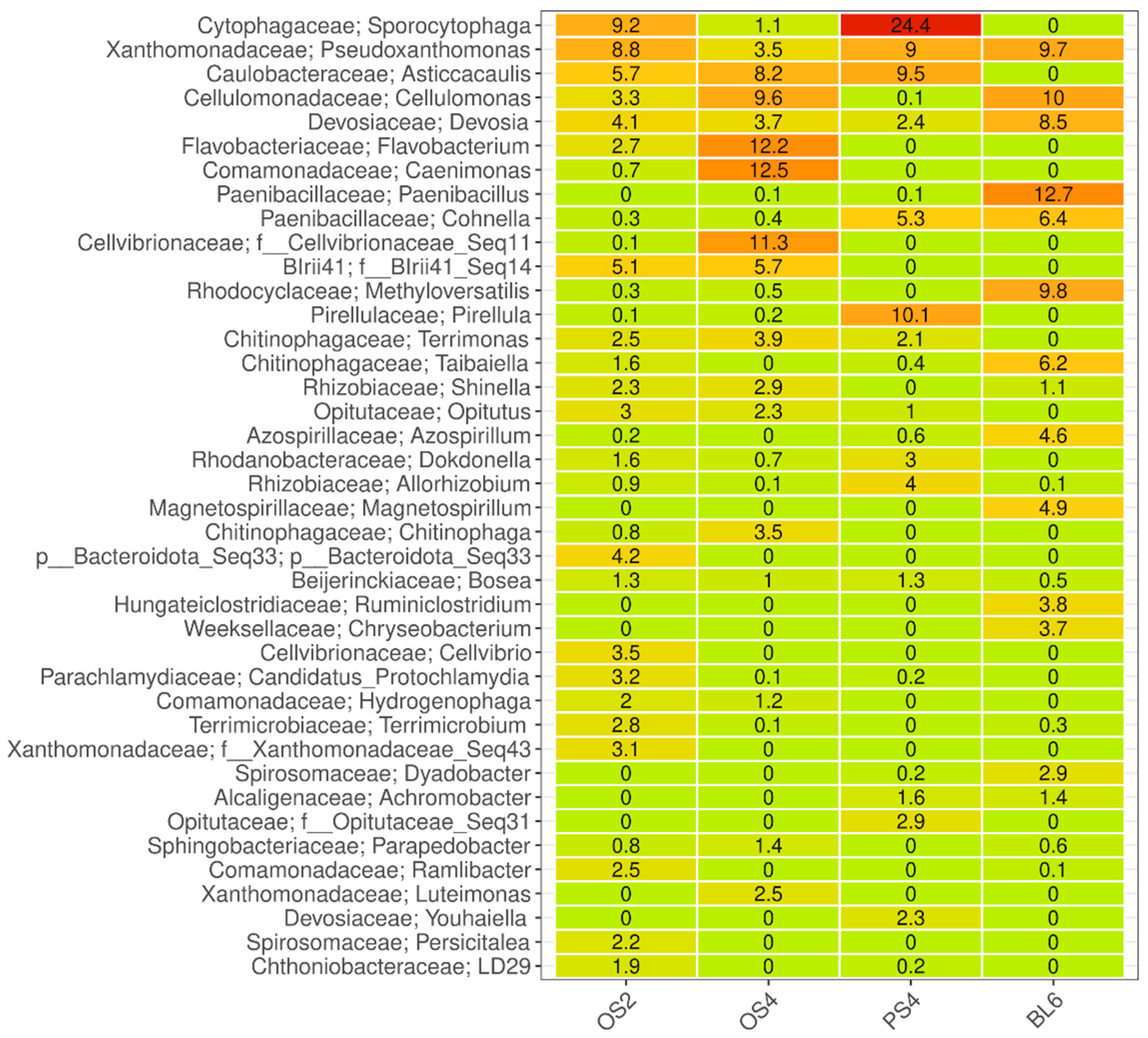
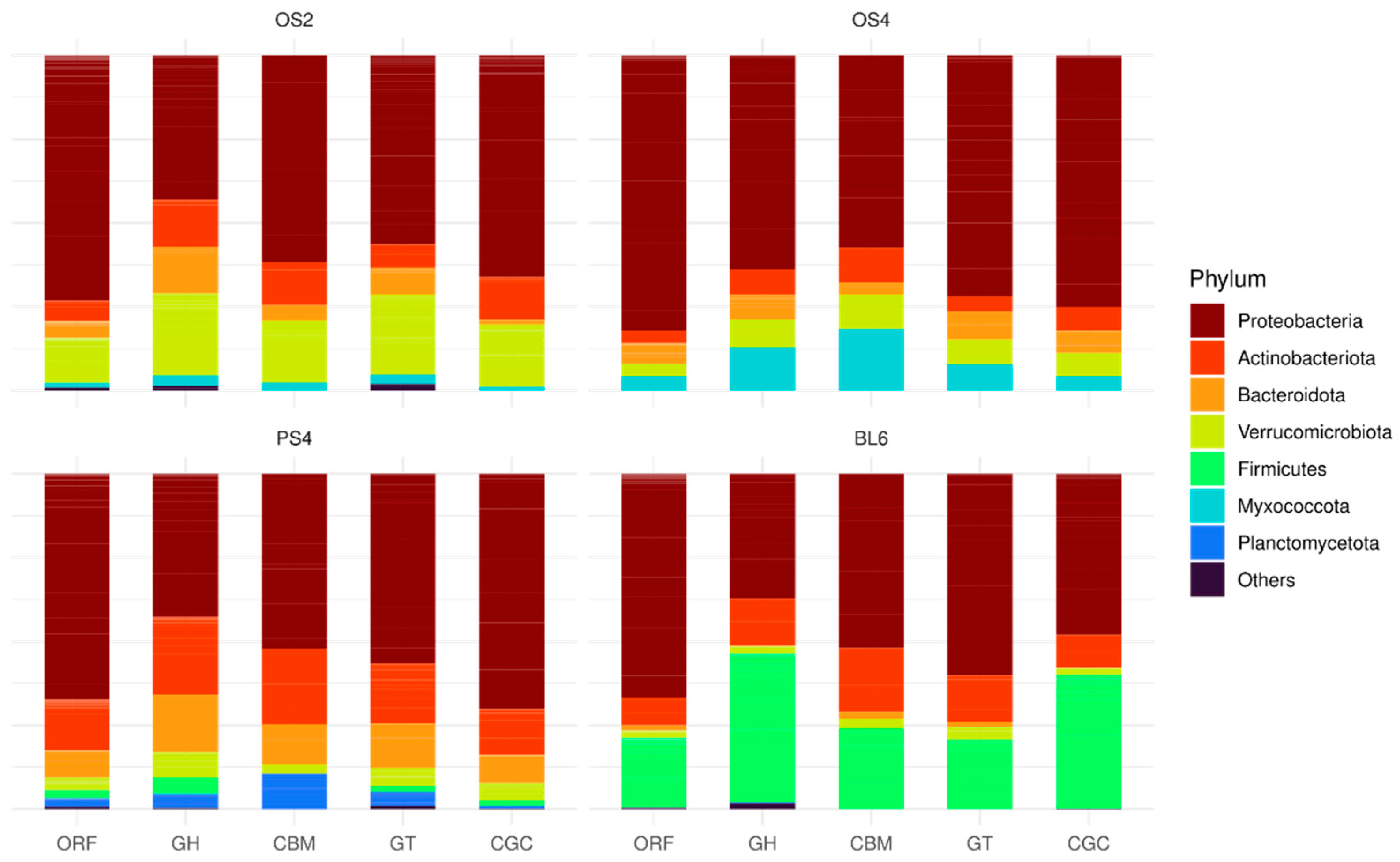
2.1.2. Taxonomic Composition of Consortia
2.2. Functional Analysis of Cellulolytic Consortia
2.2.1. Metagenome Taxonomy
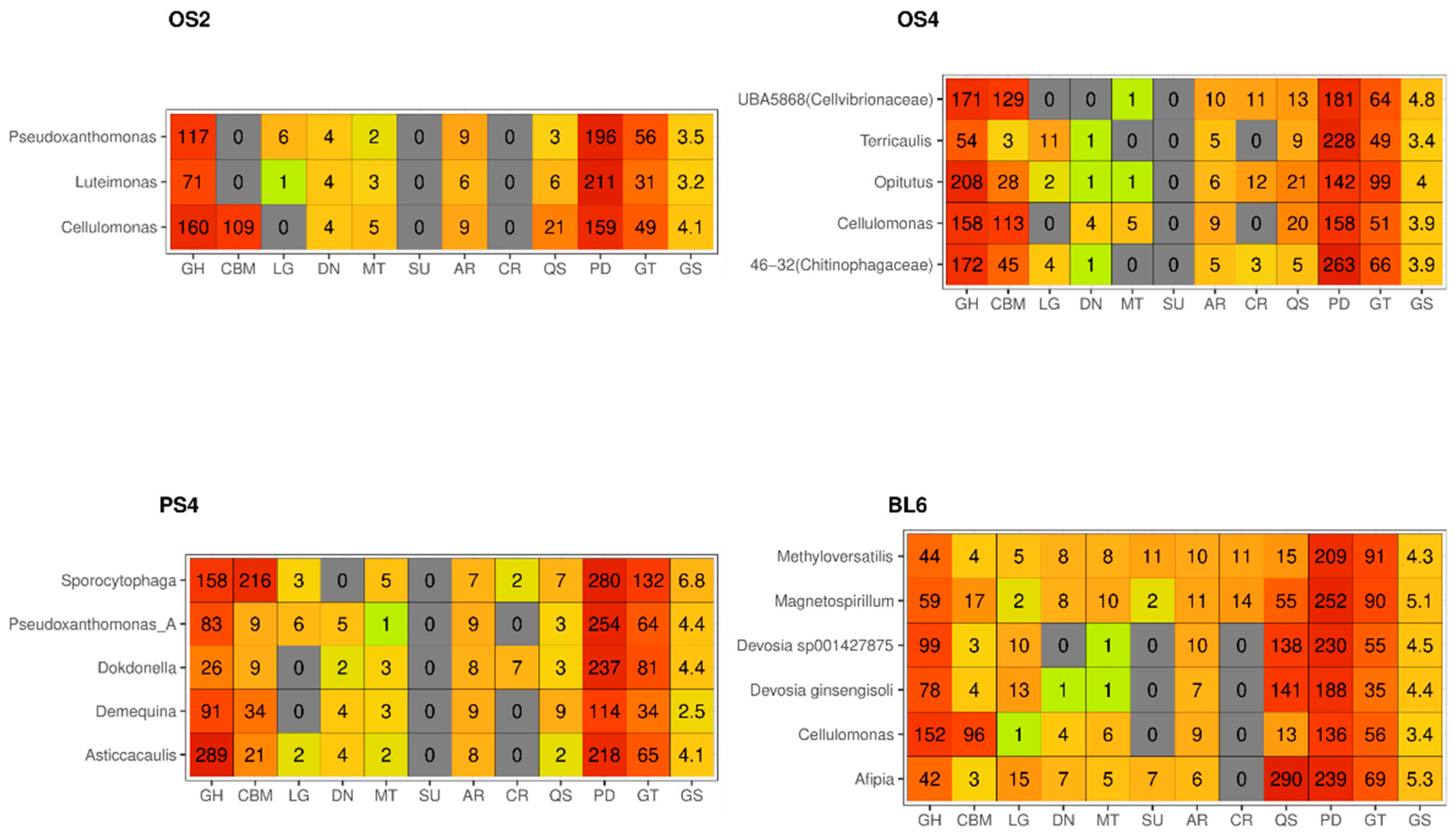
2.2.2. MAGs
2.2.3. CAZy Composition
2.2.4. Gene Set Enrichment Analysis
2.2.5. Taxonomy Evenness in KEGGs
3. Discussion
3.1. Diversity of Members of the Cellulolytic Consortia
3.2. Functional Metagenome of the Cellulolytic Consortia
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naik, S.N.; Goud, V.V.; Rout, P.K.; Dalai, A.K. Production of First and Second Generation Biofuels: A Comprehensive Review. Renew. Sustain. Energy Rev. 2010, 14, 578–597. [Google Scholar] [CrossRef]
- Howard, R.L.; Abotsi, E.; van Rensburg, E.J.; Howard, S. Lignocellulose Biotechnology: Issues of Bioconversion and Enzyme Production. Afr. J. Biotechnol. 2003, 2, 602–619. [Google Scholar] [CrossRef]
- Martins, D.A.B.; do Prado, H.F.A.; Leite, R.S.R.; Ferreira, H.; de Moretti, M.M.S.; da Silva, R.; Gomes, E. Agroindustrial Wastes as Substrates for Microbial Enzymes Production and Source of Sugar for Bioethanol Production; IntechOpen: London, UK, 2011; pp. 319–360. [Google Scholar]
- Abdeshahian, P.; Kadier, A.; Rai, P.K.; da Silva, S.S. Lignocellulose as a Renewable Carbon Source for Microbial Synthesis of Different Enzymes. In Lignocellulosic Biorefining Technologies; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2020; pp. 185–202. [Google Scholar]
- Fellows, C.M.; Brown, T.C.; Doherty, W.O.S. Lignocellulosics as a Renewable Feedstock for Chemical Industry: Chemicals from Lignin. In Green Chemistry for Environmental Remediation; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; pp. 561–610. [Google Scholar]
- Sánchez, C. Lignocellulosic Residues: Biodegradation and Bioconversion by Fungi. Biotechnol. Adv. 2009, 27, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Weidener, D.; Dama, M.; Dietrich, S.K.; Ohrem, B.; Pauly, M.; Leitner, W.; Domínguez de María, P.; Grande, P.M.; Klose, H. Multiscale Analysis of Lignocellulose Recalcitrance towards OrganoCat Pretreatment and Fractionation. Biotechnol. Biofuels 2020, 13, 155. [Google Scholar] [CrossRef]
- Pauly, M.; Keegstra, K. Cell-Wall Carbohydrates and Their Modification as a Resource for Biofuels. Plant J. 2008, 54, 559–568. [Google Scholar] [CrossRef]
- Xu, F.; Li, Y. Biomass Digestion. In Encyclopedia of Sustainable Technologies; Abraham, M.A., Ed.; Elsevier: Oxford, UK, 2017; pp. 197–204. [Google Scholar]
- Saake, B.; Lehnen, R. Lignin. In Ullmann’s Encyclopedia of Industrial Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2007. [Google Scholar]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V. Novel Enzymes for the Degradation of Cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef]
- Várnai, A.; Huikko, L.; Pere, J.; Siika-aho, M.; Viikari, L. Synergistic Action of Xylanase and Mannanase Improves the Total Hydrolysis of Softwood. Bioresour. Technol. 2011, 102, 9096–9104. [Google Scholar] [CrossRef]
- Bourne, Y.; Henrissat, B. Glycoside Hydrolases and Glycosyltransferases: Families and Functional Modules. Curr. Opin. Struct. Biol. 2001, 11, 593–600. [Google Scholar] [CrossRef]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-Binding Modules: Fine-Tuning Polysaccharide Recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef]
