Human IRP1 Translocates to the Nucleus in a Cell-Specific and Iron-Dependent Manner
Abstract
1. Introduction
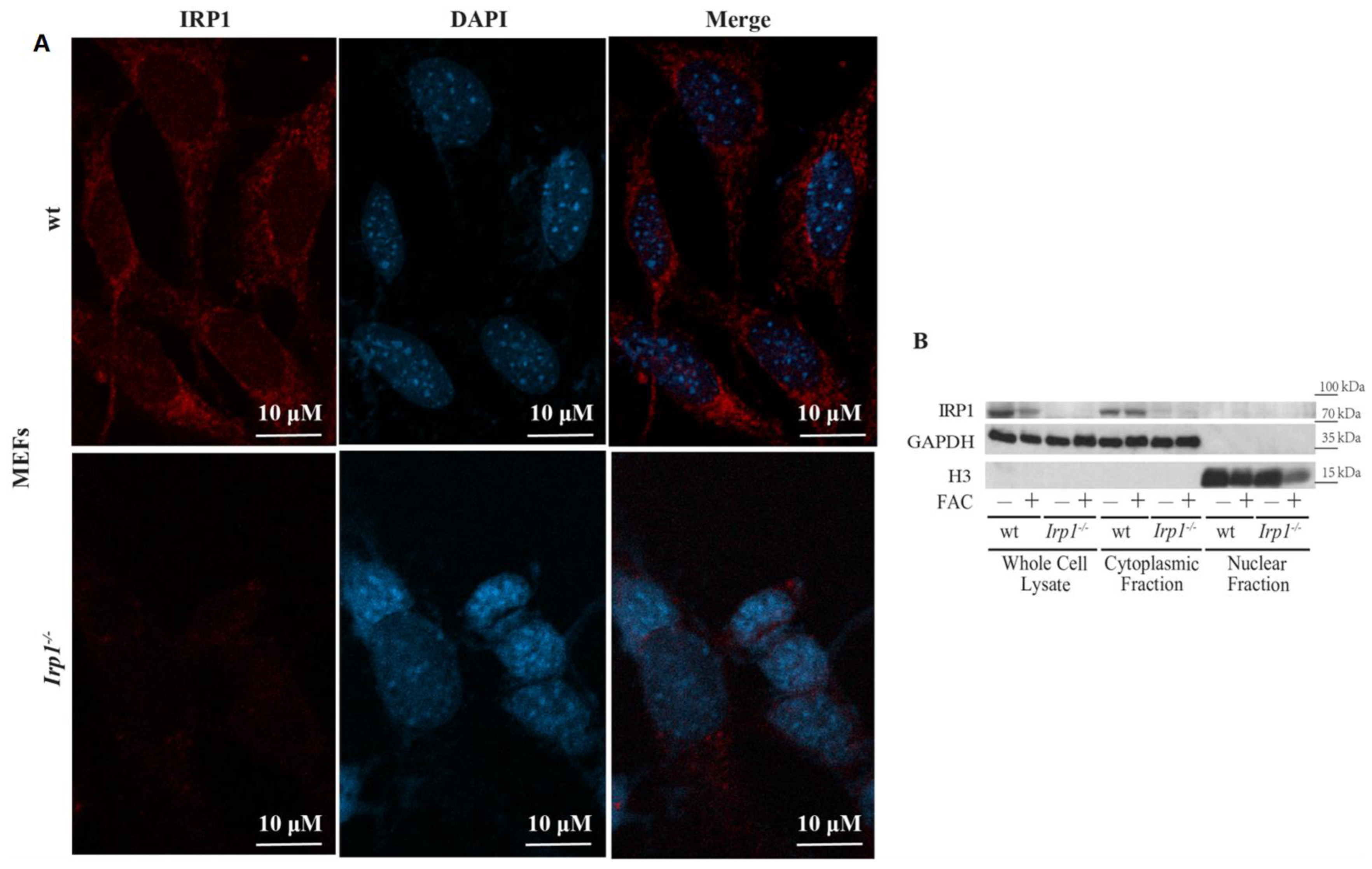
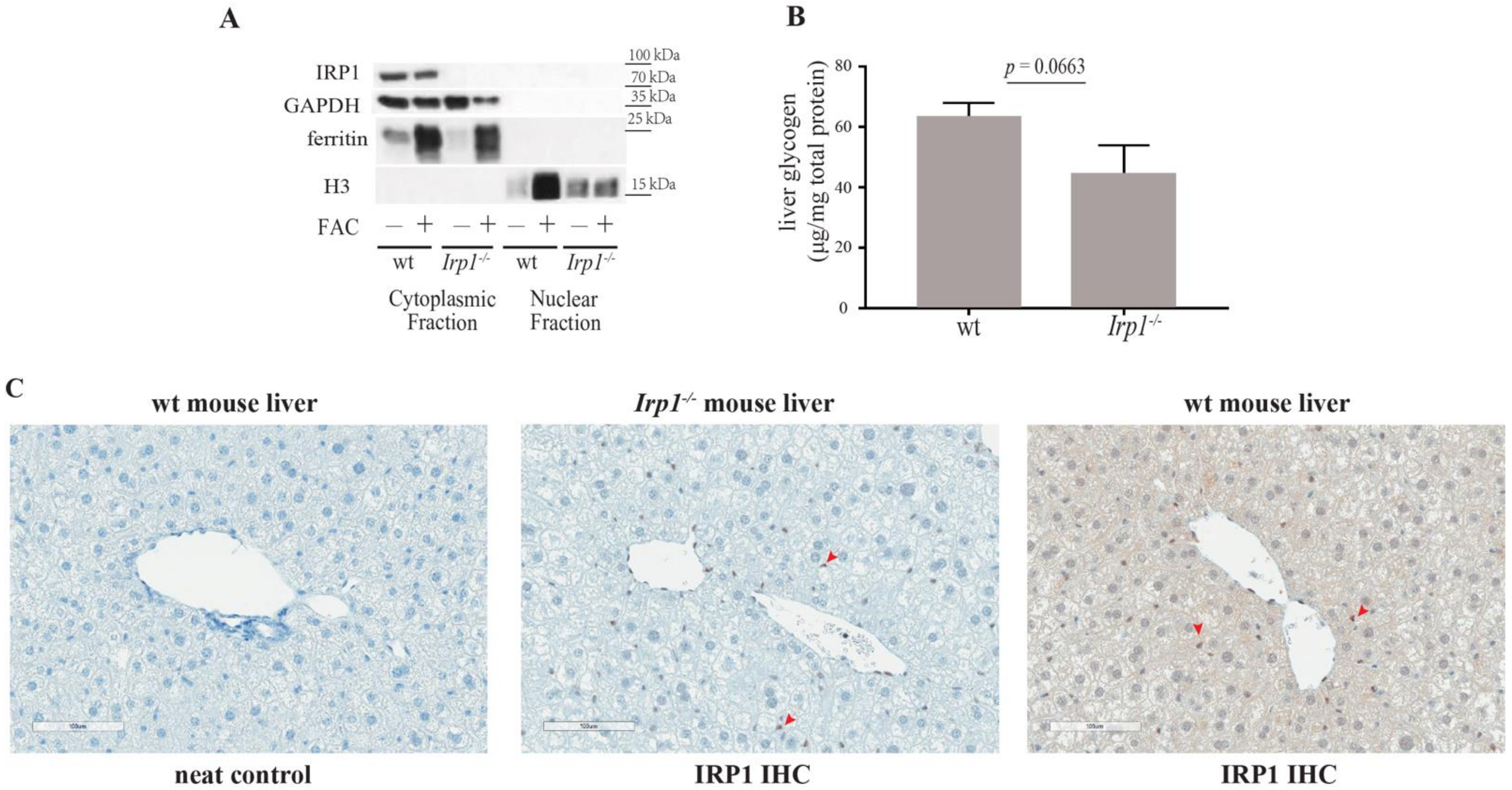
2. Results and Discussion
3. Materials and Methods
3.1. Mice
3.2. Cell Culture
3.3. Biochemical Fractionation and Western Blotting
3.4. Confocal Microscopy
3.5. Immunohistochemistry
3.6. Glycogen Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rouault, T.A. The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. Biometals 2019, 32, 343–353. [Google Scholar] [CrossRef]
- Kuhn, L.C. Iron regulatory proteins and their role in controlling iron metabolism. Metallomics 2015, 7, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.; Ou, Q.; Cox, P.; Lill, R.; King-Jones, K. Glycogen branching enzyme controls cellular iron homeostasis via Iron Regulatory Protein 1 and mitoNEET. Nat. Commun. 2019, 10, 5463. [Google Scholar] [CrossRef] [PubMed]
- Patton, S.M.; Pinero, D.J.; Surguladze, N.; Beard, J.; Connor, J.R. Subcellular localization of iron regulatory proteins to Golgi and ER membranes. J. Cell. Sci. 2005, 118, 4365–4373. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jensen, J.; Rustad, P.I.; Kolnes, A.J.; Lai, Y.C. The role of skeletal muscle glycogen breakdown for regulation of insulin sensitivity by exercise. Front. Physiol. 2011, 2, 112. [Google Scholar] [CrossRef] [PubMed]
- Adeva-Andany, M.M.; Gonzalez-Lucan, M.; Donapetry-Garcia, C.; Fernandez-Fernandez, C.; Ameneiros-Rodriguez, E. Glycogen metabolism in humans. BBA Clin. 2016, 5, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Gallardo, A.K.; Missirlis, F. Cellular iron sensing and regulation: Nuclear IRP1 extends a classic paradigm. Biochim. Biophys. Acta Mol. Cell. Res. 2020, 1867, 118705. [Google Scholar] [CrossRef] [PubMed]
