Hepatic Stellate Cell Modulates the Immune Microenvironment in the Progression of Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
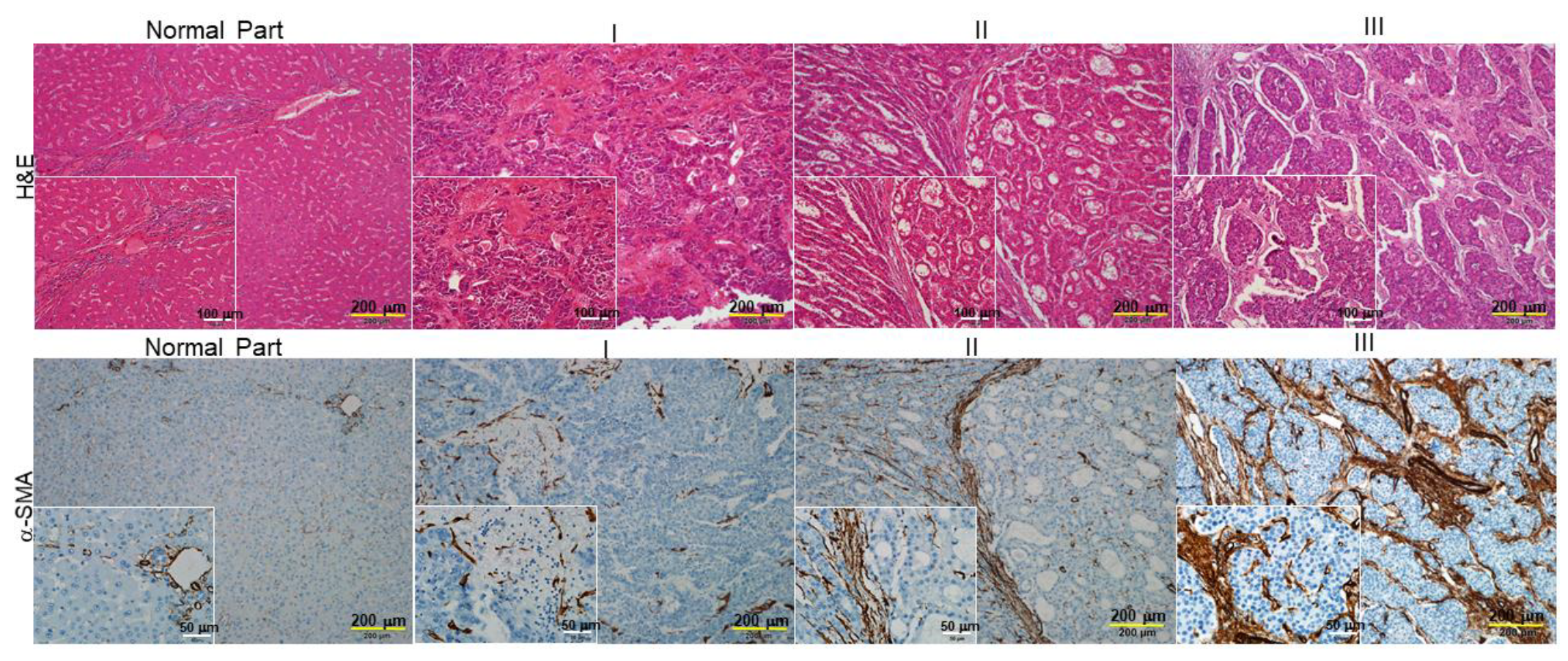
2.1. The Levels of α-Smooth Muscle Actin (SMA) Implicate the Association between the Degrees of Hepatic Fibrosis and HCC Stage
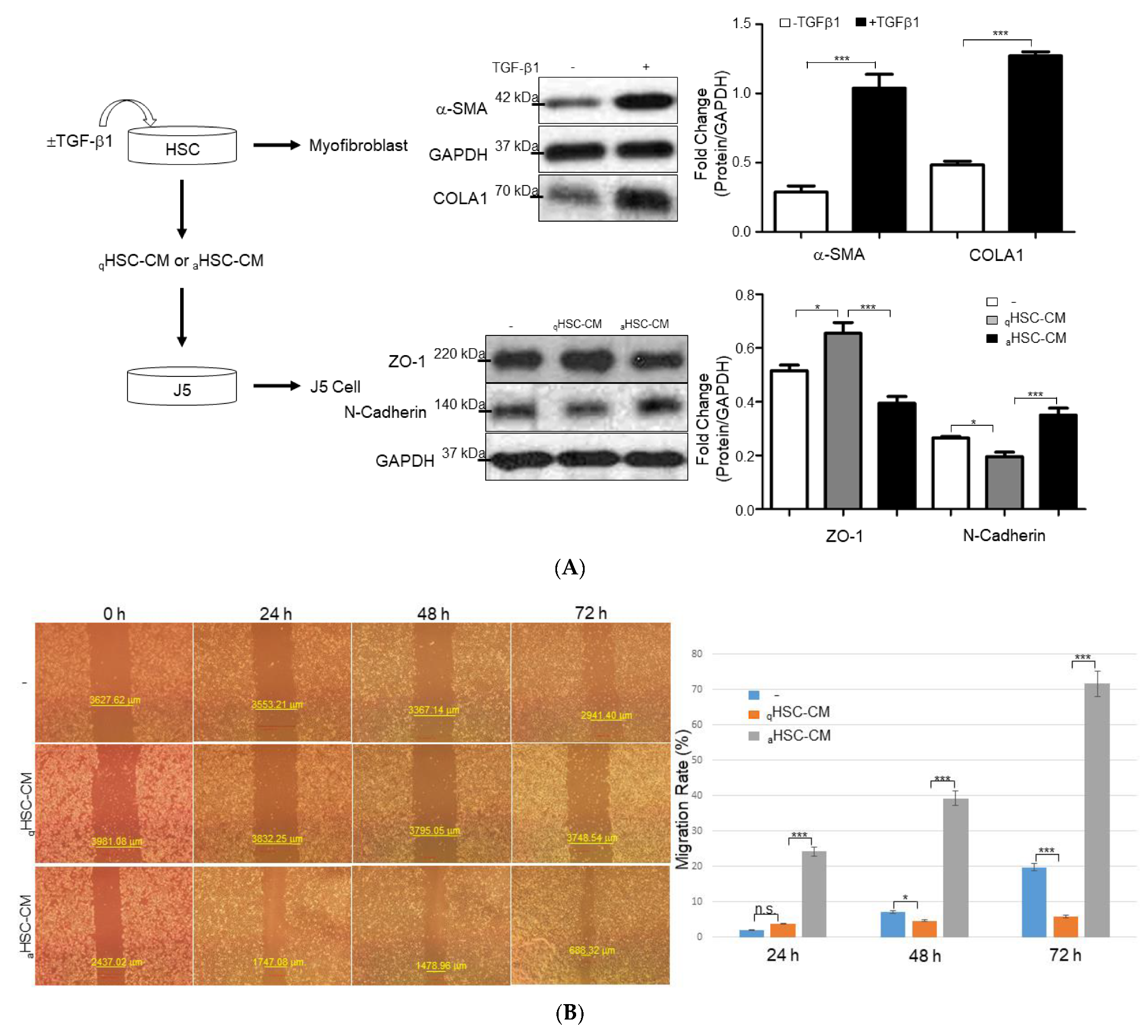
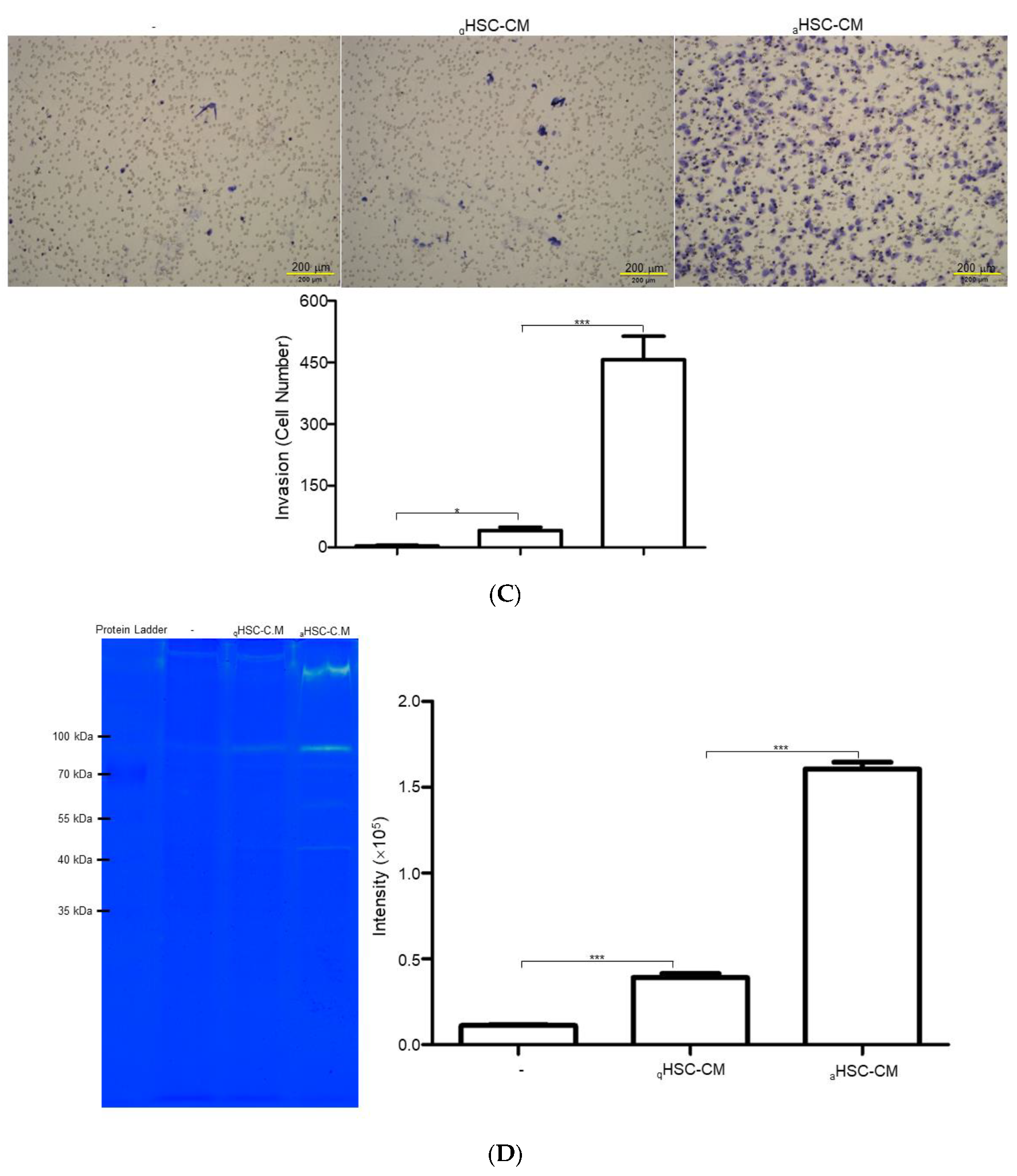
2.2. HSC-CM Stimulated Migration and Invasion of J5 Cells under Application of TGF-β1
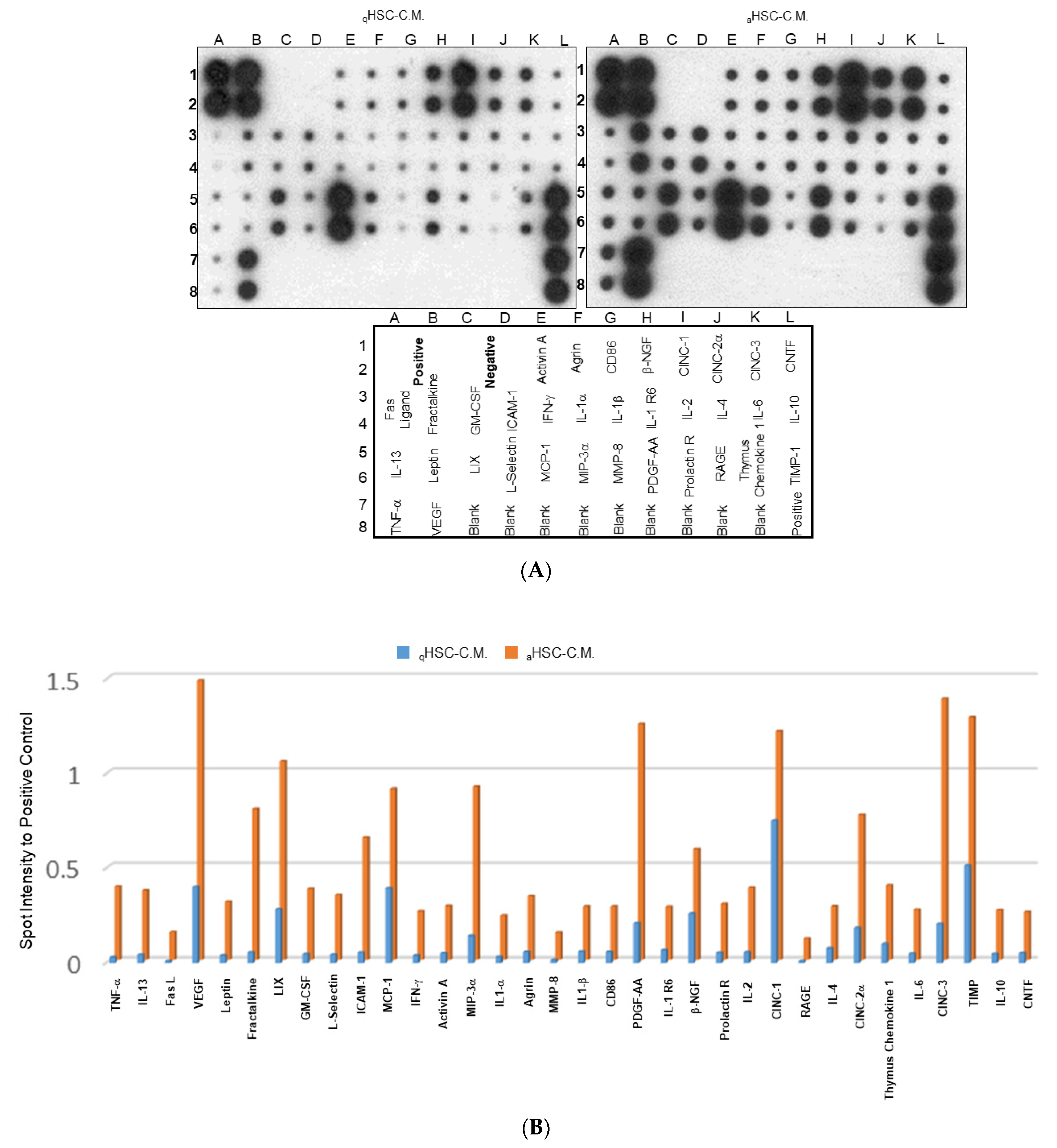
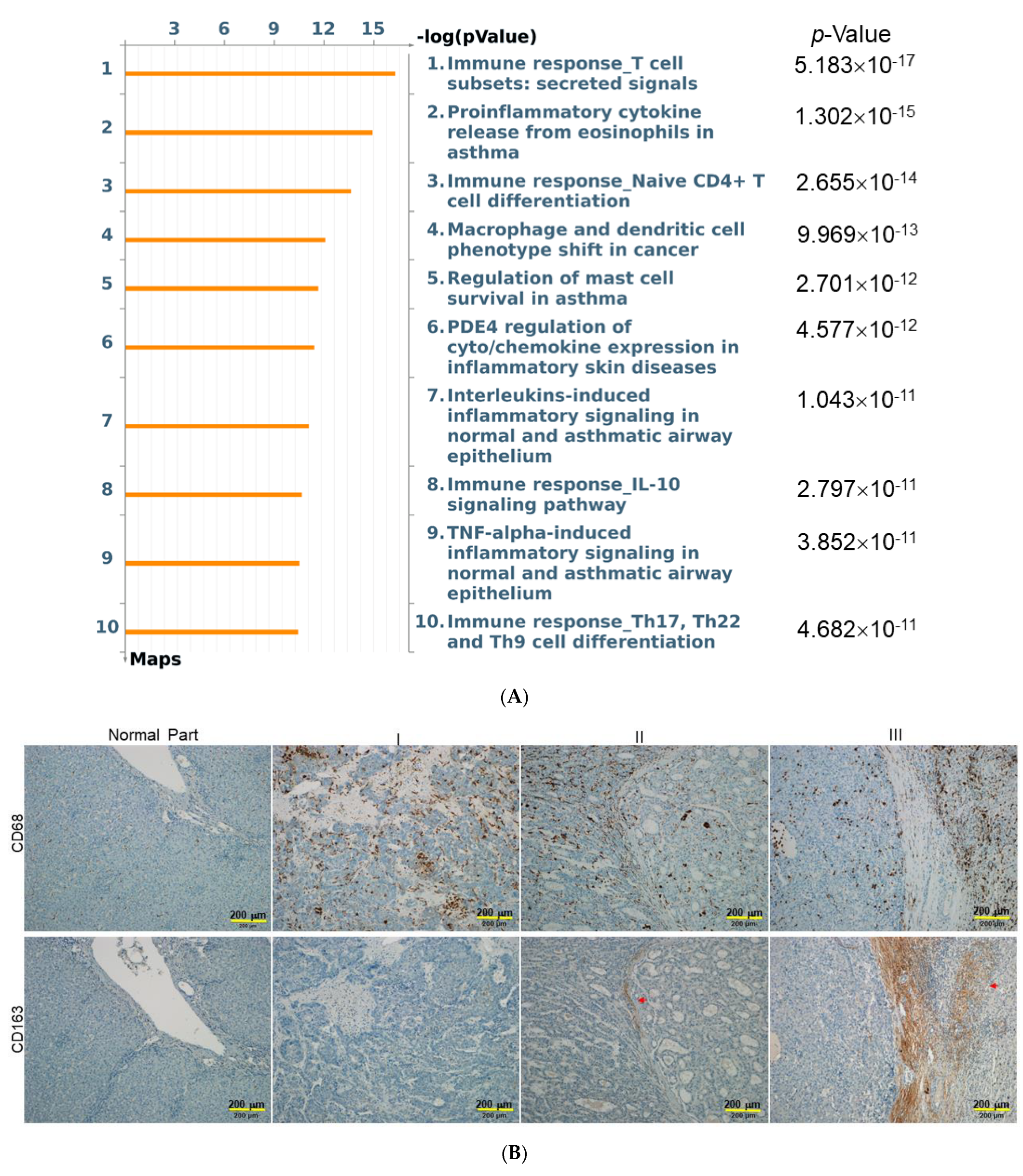
2.3. Characterization of the Changes in Global Cytokine/Chemokine Profile of Different HSC-CM
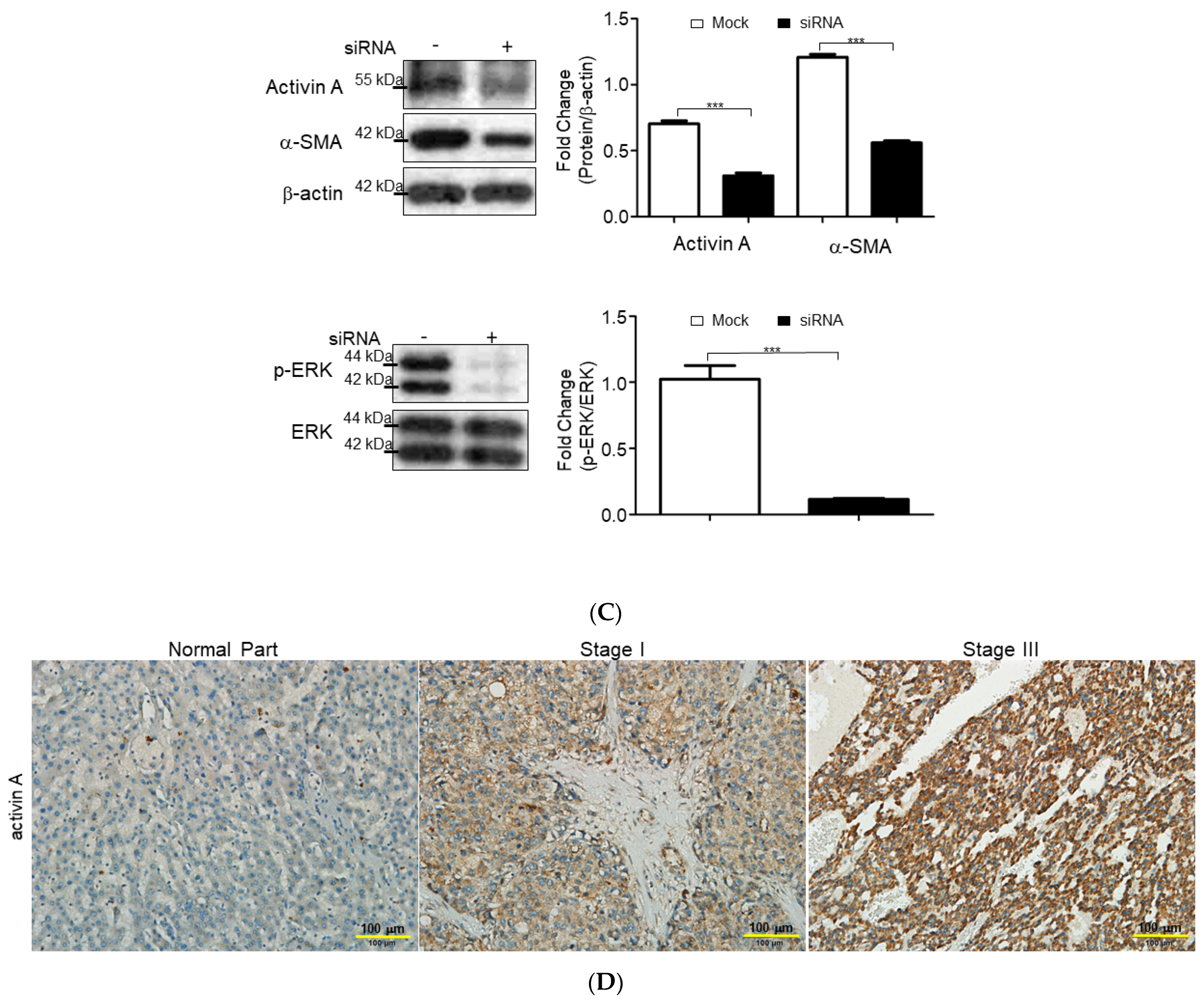
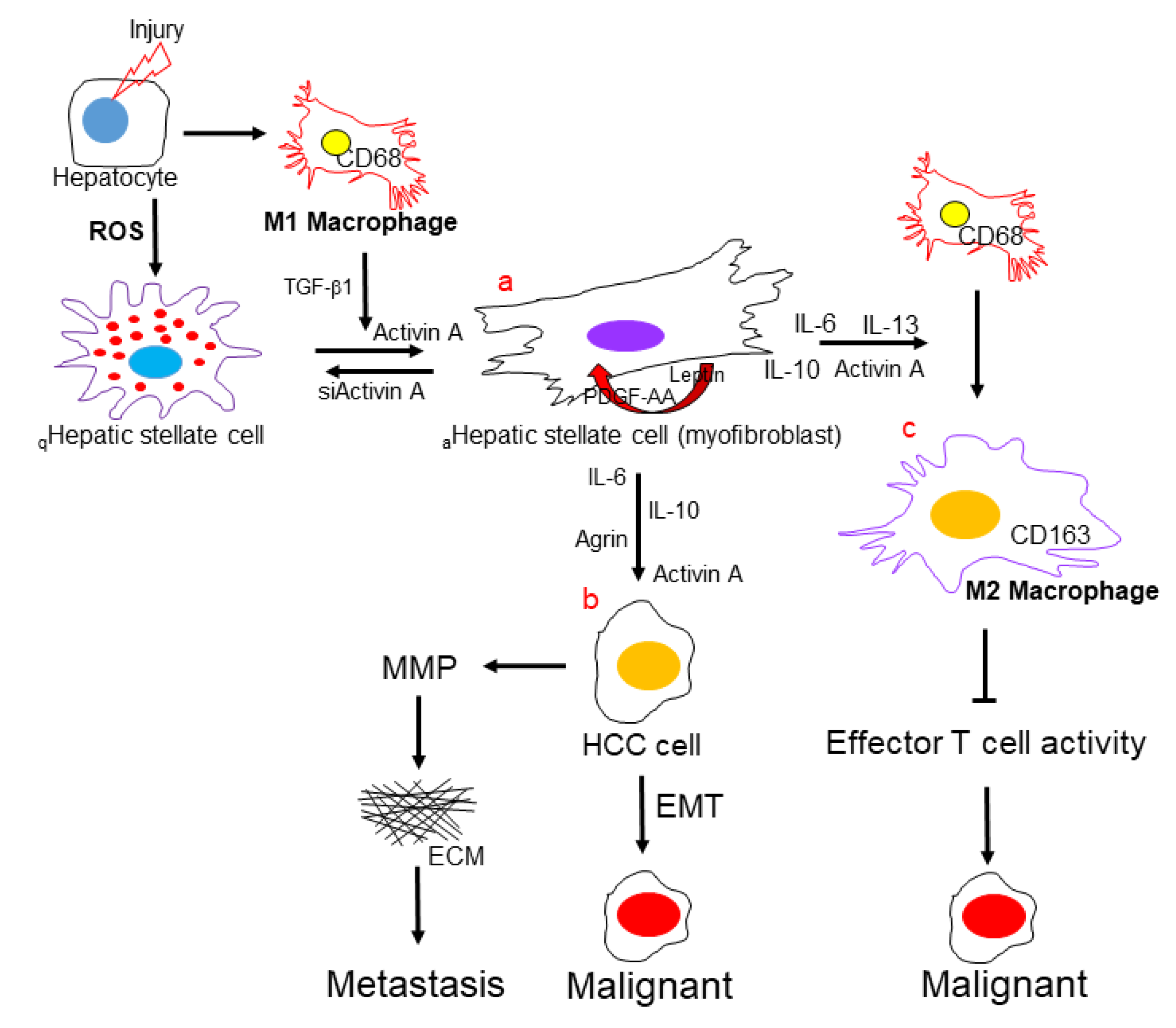
2.4. Functional Impact of Activated HSCs on HCC Microenvironment
3. Discussion
4. Materials and Methods
4.1. Tissue Array
4.2. Cell Culture
4.3. Collection of Conditioning Medium (CM) from HSCs
4.4. Wound-Migration Assay
4.5. Invasion Assay
4.6. Gelatin Zymography
4.7. Cytokine Protein Array and Functional Analysis
4.8. Western Blot Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wallace, M.C.; Friedman, S.L. Hepatic fibrosis and the microenvironment: Fertile soil for hepatocellular carcinoma development. Gene Expr. 2014, 16, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Intercellular crosstalk of liver sinusoidal endothelial cells in liver fibrosis, cirrhosis and hepatocellular carcinoma. Dig. Liver Dis. 2022, 54, 598–613. [Google Scholar] [CrossRef]