- Carrard, G.; Koivula, A.; Söderlund, H.; Béguin, P. Cellulose-Binding Domains Promote Hydrolysis of Different Sites on Crystalline Cellulose. Proc. Natl. Acad. Sci. USA 2000, 97, 10342–10347. [Google Scholar] [CrossRef]
- Linder, M.; Teeri, T.T. The Roles and Function of Cellulose-Binding Domains. J. Biotechnol. 1997, 57, 15–28. [Google Scholar] [CrossRef]
- Williams, G.J.; Thorson, J.S. Natural Product Glycosyltransferases: Properties and Applications. In Advances in Enzymology and Related Areas of Molecular Biology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2008; pp. 55–119. [Google Scholar]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An Evolving Hierarchical Family Classification for Glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Martínez, A.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martínez, M.J.; Gutiérrez, A.; del Río, J.C. Biodegradation of Lignocellulosics: Microbial, Chemical, and Enzymatic Aspects of the Fungal Attack of Lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Henrissat, B. A Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1991, 280 Pt 2, 309–316. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the Enzymatic Repertoire of the CAZy Database to Integrate Auxiliary Redox Enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Andlar, M.; Rezic, T.; Mardetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose Degradation: An Overview of Fungi and Fungal Enzymes Involved in Lignocellulose Degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
- Zang, X.; Liu, M.; Wang, H.; Fan, Y.; Zhang, H.; Liu, J.; Xing, E.; Xu, X.; Li, H. The Distribution of Active β-Glucosidase-Producing Microbial Communities in Composting. Can. J. Microbiol. 2017, 63, 998–1008. [Google Scholar] [CrossRef]
- Chukwuma, O.B.; Rafatullah, M.; Tajarudin, H.A.; Ismail, N. A Review on Bacterial Contribution to Lignocellulose Breakdown into Useful Bio-Products. Int. J. Environ. Res. Public Health 2021, 18, 6001. [Google Scholar] [CrossRef]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Metagenomic Analysis of Microbial Consortia Enriched from Compost: New Insights into the Role of Actinobacteria in Lignocellulose Decomposition. Biotechnol. Biofuels 2016, 9, 22. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Sharma, D.; Varney, R.; Simmons, B.; Isern, N.G.; Markilllie, L.M.; Nicora, C.; Norbeck, A.D.; Taylor, R.C.; Aldrich, J.T.; et al. Evidence Supporting Dissimilatory and Assimilatory Lignin Degradation in Enterobacter Lignolyticus SCF1. Front. Microbiol. 2013, 4, 280. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.Q.; Oates, N.C.; Thevarajoo, S.; Tokiman, L.; Goh, K.M.; McQueen-Mason, S.J.; Bruce, N.C.; Chong, C.S. Genomic Analysis of a Lignocellulose Degrading Strain from the Underexplored Genus Meridianimaribacter. Genomics 2020, 112, 952–960. [Google Scholar] [CrossRef]
