Dynamic Change in Starch Biosynthetic Enzymes Complexes during Grain-Filling Stages in BEIIb Active and Deficient Rice
Abstract
:1. Introduction
2. Results
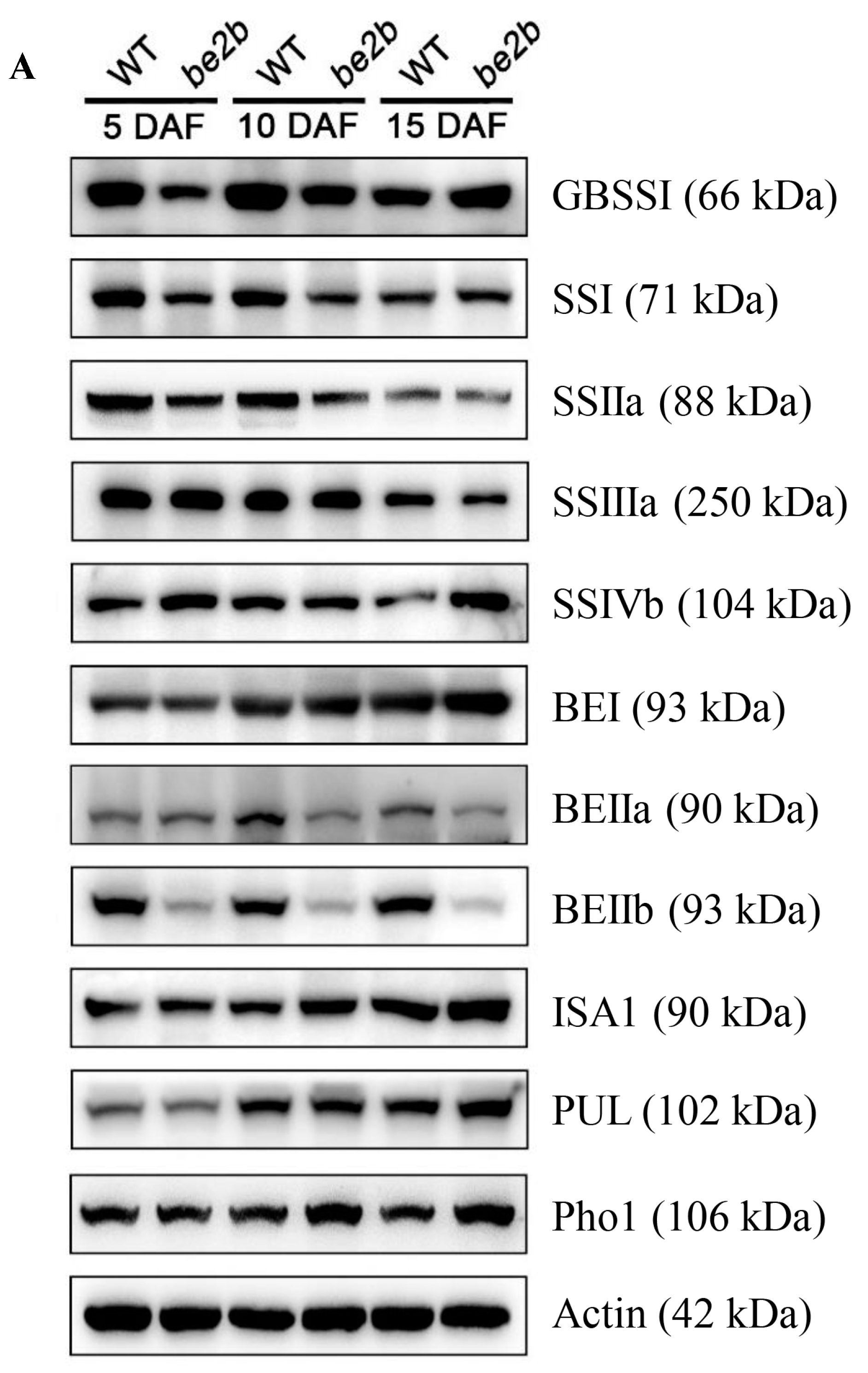
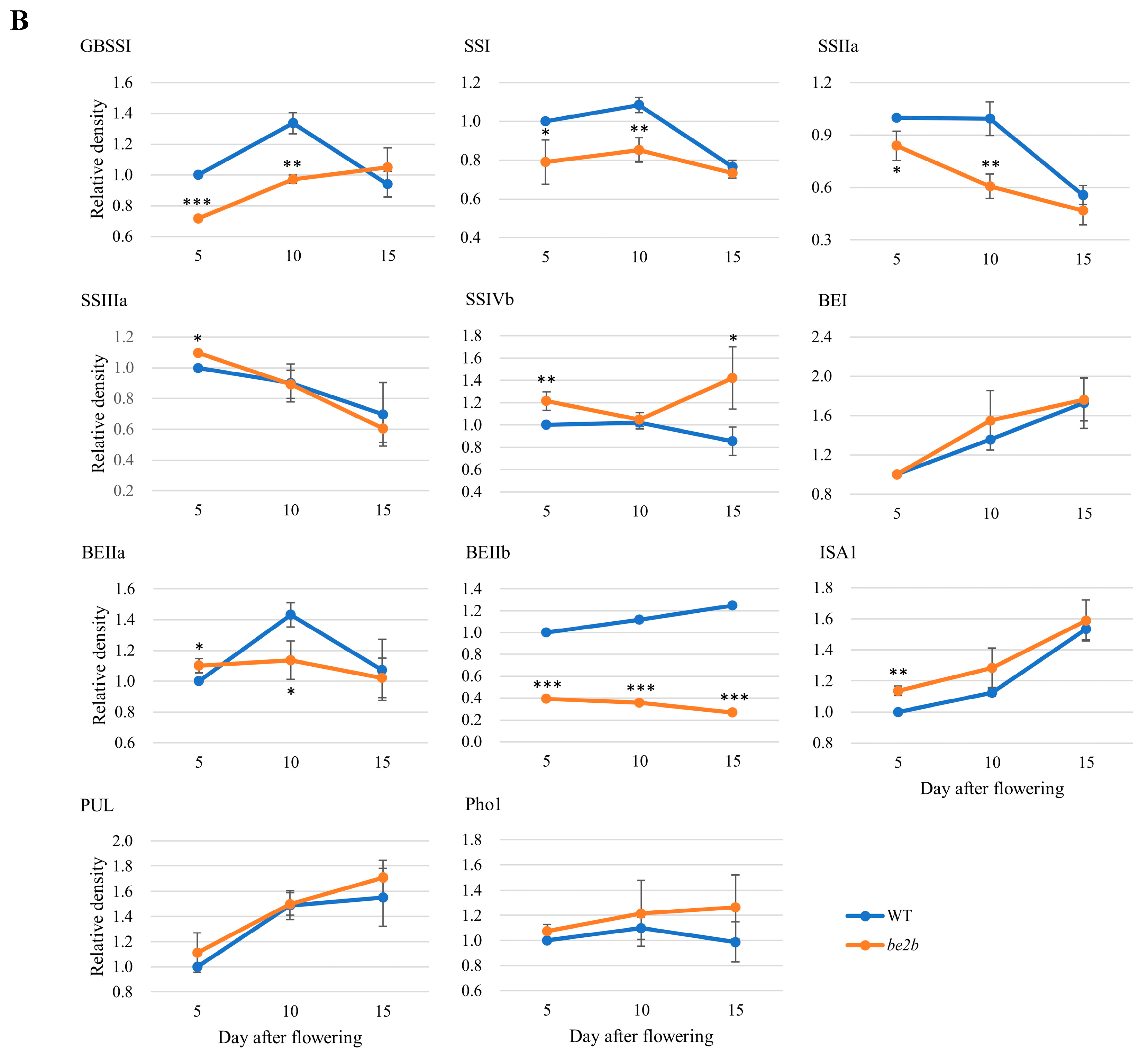
2.1. Accumulation of Starch Biosynthetic Related Enzymes (SSREs)
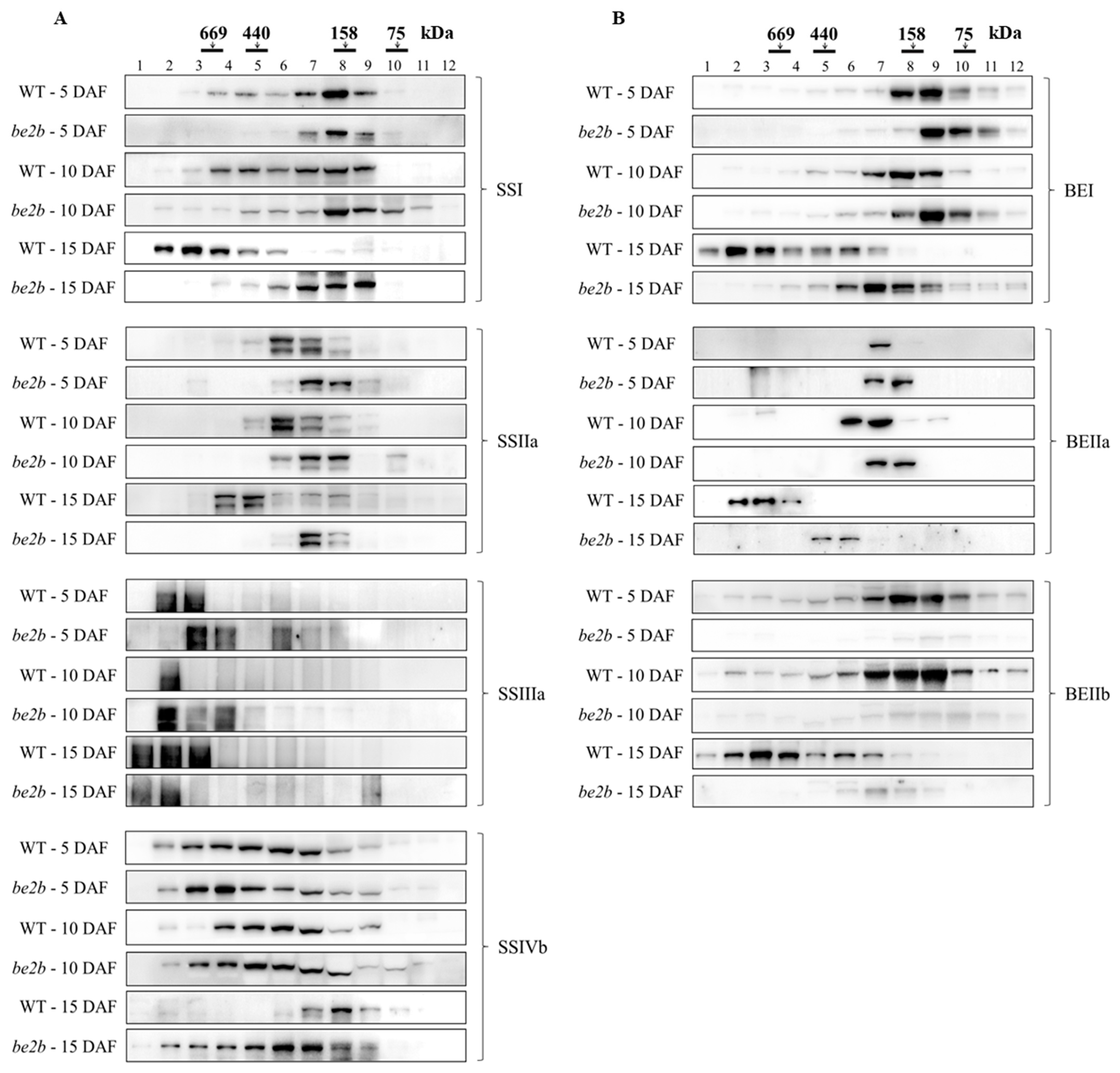
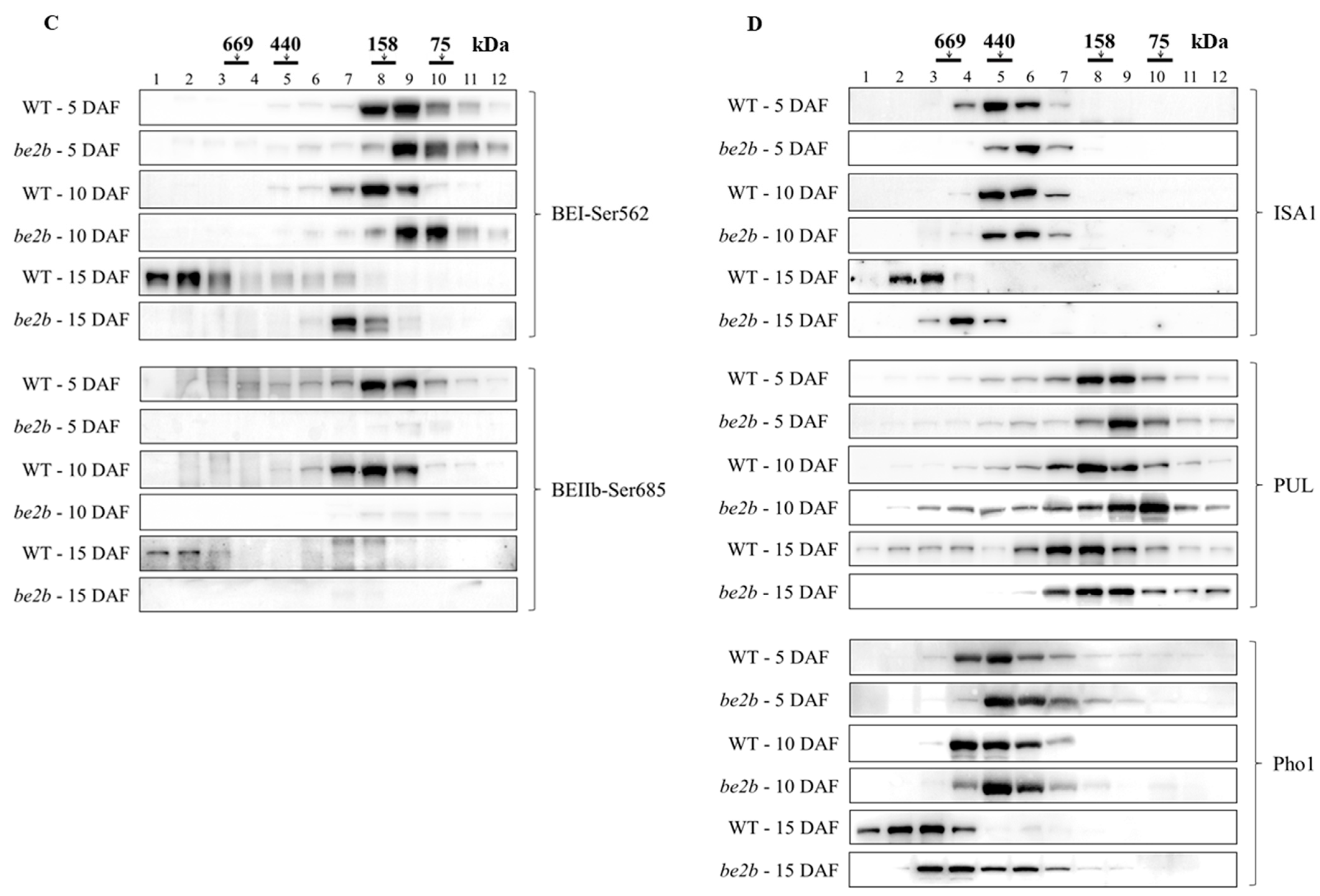
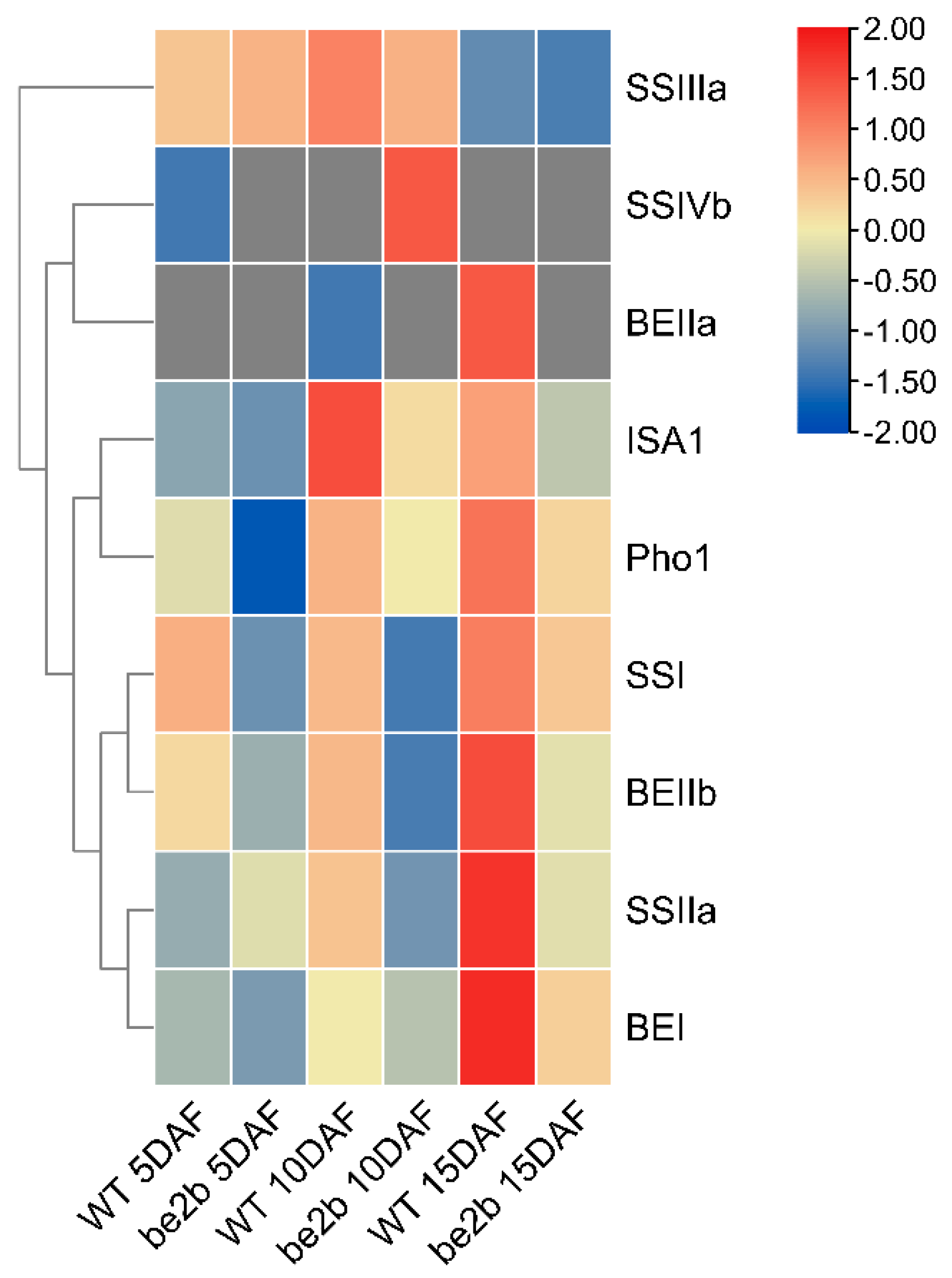
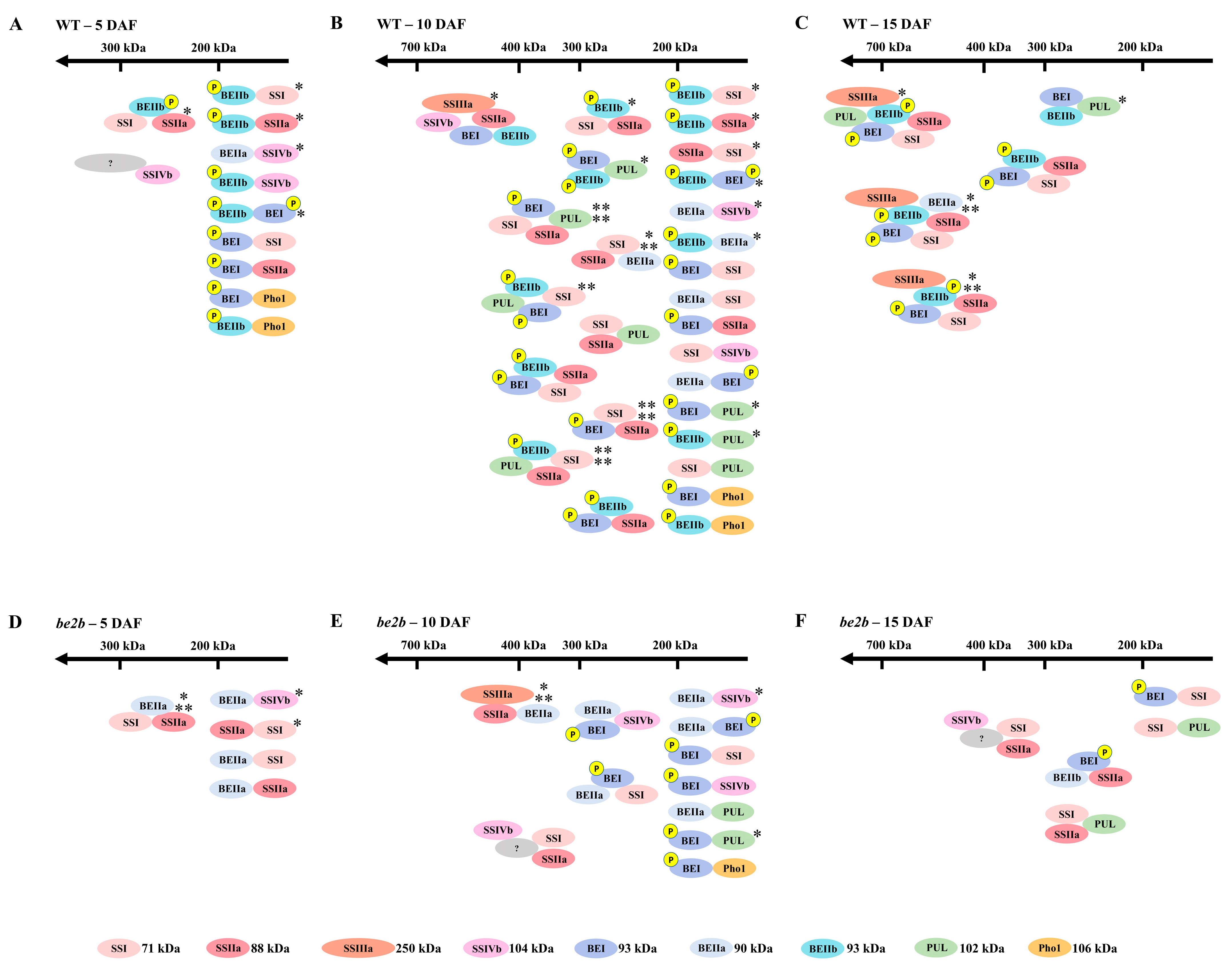
2.2. Molecular Weight Distribution of SSREs
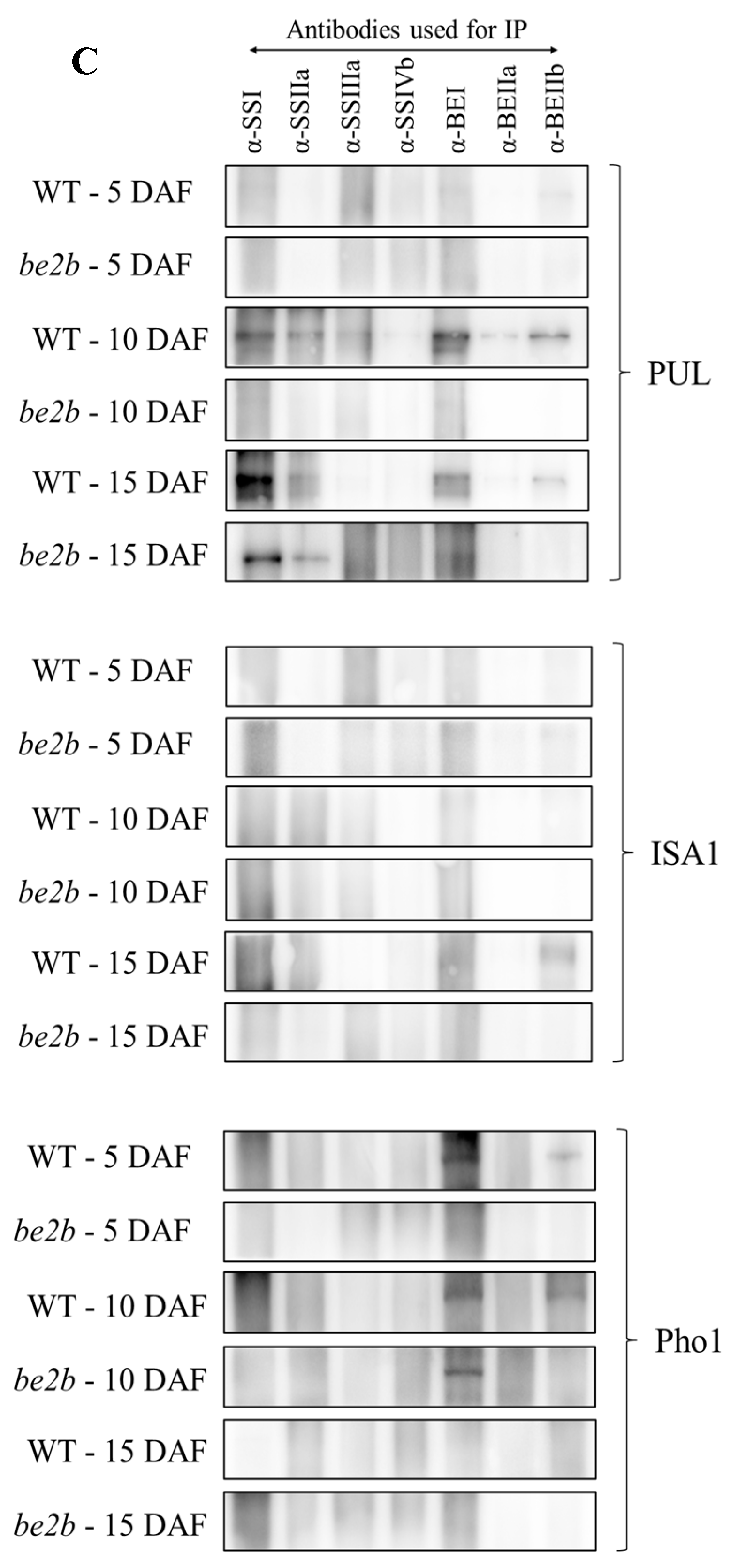
2.3. Co-Immunoprecipitation
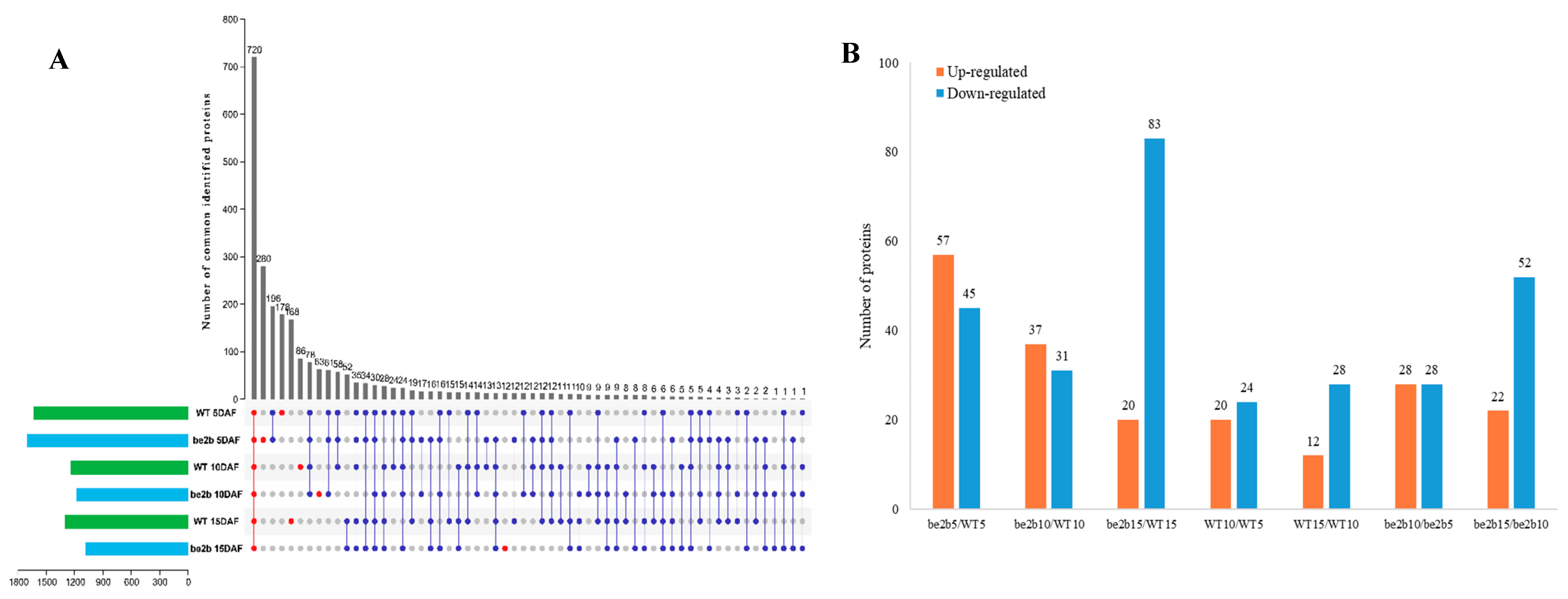
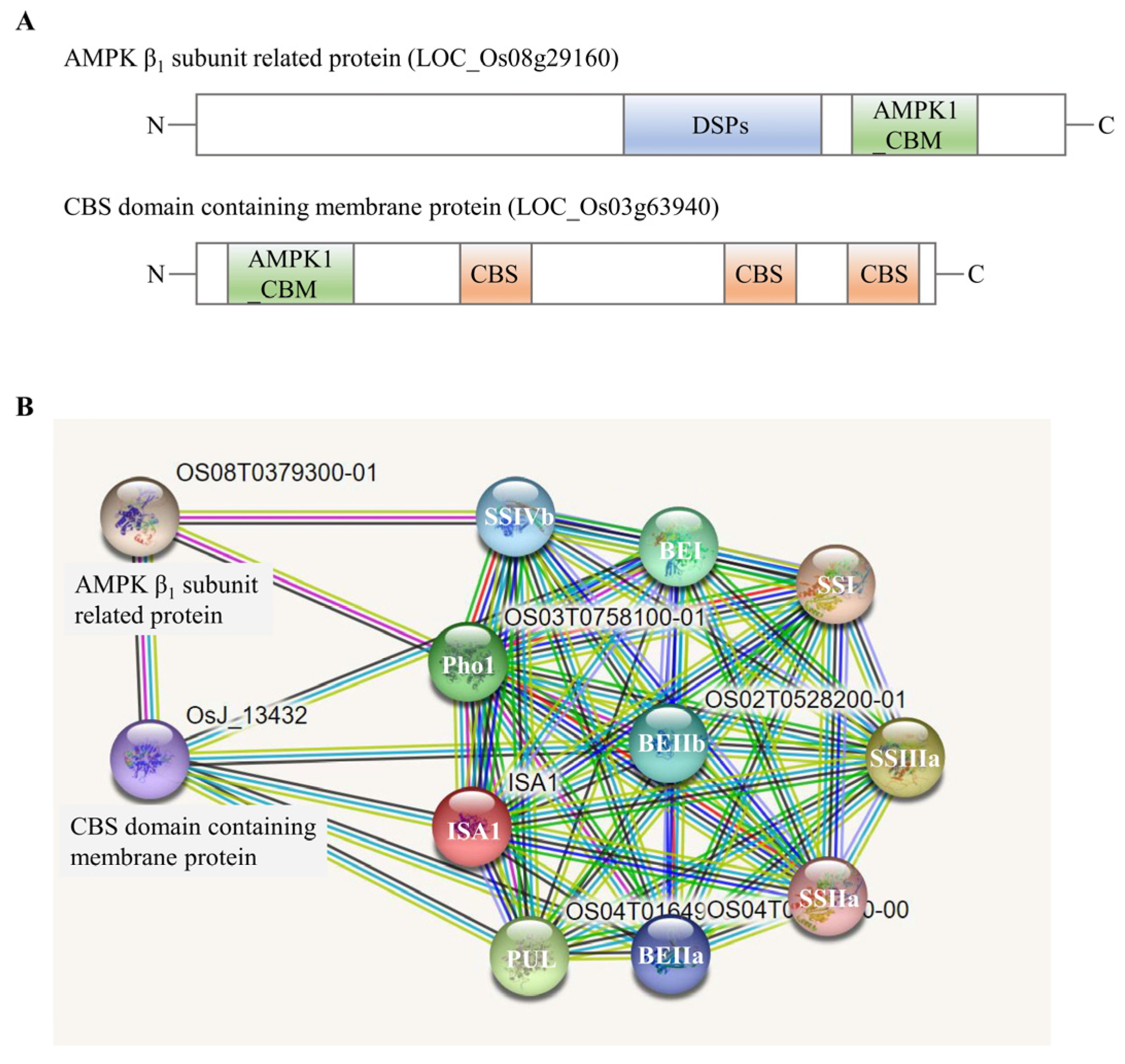
2.4. Identification of Large Protein Complex Components
3. Discussion
3.1. Components of Multi-Protein Complexes Vary at Different Seed Development Stages
3.2. Effects of BEIIb Deficiency on Multi-Enzyme Complex Formation in the Developing Rice Seed
3.3. The Essential Role of Protein Phosphorylation and Protein Kinases in Multi-Protein Complexes Formation
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Protein Extraction
4.3. Gel Permeation Chromatography
4.4. Co-Immunoprecipitation
4.5. Western Blotting
4.6. Protein Preparation, Digestion, LC-MS/MS, and Data Analysis