- Aoki, T.; Nishida, N.; Kudo, M. Current Perspectives on the Immunosuppressive Niche and Role of Fibrosis in Hepatocellular Carcinoma and the Development of Antitumor Immunity. J. Histochem. Cytochem. 2022, 70, 53–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hassan, W.; Jabeen, Q.; Khan, G.J.; Iqbal, F. Interdependent and independent multidimensional role of tumor microenvironment on hepatocellular carcinoma. Cytokine 2018, 103, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.I.; Conroy, K.P.; Henderson, N.C. Hepatic stellate cells: Central modulators of hepatic carcinogenesis. BMC Gastroenterol. 2015, 15, 63. [Google Scholar] [CrossRef]
- Shiraha, H.; Iwamuro, M.; Okada, H. Hepatic Stellate Cells in Liver Tumor. Adv. Exp. Med. Biol. 2020, 1234, 43–56. [Google Scholar]
- Barry, A.E.; Baldeosingh, R.; Lamm, R.; Patel, K.; Zhang, K.; Dominguez, D.A.; Kirton, K.J.; Shah, A.P.; Dang, H. Hepatic Stellate Cells and Hepatocarcinogenesis. Front. Cell Dev. Biol. 2020, 8, 709. [Google Scholar] [CrossRef]
- Fabregat, I.; Caballero-Díaz, D. Transforming Growth Factor-β-Induced Cell Plasticity in Liver Fibrosis and Hepatocarcinogenesis. Front. Oncol. 2018, 8, 357. [Google Scholar] [CrossRef]
- Carloni, V.; Luong, T.V.; Rombouts, K. Hepatic stellate cells and extracellular matrix in hepatocellular carcinoma: More complicated than ever. Liver Int. 2014, 34, 834–843. [Google Scholar] [CrossRef]
- Coulouarn, C.; Clément, B. Stellate cells and the development of liver cancer: Therapeutic potential of targeting the stroma. J. Hepatol. 2014, 60, 1306–1309. [Google Scholar] [CrossRef]