- Ventorino, V.; Ionata, E.; Birolo, L.; Montella, S.; Marcolongo, L.; de Chiaro, A.; Espresso, F.; Faraco, V.; Pepe, O. Lignocellulose-Adapted Endo-Cellulase Producing Streptomyces Strains for Bioconversion of Cellulose-Based Materials. Front. Microbiol. 2016, 7, 2061. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Tolalpa, L.; Jiménez, D.J.; de Lima Brossi, M.J.; Salles, J.F.; van Elsas, J.D. Different Inocula Produce Distinctive Microbial Consortia with Similar Lignocellulose Degradation Capacity. Appl. Microbiol. Biotechnol. 2016, 100, 7713–7725. [Google Scholar] [CrossRef]
- Cortes-Tolalpa, L.; Wang, Y.; Salles, J.F.; van Elsas, J.D. Comparative Genome Analysis of the Lignocellulose Degrading Bacteria Citrobacter freundii So4 and Sphingobacterium multivorum W15. Front. Microbiol. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wongwilaiwalin, S.; Rattanachomsri, U.; Laothanachareon, T.; Eurwilaichitr, L.; Igarashi, Y.; Champreda, V. Analysis of a Thermophilic Lignocellulose Degrading Microbial Consortium and Multi-Species Lignocellulolytic Enzyme System. Enzyme Microb. Technol. 2010, 47, 283–290. [Google Scholar] [CrossRef]
- Zuroff, T.R.; Xiques, S.B.; Curtis, W.R. Consortia-Mediated Bioprocessing of Cellulose to Ethanol with a Symbiotic Clostridium Phytofermentans/Yeast Co-Culture. Biotechnol. Biofuels 2013, 6, 59. [Google Scholar] [CrossRef]
- Cheng, Y.F.; Edwards, J.E.; Allison, G.G.; Zhu, W.-Y.; Theodorou, M.K. Diversity and Activity of Enriched Ruminal Cultures of Anaerobic Fungi and Methanogens Grown Together on Lignocellulose in Consecutive Batch Culture. Bioresour. Technol. 2009, 100, 4821–4828. [Google Scholar] [CrossRef]
- Poszytek, K.; Ciezkowska, M.; Sklodowska, A.; Drewniak, L. Microbial Consortium with High Cellulolytic Activity (MCHCA) for Enhanced Biogas Production. Front. Microbiol. 2016, 7, 324. [Google Scholar] [CrossRef]
- Gavande, P.V.; Basak, A.; Sen, S.; Lepcha, K.; Murmu, N.; Rai, V.; Mazumdar, D.; Saha, S.P.; Das, V.; Ghosh, S. Functional Characterization of Thermotolerant Microbial Consortium for Lignocellulolytic Enzymes with Central Role of Firmicutes in Rice Straw Depolymerization. Sci. Rep. 2021, 11, 3032. [Google Scholar] [CrossRef]
- Medie, F.M.; Davies, G.J.; Drancourt, M.; Henrissat, B. Genome Analyses Highlight the Different Biological Roles of Cellulases. Nat. Rev. Microbiol. 2012, 10, 227–234. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. DbCAN2: A Meta Server for Automated Carbohydrate-Active Enzyme Annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Puentes-Téllez, P.E.; Falcao Salles, J. Construction of Effective Minimal Active Microbial Consortia for Lignocellulose Degradation. Microb. Ecol. 2018, 76, 419–429. [Google Scholar] [CrossRef]
- Liu, L.; Gao, P.; Chen, G.; Wang, L. Draft Genome Sequence of Cellulose-Digesting Bacterium Sporocytophaga myxococcoides PG-01. Genome Announc. 2014, 2, e01154-14. [Google Scholar] [CrossRef]
- Taillefer, M.; Arntzen, M.Ø.; Henrissat, B.; Pope, P.B.; Larsbrink, J. Proteomic Dissection of the Cellulolytic Machineries Used by Soil-Dwelling Bacteroidetes. mSystems 2018, 3, e00240-18. [Google Scholar] [CrossRef]