- Thermann, R.; Neu-Yilik, G.; Deters, A.; Frede, U.; Wehr, K.; Hagemeier, C.; Hentze, M.W.; Kulozik, A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998, 17, 3484–3494. [Google Scholar] [CrossRef] [PubMed]
- Christova, T.; Templeton, D.M. Effect of hypoxia on the binding and subcellular distribution of iron regulatory proteins. Mol. Cell. Biochem. 2007, 301, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Meyron-Holtz, E.G.; Ghosh, M.C.; Iwai, K.; LaVaute, T.; Brazzolotto, X.; Berger, U.V.; Land, W.; Ollivierre-Wilson, H.; Grinberg, A.; Love, P.; et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004, 23, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Sutendra, G.; Kinnaird, A.; Dromparis, P.; Paulin, R.; Stenson, T.H.; Haromy, A.; Hashimoto, K.; Zhang, N.; Flaim, E.; Michelakis, E.D. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 2014, 158, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Y.R.; Liu, K.; Yin, Z.; Liu, R.; Xia, Y.; Tan, L.; Yang, P.; Lee, J.H.; Li, X.J.; et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 2017, 552, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Kafkia, E.; Andres-Pons, A.; Ganter, K.; Seiler, M.; Smith, T.S.; Andrejeva, A.; Jouhten, P.; Pereira, F.; Franco, C.; Kuroshchenkova, A.; et al. Operation of a TCA cycle subnetwork in the mammalian nucleus. Sci. Adv. 2022, 8, eabq5206. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, N.; Pantopoulos, K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2alpha mRNA translation. Blood 2013, 122, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.S.; Lei, Y.L. Generation and Culture of Mouse Embryonic Fibroblasts. Methods Mol. Biol. 2019, 1960, 85–91. [Google Scholar] [PubMed]