- Seki, E.; Schwabe, R.F. Hepatic inflammation and fibrosis: Functional links and key pathways. Hepatology 2015, 61, 1066–1079. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Rong, D.; Zhang, B.; Zheng, W.; Wang, X.; Chen, Z.; Tang, W. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: Challenges and opportunities. Mol. Cancer 2019, 18, 130. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; He, K.; Pan, Y.; Wang, H.; Luo, Y.; Xia, Q. The role of tumor-associated macrophages in primary hepatocellular carcinoma and its related targeting therapy. Int. J. Med. Sci. 2021, 18, 2109–2116. [Google Scholar] [CrossRef]
- Zhou, D.; Luan, J.; Huang, C.; Li, J. Tumor-Associated Macrophages in Hepatocellular Carcinoma: Friend or Foe? Gut Liver 2021, 15, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Degroote, H.; Van Dierendonck, A.; Geerts, A.; Van Vlierberghe, H.; Devisscher, L. Preclinical and Clinical Therapeutic Strategies Affecting Tumor-Associated Macrophages in Hepatocellular Carcinoma. J. Immunol. Res. 2018, 2018, 7819520. [Google Scholar] [CrossRef]
- Harding, J.J.; Khalil, D.N.; Abou-Alfa, G.K. Biomarkers: What Role Do They Play (If Any) for Diagnosis, Prognosis and Tumor Response Prediction for Hepatocellular Carcinoma? Dig. Dis. Sci. 2019, 64, 918–927. [Google Scholar] [CrossRef]
- Yang, P.; Li, X.; Wu, M.; Kwoh, C.K.; Ng, S.K. Inferring gene-phenotype associations via global protein complex network propagation. PLoS ONE 2011, 6, e21502. [Google Scholar] [CrossRef]