- Elberson, M.A.; Malekzadeh, F.; Yazdi, M.T.; Kameranpour, N.; Noori-Daloii, M.R.; Matte, M.H.; Shahamat, M.; Colwell, R.R.; Sowers, K.R. Cellulomonas persica sp. nov. and Cellulomonas iranensis sp. nov., Mesophilic Cellulose-Degrading Bacteria Isolated from Forest Soils. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 3, 993–996. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poulsen, H.V.; Willink, F.W.; Ingvorsen, K. Aerobic and Anaerobic Cellulase Production by Cellulomonas Uda. Arch. Microbiol. 2016, 198, 725–735. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Khawdas, W.; Aso, Y.; Ohara, H. Microbial Fuel Cells Using Cellulomonas spp. with Cellulose as Fuel. J. Biosci. Bioeng. 2017, 123, 358–363. [Google Scholar] [CrossRef]
- Vladut-Talor, M.; Kauri, T.; Kushner, D.J. Effects of Cellulose on Growth, Enzyme Production, and Ultrastructure of a Cellulomonas Species. Arch. Microbiol. 1986, 144, 191–195. [Google Scholar] [CrossRef]
- Herrera, L.M.; Braña, V.; Franco Fraguas, L.; Castro-Sowinski, S. Characterization of the Cellulase-Secretome Produced by the Antarctic Bacterium Flavobacterium sp. AUG42. Microbiol. Res. 2019, 223–225, 13–21. [Google Scholar] [CrossRef]
- Kim, H.; Yu, S.M. Flavobacterium nackdongense sp. nov., a Cellulose-Degrading Bacterium Isolated from Sediment. Arch. Microbiol. 2020, 202, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Jiang, J.; Xu, Z.; Zhou, Y.; Leung, F.C.-C. Complete Genome Sequence of Pseudoxanthomonas suwonensis Strain J1, a Cellulose-Degrading Bacterium Isolated from Leaf- and Wood-Enriched Soil. Genome Announc. 2015, 3, e00614-15. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Revathi, K.; Khanna, S. Biodegradation of Cellulosic and Lignocellulosic Waste by Pseudoxanthomonas sp R-28. Carbohydr. Polym. 2015, 134, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Eichorst, S.A.; Kuske, C.R. Identification of Cellulose-Responsive Bacterial and Fungal Communities in Geographically and Edaphically Different Soils by Using Stable Isotope Probing. Appl. Environ. Microbiol. 2012, 78, 2316–2327. [Google Scholar] [CrossRef]
- Stursová, M.; Zifčáková, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose Utilization in Forest Litter and Soil: Identification of Bacterial and Fungal Decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Zühlke, D.; Větrovský, T.; Becher, D.; Riedel, K.; Baldrian, P. Decoding the Complete Arsenal for Cellulose and Hemicellulose Deconstruction in the Highly Efficient Cellulose Decomposer Paenibacillus O199. Biotechnol. Biofuels 2016, 9, 104. [Google Scholar] [CrossRef]
- Kim, E.S.; Kim, B.-S.; Kim, K.-Y.; Woo, H.-M.; Lee, S.-M.; Um, Y. Aerobic and Anaerobic Cellulose Utilization by Paenibacillus sp. CAA11 and Enhancement of Its Cellulolytic Ability by Expressing a Heterologous Endoglucanase. J. Biotechnol. 2018, 268, 21–27. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Pepe-Ranney, C.; Weisenhorn, P.; Lipton, M.; Buckley, D.H. Competitive Exclusion and Metabolic Dependency among Microorganisms Structure the Cellulose Economy of an Agricultural Soil. mBio 2021, 12, e03099-20. [Google Scholar] [CrossRef]
- Dahal, R.H.; Lee, H.; Chaudhary, D.K.; Kim, D.-Y.; Son, J.; Kim, J.; Ka, J.-O.; Kim, D.-U. Caenimonas soli sp. Nov., Isolated from Soil. Arch. Microbiol. 2021, 203, 1123–1129. [Google Scholar] [CrossRef]
- Kim, S.-J.; Weon, H.-Y.; Kim, Y.-S.; Moon, J.Y.; Seok, S.J.; Hong, S.-B.; Kwon, S.-W. Caenimonas terrae sp. Nov., Isolated from a Soil Sample in Korea, and Emended Description of the Genus Caenimonas Ryu et al. 2008. J. Microbiol. 2012, 50, 864–868. [Google Scholar] [CrossRef]