4.7. Statistical and Bioinformatic Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohdan, T.; Francisco, P.B., Jr.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef]
- Ishimaru, T.; Matsuda, T.; Ohsugi, R.; Yamagishi, T. Morphological development of rice caryopses located at the different positions in a panicle from early to middle stage of grain filling. Funct. Plant Biol. 2003, 30, 1139–1149. [Google Scholar] [CrossRef]
- Xu, S.B.; Li, T.; Deng, Z.Y.; Chong, K.; Xue, Y.; Wang, T. Dynamic proteomic analysis reveals a switch between central carbon metabolism and alcoholic fermentation in rice filling grains. Plant Physiol. 2008, 148, 908–925. [Google Scholar] [CrossRef]
- Rösti, S.; Fahy, B.; Denyer, K. A mutant of rice lacking the leaf large subunit of ADP-glucose pyrophosphorylase has drastically reduced leaf starch content but grows normally. Funct. Plant Biol. 2007, 34, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Eom, J.S.; Hwang, S.K.; Shin, D.; An, G.; Okita, T.W.; Jeon, J.S. Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 2016, 67, 5557–5569. [Google Scholar] [CrossRef] [PubMed]
- Cook, F.R.; Fahy, B.; Trafford, K. A rice mutant lacking a large subunit of ADP-glucose pyrophosphorylase has drastically reduced starch content in the culm but normal plant morphology and yield. Funct. Plant Biol. 2012, 39, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Okamura, M.; Hirose, T.; Hashida, Y.; Yamagishi, T.; Ohsugi, R.; Aoki, N. Starch reduction in rice stems due to a lack of OsAGPL1 or OsAPL3 decreases grain yield under low irradiance during ripening and modifies plant architecture. Funct. Plant Biol. 2013, 40, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Irshad, A.; Guo, H.; Rehman, S.U.; Wang, X.; Wang, C.; Raza, A.; Zhou, C.; Li, Y.; Liu, L. Soluble starch synthase enzymes in cereals: An updated review. Agronomy 2021, 11, 1983. [Google Scholar] [CrossRef]
- Pandey, M.K.; Rani, N.S.; Madhav, M.S.; Sundaram, R.M.; Varaprasad, G.S.; Sivaranjani, A.K.; Bohra, A.; Kumar, G.R.; Kumar, A. Different isoforms of starch-synthesizing enzymes controlling amylose and amylopectin content in rice (Oryza sativa L.). Biotechnol. Adv. 2012, 30, 1697–1706. [Google Scholar] [CrossRef]
- Nakamura, Y.; Utsumi, Y.; Sawada, T.; Aihara, S.; Utsumi, C.; Yoshida, M.; Kitamura, S. Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol. 2010, 51, 776–794. [Google Scholar] [CrossRef]
- Satoh, H.; Nishi, A.; Yamashita, K.; Takemoto, Y.; Tanaka, Y.; Hosaka, Y.; Sakurai, A.; Fujita, N.; Nakamura, Y. Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 2003, 133, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Nishi, A.; Nakamura, Y.; Tanaka, N.; Satoh, H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 2001, 127, 459–472. [Google Scholar] [CrossRef]