- Saviano, A.; Henderson, N.C.; Baumert, T.F. Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J. Hepatol. 2020, 73, 1219–1230. [Google Scholar] [CrossRef]
- Baglieri, J.; Brenner, D.A.; Kisseleva, T. The Role of Fibrosis and Liver-Associated Fibroblasts in the Pathogenesis of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 1723. [Google Scholar] [CrossRef]
- Dhar, D.; Baglieri, J.; Kisseleva, T.; Brenner, D.A. Mechanisms of liver fibrosis and its role in liver cancer. Exp. Biol. Med. 2020, 245, 96–108. [Google Scholar] [CrossRef]
- Ezhilarasan, D. Hepatic stellate cells in the injured liver: Perspectives beyond hepatic fibrosis. J. Cell. Physiol. 2022, 237, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Wang, H.; Burke, L.J.; Bridle, K.R.; Li, X.; Zhao, C.X.; Crawford, D.H.G.; Roberts, M.S.; Liang, X. Therapeutic modulators of hepatic stellate cells for hepatocellular carcinoma. Int. J. Cancer 2020, 147, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.C.; Lin, S.M.; Chen, M.F.; Pan, T.L.; Wang, P.W.; Yeh, C.T. Evaluation of serum matrix metalloproteinase (MMP)-9 to MMP-2 ratio as a biomarker in hepatocellular carcinoma. Hepatogastroenterology 2010, 57, 98–102. [Google Scholar]
- Song, J.; Ge, Z.; Yang, X.; Luo, Q.; Wang, C.; You, H.; Ge, T.; Deng, Y.; Lin, H.; Cui, Y.; et al. Hepatic stellate cells activated by acidic tumor microenvironment promote the metastasis of hepatocellular carcinoma via osteopontin. Cancer Lett. 2015, 356, 713–720. [Google Scholar] [CrossRef]