- Johnke, J.; Fraune, S.; Bosch, T.C.G.; Hentschel, U.; Schulenburg, H. Bdellovibrio and Like Organisms Are Predictors of Microbiome Diversity in Distinct Host Groups. Microb. Ecol. 2020, 79, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.L.; Yang, R.; Decker, T.; Cavalliere, C.; Andreev, V.; Bircher, N.; Cornell, J.; Dohmen, R.; Pratt, C.J.; Grinnell, A.; et al. Genomes of Novel Myxococcota Reveal Severely Curtailed Machineries for Predation and Cellular Differentiation. Appl. Environ. Microbiol. 2021, 87, e0170621. [Google Scholar] [CrossRef] [PubMed]
- Mille-Lindblom, C.; Tranvik, L.J. Antagonism between Bacteria and Fungi on Decomposing Aquatic Plant Litter. Microb. Ecol. 2003, 45, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Pajdak-Stós, A.; Sobczyk, M.; Fiałkowska, E.; Kocerba-Soroka, W.; Fyda, J. The Effect of Three Different Predatory Ciliate Species on Activated Sludge Microfauna. Eur. J. Protistol. 2017, 58, 87–93. [Google Scholar] [CrossRef]
- Lee, J.A.; Baugh, A.C.; Shevalier, N.J.; Strand, B.; Stolyar, S.; Marx, C.J. Cross-Feeding of a Toxic Metabolite in a Synthetic Lignocellulose-Degrading Microbial Community. Microorganisms 2021, 9, 321. [Google Scholar] [CrossRef] [PubMed]
- Dumitrache, A.; Wolfaardt, G.M.; Allen, D.G.; Liss, S.N.; Lynd, L.R. Tracking the Cellulolytic Activity of Clostridium Thermocellum Biofilms. Biotechnol. Biofuels 2013, 6, 175. [Google Scholar] [CrossRef]
- Betts, G. 23—Other Spoilage Bacteria. In Food Spoilage Microorganisms; de Blackburn, C.W., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Sawston, UK, 2006; pp. 668–693. [Google Scholar]
- Blakemore, R. Magnetotactic Bacteria. Science 1975, 190, 377–379. [Google Scholar] [CrossRef]
- Smalley, N.E.; Taipale, S.; De Marco, P.; Doronina, N.V.; Kyrpides, N.; Shapiro, N.; Woyke, T.; Kalyuzhnaya, M.G. Functional and Genomic Diversity of Methylotrophic Rhodocyclaceae: Description of Methyloversatilis discipulorum sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 2227–2233. [Google Scholar] [CrossRef]
- Koeck, D.E.; Ludwig, W.; Wanner, G.; Zverlov, V.V.; Liebl, W.; Schwarz, W.H. Herbinix hemicellulosilytica gen. nov., sp. nov., a Thermophilic Cellulose-Degrading Bacterium Isolated from a Thermophilic Biogas Reactor. Int. J. Syst. Evol. Microbiol. 2015, 65, 2365–2371. [Google Scholar] [CrossRef]
- Ueki, A.; Ohtaki, Y.; Kaku, N.; Ueki, K. Descriptions of Anaerotaenia torta gen. nov., sp. nov. and Anaerocolumna cellulosilytica gen. nov., sp. nov. Isolated from a Methanogenic Reactor of Cattle Waste and Reclassification of Clostridium aminovalericum, Clostridium jejuense and Clostridium xylanovorans as Anaerocolumna Species. Int. J. Syst. Evol. Microbiol. 2016, 66, 2936–2943. [Google Scholar] [CrossRef]
- Wu, K.; Cheng, L. Ruminiclostridium. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2021; pp. 1–11. [Google Scholar]
- Brethauer, S.; Shahab, R.L.; Studer, M.H. Impacts of Biofilms on the Conversion of Cellulose. Appl. Microbiol. Biotechnol. 2020, 104, 5201–5212. [Google Scholar] [CrossRef] [PubMed]