- Utsumi, Y.; Utsumi, C.; Sawada, T.; Fujita, N.; Nakamura, Y. Functional diversity of isoamylase oligomers: The ISA1 homo-oligomer is essential for amylopectin biosynthesis in rice endosperm. Plant Physiol. 2011, 156, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.-S.; Umemoto, T.; Kawagoe, Y. Rice debranching enzyme isoamylase3 facilitates starch metabolism and affects plastid morphogenesis. Plant Cell Physiol. 2011, 52, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Kubo, A.; Shimamune, T.; Matsuda, T.; Harada, K.; Satoh, H. Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 1997, 12, 143–153. [Google Scholar] [CrossRef]
- Satoh, H.; Shibahara, K.; Tokunaga, T.; Nishi, A.; Tasaki, M.; Hwang, S.K.; Okita, T.W.; Kaneko, N.; Fujita, N.; Yoshida, M.; et al. Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 2008, 20, 1833–1849. [Google Scholar] [CrossRef]
- Hwang, S.-K.; Nishi, A.; Satoh, H.; Okita, T. Rice endosperm-specific plastidial α-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Arch. Biochem. Biophys. 2010, 495, 82–92. [Google Scholar] [CrossRef]
- Hwang, S.-K.; Koper, K.; Satoh, H.; Okita, T.W. Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J. Biol. Chem. 2016, 291, 19994–20007. [Google Scholar] [CrossRef]
- Zhang, G.; Cheng, Z.; Zhang, X.; Guo, X.; Su, N.; Jiang, L.; Mao, L.; Wan, J. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L.) uncovers interactive effects on the physicochemical properties of starch. Genome 2011, 54, 448–459. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Wait, R.; Lu, Z.; Akkasaeng, R.; Bowsher, C.G.; Esposito, S.; Kosar-Hashemi, B.; Morell, M.K.; Emes, M.J. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 2004, 16, 694–708. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Beisel, K.G.; Cameron, S.; Makhmoudova, A.; Liu, F.; Bresolin, N.S.; Wait, R.; Morell, M.K.; Emes, M.J. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 2008, 146, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Hennen-Bierwagen, T.A.; Liu, F.; Marsh, R.S.; Kim, S.; Gan, Q.; Tetlow, I.J.; Emes, M.J.; James, M.G.; Myers, A.M. Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 2008, 146, 1892–1908. [Google Scholar] [CrossRef] [PubMed]
- Hennen-Bierwagen, T.A.; Lin, Q.; Grimaud, F.; Planchot, V.; Keeling, P.L.; James, M.G.; Myers, A.M. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: A model for regulation of carbon allocation in maize amyloplasts. Plant Physiol. 