- Chang, W.S.; Tsai, C.W.; Yang, J.S.; Hsu, Y.M.; Shih, L.C.; Chiu, H.Y.; Bau, D.T.; Tsai, F.J. Resveratrol inhibited the metastatic behaviors of cisplatin-resistant human oral cancer cells via phosphorylation of ERK/p-38 and suppression of MMP-2/9. J. Food Biochem. 2021, 45, e13666. [Google Scholar] [CrossRef]
- Hrckova, G.; Velebný, S.; Solár, P. Dynamics of hepatic stellate cells, collagen types I and III synthesis and gene expression of selected cytokines during hepatic fibrogenesis following Mesocestoides vogae (Cestoda) infection in mice. Int. J. Parasitol. 2010, 40, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Aishima, S.; Fujita, N.; Tanaka, Y.; Kubo, Y.; Motomura, T.; Taketomi, A.; Shirabe, K.; Maehara, Y.; Oda, Y. Tumor-associated macrophage promotes tumor progression via STAT3 signaling in hepatocellular carcinoma. Pathobiology 2013, 80, 146–154. [Google Scholar] [CrossRef]
- Zhao, Z.; Xiong, S.; Wang, R.; Li, Y.; Wang, X.; Wang, Y.; Bai, S.; Chen, W.; Zhao, Y.; Cheng, B. Peri-tumor fibroblasts promote tumorigenesis and metastasis of hepatocellular carcinoma via Interleukin6/STAT3 signaling pathway. Cancer Manag. Res. 2019, 11, 2889–2901. [Google Scholar] [CrossRef]
- Hsieh, C.C.; Hung, C.H.; Chiang, M.; Tsai, Y.C.; He, J.T. Hepatic Stellate Cells Enhance Liver Cancer Progression by Inducing Myeloid-Derived Suppressor Cells through Interleukin-6 Signaling. Int. J. Mol. Sci. 2019, 20, 5079. [Google Scholar] [CrossRef]