- Pollet, T.; Berdjeb, L.; Garnier, C.; Durrieu, G.; Le Poupon, C.; Misson, B.; Jean-François, B. Prokaryotic Community Successions and Interactions in Marine Biofilms: The Key Role of Flavobacteriia. FEMS Microbiol. Ecol. 2018, 94, fiy083. [Google Scholar] [CrossRef]
- Stam, M.R.; Danchin, E.G.J.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the Large Glycoside Hydrolase Family 13 into Subfamilies: Towards Improved Functional Annotations of Alpha-Amylase-Related Proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef]
- Brás, J.L.A.; Cartmell, A.; Carvalho, A.L.M.; Verzé, G.; Bayer, E.A.; Vazana, Y.; Correia, M.A.S.; Prates, J.A.M.; Ratnaparkhe, S.; Boraston, A.B.; et al. Structural Insights into a Unique Cellulase Fold and Mechanism of Cellulose Hydrolysis. Proc. Natl. Acad. Sci. USA 2011, 108, 5237–5242. [Google Scholar] [CrossRef] [PubMed]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and Hemicellulose Decomposition by Forest Soil Bacteria Proceeds by the Action of Structurally Variable Enzymatic Systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Han, Y.; Dodd, D.; Schroeder, C.M.; Mackie, R.I.; Cann, I.K.O. Chapter 1—Thermostable Enzymes as Biocatalysts in the Biofuel Industry. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2010; Volume 70, pp. 1–55. [Google Scholar]
- Aspeborg, H.; Coutinho, P.M.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, Substrate Specificity and Subfamily Classification of Glycoside Hydrolase Family 5 (GH5). BMC Evol. Biol. 2012, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, N.; Goto, T.; Takahashi, K.; Kasai, D.; Otsuka, Y.; Nakamura, M.; Katayama, Y.; Fukuda, M.; Masai, E. A Bacterial Aromatic Aldehyde Dehydrogenase Critical for the Efficient Catabolism of Syringaldehyde. Sci. Rep. 2017, 7, 44422. [Google Scholar] [CrossRef]
- Zeng, Y.; Charkowski, A.O. The Role of ATP-Binding Cassette Transporters in Bacterial Phytopathogenesis. Phytopathology 2021, 111, 600–610. [Google Scholar] [CrossRef]
- Li, B.; Maezato, Y.; Kim, S.H.; Kurihara, S.; Liang, J.; Michael, A.J. Polyamine-Independent Growth and Biofilm Formation, and Functional Spermidine/Spermine N-Acetyltransferases in Staphylococcus Aureus and Enterococcus Faecalis. Mol. Microbiol. 2019, 111, 159–175. [Google Scholar] [CrossRef]
- Campbell, A.G.; Schwientek, P.; Vishnivetskaya, T.; Woyke, T.; Levy, S.; Beall, C.J.; Griffen, A.; Leys, E.; Podar, M. The Under-Recognized Dominance of Verrucomicrobia in Soil Bacterial Communities. Environ. Microbiol. 2015, 16, 2635–2643. [Google Scholar] [CrossRef]
- Tláskal, V.; Voříšková, J.; Baldrian, P. Bacterial Succession on Decomposing Leaf Litter Exhibits a Specific Occurrence Pattern of Cellulolytic Taxa and Potential Decomposers of Fungal Mycelia. FEMS Microbiol. Ecol. 2016, 92, fiw177. [Google Scholar] [CrossRef] [PubMed]
- van der Lelie, D.; Taghavi, S.; McCorkle, S.M.; Li, L.-L.; Malfatti, S.A.; Monteleone, D.; Donohoe, B.S.; Ding, S.-Y.; Adney, W.S.; Himmel, M.E.; et al. The Metagenome of an Anaerobic Microbial Community Decomposing Poplar Wood Chips. PLoS One 2012, 7, e36740. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R. Development of Microorganisms for Cellulose-Biofuel Consolidated Bioprocessings: Metabolic Engineers’ Tricks. Comput. Struct. Biotechnol. J. 2012, 3, e201210007. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, H.B.; Clayton, J. On the Decomposition of Cellulose by an Aerobic Organism (Spirochaeta cytophaga, n. sp.). J. Agric. Sci. 1919, 9, 143–172. [Google Scholar] [CrossRef]