- Fillebeen, C.; Wilkinson, N.; Charlebois, E.; Katsarou, A.; Wagner, J.; Pantopoulos, K. Hepcidin-mediated hypoferremic response to acute inflammation requires a threshold of Bmp6/Hjv/Smad signaling. Blood 2018, 132, 1829–1841. [Google Scholar] [CrossRef] [PubMed]
- Fillebeen, C.; Chahine, D.; Caltagirone, A.; Segal, P.; Pantopoulos, K. A Phosphomimetic Mutation at Ser-138 Renders Iron Regulatory Protein 1 Sensitive to Iron-Dependent Degradation. Mol. Cell. Biol. 2003, 23, 6973–6981. [Google Scholar] [CrossRef] [PubMed]
- Katsarou, A.; Gkouvatsos, K.; Fillebeen, C.; Pantopoulos, K. Tissue-Specific Regulation of Ferroportin in Wild-Type and Hjv−/− Mice Following Dietary Iron Manipulations. Hepatol. Commun. 2021, 5, 2139–2150. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, W.; Fillebeen, C.; Pantopoulos, K. Human IRP1 Translocates to the Nucleus in a Cell-Specific and Iron-Dependent Manner. Int. J. Mol. Sci. 2022, 23, 10740. https://doi.org/10.3390/ijms231810740
Gu W, Fillebeen C, Pantopoulos K. Human IRP1 Translocates to the Nucleus in a Cell-Specific and Iron-Dependent Manner. International Journal of Molecular Sciences. 2022; 23(18):10740. https://doi.org/10.3390/ijms231810740
Chicago/Turabian StyleGu, Wen, Carine Fillebeen, and Kostas Pantopoulos. 2022. "Human IRP1 Translocates to the Nucleus in a Cell-Specific and Iron-Dependent Manner" International Journal of Molecular Sciences 23, no. 18: 10740. https://doi.org/10.3390/ijms231810740
APA StyleGu, W., Fillebeen, C., & Pantopoulos, K. (2022). Human IRP1 Translocates to the Nucleus in a Cell-Specific and Iron-Dependent Manner. International Journal of Molecular Sciences, 23(18), 10740. https://doi.org/10.3390/ijms231810740