- Lv, X.; Fang, C.; Yin, R.; Qiao, B.; Shang, R.; Wang, J.; Song, W.; He, Y.; Chen, Y. Agrin para-secreted by PDGF-activated human hepatic stellate cells promotes hepatocarcinogenesis in vitro and in vivo. Oncotarget 2017, 8, 105340–105355. [Google Scholar] [CrossRef]
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Kreidl, E.; Oztürk, D.; Metzner, T.; Berger, W.; Grusch, M. Activins and follistatins: Emerging roles in liver physiology and cancer. World J. Hepatol. 2009, 1, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F. Targeting hepatic macrophages to treat liver diseases. J. Hepatol. 2017, 66, 1300–1312. [Google Scholar] [CrossRef] [PubMed]
- Lurje, I.; Hammerich, L.; Tacke, F. Dendritic Cell and T Cell Crosstalk in Liver Fibrogenesis and Hepatocarcinogenesis: Implications for Prevention and Therapy of Liver Cancer. Int. J. Mol. Sci. 2020, 21, 7378. [Google Scholar] [CrossRef] [PubMed]
- Shirabe, K.; Mano, Y.; Muto, J.; Matono, R.; Motomura, T.; Toshima, T.; Takeishi, K.; Uchiyama, H.; Yoshizumi, T.; Taketomi, A.; et al. Role of tumor-associated macrophages in the progression of hepatocellular carcinoma. Surg. Today 2012, 42, 1–7. [Google Scholar] [CrossRef]
- Wijesundera, K.K.; Izawa, T.; Tennakoon, A.H.; Murakami, H.; Golbar, H.M.; Katou-Ichikawa, C.; Tanaka, M.; Kuwamura, M.; Yamate, J. M1- and M2-macrophage polarization in rat liver cirrhosis induced by thioacetamide (TAA), focusing on Iba1 and galectin-3. Exp. Mol. Pathol. 2014, 96, 382–392. [Google Scholar] [CrossRef]
- Li, Z.; Wu, T.; Zheng, B.; Chen, L. Individualized precision treatment: Targeting TAM in HCC. Cancer Lett. 2019, 458, 86–91. [Google Scholar] [CrossRef]