- Gladkov, G.; Kimeklis, A.; Zverev, A.; Pershina, E.; Ivanova, E.; Kichko, A.; Andronov, E.; Abakumov, E. Soil Microbiome of the Postmining Areas in Polar Ecosystems in Surroundings of Nadym, Western Siberia, Russia. Open Agric. 2019, 4, 684–696. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 12 August 2022).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 2013, 8, e61217. [Google Scholar] [CrossRef]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R Package to Analyse and Visualise 16S RRNA Amplicon Data. bioRxiv 2018. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Champaign, IL, USA, 1949. [Google Scholar]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of Two Random Variables Is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Gower, J.C. Principal Coordinates Analysis. In Wiley StatsRef: Statistics Reference Online; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 1–7. [Google Scholar]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Silverman, J.D.; Washburne, A.D.; Mukherjee, S.; David, L.A. A Phylogenetic Transform Enhances Analysis of Compositional Microbiota Data. eLife 2017, 6, e21887. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of Neural Network Basecalling Tools for Oxford Nanopore Sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Bickhart, D.M.; Behsaz, B.; Gurevich, A.; Rayko, M.; Shin, S.B.; Kuhn, K.; Yuan, J.; Polevikov, E.; Smith, T.P.L.; et al. MetaFlye: Scalable Long-Read Metagenome Assembly Using Repeat Graphs. Nat. Methods 2020, 17, 1103–1110. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de Novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Medaka: Sequence Correction Provided by ONT Research. Available online: https://github.com/nanoporetech/medaka (accessed on 22 February 2022).
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Struo2: A Pipeline for Building Custom Databases for Common Metagenome Profilers. Available online: https://github.com/leylabmpi/Struo2 (accessed on 12 August 2022).
- Loman Lab Mock Community Experiments. Databases. Available online: https://lomanlab.github.io/mockcommunity/mc_databases.html (accessed on 12 August 2022).
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. DbCAN: A Web Resource for Automated Carbohydrate-Active Enzyme Annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Busk, P.K.; Pilgaard, B.; Lezyk, M.J.; Meyer, A.S.; Lange, L. Homology to Peptide Pattern for Annotation of Carbohydrate-Active Enzymes and Prediction of Function. BMC Bioinform. 2017, 18, 214. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, H.; Wu, P.; Entwistle, S.; Li, X.; Yohe, T.; Yi, H.; Yang, Z.; Yin, Y. DbCAN-Seq: A Database of Carbohydrate-Active Enzyme (CAZyme) Sequence and Annotation. Nucleic Acids Res. 2018, 46, D516–D521. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. Statistical Methods for Research Workers; Oliver and Boyd: Edinburgh, UK, 1938. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.2-0. 2014. Available online: http://CRAN.Rproject.org/package=vegan (accessed on 12 August 2022).