2009, 149, 1541–1559. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Makhmoudova, A.; Lee, E.A.; Wait, R.; Emes, M.J.; Tetlow, I.J. The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 2009, 60, 4423–4440. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ahmed, Z.; Lee, E.A.; Donner, E.; Liu, Q.; Ahmed, R.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J. Exp. Bot. 2012, 63, 1167–1183. [Google Scholar] [CrossRef]
- Makhmoudova, A.; Williams, D.; Brewer, D.; Massey, S.; Patterson, J.; Silva, A.; Vassall, K.A.; Liu, F.; Subedi, S.; Harauz, G.; et al. Identification of multiple phosphorylation sites on maize endosperm starch branching enzyme IIb, a key enzyme in amylopectin biosynthesis. J. Biol. Chem. 2014, 289, 9233–9246. [Google Scholar] [CrossRef]
- Liu, F.; Romanova, N.; Lee, E.A.; Ahmed, R.; Evans, M.; Gilbert, E.P.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules. Biochem. J. 2012, 448, 373–387. [Google Scholar] [CrossRef]
- Ahmed, Z.; Tetlow, I.J.; Ahmed, R.; Morell, M.K.; Emes, M.J. Protein–protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 2015, 233, 95–106. [Google Scholar] [CrossRef]
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J.; Emes, M.J.; Nakamura, Y.; Fujita, N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482. [Google Scholar] [CrossRef]