- Sugimura, K.; Miyata, H.; Tanaka, K.; Takahashi, T.; Kurokawa, Y.; Yamasaki, M.; Nakajima, K.; Takiguchi, S.; Mori, M.; Doki, Y. High infiltration of tumor-associated macrophages is associated with a poor response to chemotherapy and poor prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. J. Surg. Oncol. 2015, 111, 752–759. [Google Scholar] [CrossRef]
- Ma, P.F.; Gao, C.C.; Yi, J.; Zhao, J.L.; Liang, S.Q.; Zhao, Y.; Ye, Y.C.; Bai, J.; Zheng, Q.J.; Dou, K.F.; et al. Cytotherapy with M1-polarized macrophages ameliorates liver fibrosis by modulating immune microenvironment in mice. J. Hepatol. 2017, 67, 770–779. [Google Scholar] [CrossRef]
- Lin, T.Y.; Wang, P.W.; Huang, C.H.; Yang, P.M.; Pan, T.L. Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. Int. J. Mol. Sci. 2020, 21, 6077. [Google Scholar] [CrossRef]
- Pan, T.L.; Wang, P.W.; Huang, C.C.; Yeh, C.T.; Hu, T.H.; Yu, J.S. Network analysis and proteomic identification of vimentin as a key regulator associated with invasion and metastasis in human hepatocellular carcinoma cells. J. Proteomics 2012, 75, 4676–4692. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Wu, T.H.; Pan, T.L.; Chen, M.H.; Goto, S.; Chen, C.L. Integrated Proteome and Cytokine Profiles Reveal Ceruloplasmin Eliciting Liver Allograft Tolerance via Antioxidant Cascades. Front. Immunol. 2018, 9, 2216. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Y.; Wang, P.W.; Huang, C.H.; Chen, M.H.; Wu, Y.R.; Pan, T.L. Skin aging caused by intrinsic or extrinsic processes characterized with functional proteomics. Proteomics 2016, 16, 2718–2731. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.H.; Wang, P.W.; Lin, T.Y.; Yang, P.M.; Li, W.T.; Yeh, C.T.; Pan, T.L. Antioxidant properties of red raspberry extract alleviate hepatic fibrosis via inducing apoptosis and transdifferentiation of activated hepatic stellate cells. Biomed. Pharmacother. 2021, 144, 112284. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.-W.; Lin, T.-Y.; Yang, P.-M.; Yeh, C.-T.; Pan, T.-L. Hepatic Stellate Cell Modulates the Immune Microenvironment in the Progression of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2022, 23, 10777. https://doi.org/10.3390/ijms231810777
Wang P-W, Lin T-Y, Yang P-M, Yeh C-T, Pan T-L. Hepatic Stellate Cell Modulates the Immune Microenvironment in the Progression of Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2022; 23(18):10777. https://doi.org/10.3390/ijms231810777
Chicago/Turabian StyleWang, Pei-Wen, Tung-Yi Lin, Pei-Ming Yang, Chau-Ting Yeh, and Tai-Long Pan. 2022. "Hepatic Stellate Cell Modulates the Immune Microenvironment in the Progression of Hepatocellular Carcinoma" International Journal of Molecular Sciences 23, no. 18: 10777. https://doi.org/10.3390/ijms231810777
APA StyleWang, P.-W., Lin, T.-Y., Yang, P.-M., Yeh, C.-T., & Pan, T.-L. (2022). Hepatic Stellate Cell Modulates the Immune Microenvironment in the Progression of Hepatocellular Carcinoma. International Journal of Molecular Sciences, 23(18), 10777. https://doi.org/10.3390/ijms231810777