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.-A.; Hugenholtz, P. GTDB: An Ongoing Census of Bacterial and Archaeal Diversity through a Phylogenetically Consistent, Rank Normalized and Complete Genome-Based Taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef]
- Shaffer, M.; Borton, M.A.; McGivern, B.B.; Zayed, A.A.; La Rosa, S.L.; Solden, L.M.; Liu, P.; Narrowe, A.B.; Rodríguez-Ramos, J.; Bolduc, B.; et al. DRAM for Distilling Microbial Metabolism to Automate the Curation of Microbiome Function. Nucleic Acids Res. 2020, 48, 8883–8900. [Google Scholar] [CrossRef] [PubMed]
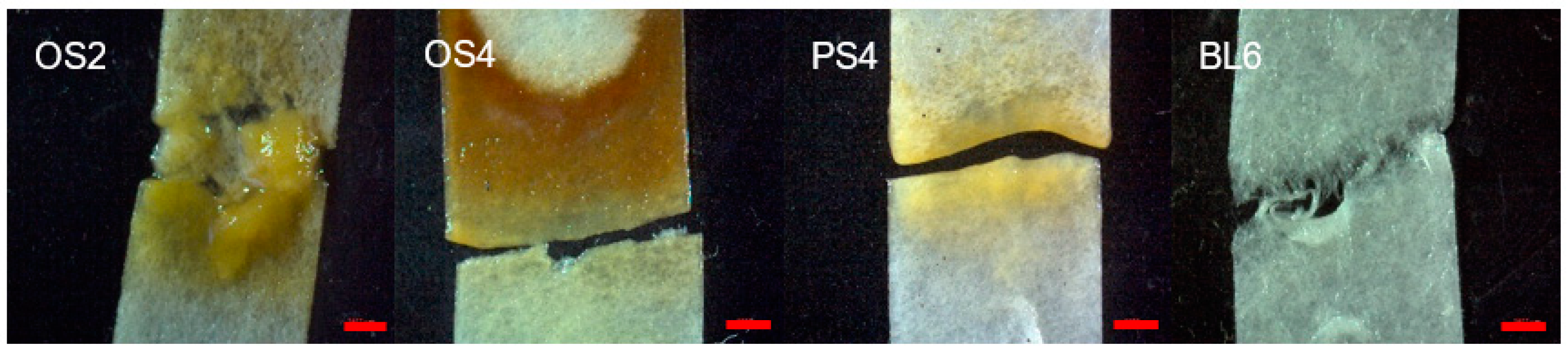

| Consortia ID | Substrate | Composting Time, Months | Substrate Dilution | Color of Consortia | Gas Formation |
|---|---|---|---|---|---|
| OS2 | oat straw | 2 | 106 | yellow | no |
| OS4 | oat straw | 4 | 106 | brown | yes |
| PS4 | pine sawdust | 4 | 103 | yellow | no |
| BL6 | birch leaf litter | 6 | 104 | transparent | yes |
| Consortium ID | Contigs | Longest Contig, bp | Total Contigs Length | N50 |
|---|---|---|---|---|
| OS2 | 1879 | 1,582,432 | 86,161,676 | 72,582 |
| OS4 | 1612 | 5,307,558 | 96,558,959 | 131,290 |
| PS4 | 1466 | 2,705,151 | 87,318,114 | 112,897 |
| BL6 | 1748 | 3,656,276 | 108,520,437 | 186,463 |
| Consortium ID | AAs | CBMs | CEs | GHs | GTs | PLs |
|---|---|---|---|---|---|---|
| OS2 | 4.1 | 10.8 | 10.7 | 43.1 | 29.2 | 2.2 |
| OS4 | 3.7 | 11.3 | 11.1 | 41.9 | 30.6 | 1.4 |
| PS4 | 2.6 | 10.2 | 11.1 | 44.3 | 30.0 | 1.8 |
| BL6 | 2.7 | 12.0 | 9.5 | 47.0 | 26.1 | 2.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gladkov, G.V.; Kimeklis, A.K.; Afonin, A.M.; Lisina, T.O.; Orlova, O.V.; Aksenova, T.S.; Kichko, A.A.; Pinaev, A.G.; Andronov, E.E. The Structure of Stable Cellulolytic Consortia Isolated from Natural Lignocellulosic Substrates. Int. J. Mol. Sci. 2022, 23, 10779. https://doi.org/10.3390/ijms231810779
Gladkov GV, Kimeklis AK, Afonin AM, Lisina TO, Orlova OV, Aksenova TS, Kichko AA, Pinaev AG, Andronov EE. The Structure of Stable Cellulolytic Consortia Isolated from Natural Lignocellulosic Substrates. International Journal of Molecular Sciences. 2022; 23(18):10779. https://doi.org/10.3390/ijms231810779
Chicago/Turabian StyleGladkov, Grigory V., Anastasiia K. Kimeklis, Alexey M. Afonin, Tatiana O. Lisina, Olga V. Orlova, Tatiana S. Aksenova, Arina A. Kichko, Alexander G. Pinaev, and Evgeny E. Andronov. 2022. "The Structure of Stable Cellulolytic Consortia Isolated from Natural Lignocellulosic Substrates" International Journal of Molecular Sciences 23, no. 18: 10779. https://doi.org/10.3390/ijms231810779
APA StyleGladkov, G. V., Kimeklis, A. K., Afonin, A. M., Lisina, T. O., Orlova, O. V., Aksenova, T. S., Kichko, A. A., Pinaev, A. G., & Andronov, E. E. (2022). The Structure of Stable Cellulolytic Consortia Isolated from Natural Lignocellulosic Substrates. International Journal of Molecular Sciences, 23(18), 10779. https://doi.org/10.3390/ijms231810779