- Crofts, N.; Iizuka, Y.; Abe, N.; Miura, S.; Kikuchi, K.; Matsushima, R.; Fujita, N. Rice mutants lacking starch synthase I or branching enzyme IIb activity altered starch biosynthetic protein complexes. Front. Plant Sci. 2018, 9, 1817. [Google Scholar] [CrossRef]
- Miura, S.; Crofts, N.; Saito, Y.; Hosaka, Y.; Oitome, N.F.; Watanabe, T.; Kumamaru, T.; Fujita, N. Starch synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than japonica rice cultivars. Front. Plant Sci. 2018, 9, 645. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Crofts, N.; Oitome, N.F.; Fujita, N. Analyses of starch biosynthetic protein complexes and starch properties from developing mutant rice seeds with minimal starch synthase activities. BMC Plant Biol. 2018, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Nakamura, Y.; Fujita, N. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci. 2017, 262, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. Towards a Better Understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model Tissue. Plant Cell Physiol. 2002, 43, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, H.; Nakamura, Y. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol. 1992, 33, 985–991. [Google Scholar]
- Tanaka, N.; Fujita, N.; Nishi, A.; Satoh, H.; Hosaka, Y.; Ugaki, M.; Kawasaki, S.; Nakamura, Y. The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2004, 2, 507–516. [Google Scholar] [CrossRef]
- Kubo, A.; Akdogan, G.; Nakaya, M.; Shojo, A.; Suzuki, S.; Satoh, H.; Kitamura, S. Structure, physical, and digestive properties of starch from wx ae double-mutant rice. J. Agric. Food Chem. 2010, 58, 4463–4469. [Google Scholar] [CrossRef]
- Tsuiki, K.; Fujisawa, H.; Itoh, A.; Sato, M.; Fujita, N. Alterations of starch structure lead to increased resistant starch of steamed rice: Identification of high resistant starch rice lines. J. Cereal Sci. 2016, 68, 88–92. [Google Scholar] [CrossRef]
- Sawada, T.; Itoh, M.; Nakamura, Y. Contributions of three starch branching enzyme isozymes to the fine structure of amylopectin in rice endosperm. Front. Plant Sci. 2018, 9, 1536. [Google Scholar] [CrossRef]
- Nakamura, Y.; Francisco, P.B.; Hosaka, Y.; Sato, A.; Sawada, T.; Kubo, A.; Fujita, N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 2005, 58, 213–227. [Google Scholar] [CrossRef]
- Ying, Y.; Zhang, Z.; Tappiban, P.; Xu, F.; Deng, G.; Dai, G.; Bao, J. Starch fine structure and functional properties during seed development in BEIIb active and deficient rice. Carbohydr. Polym. 2022, 292, 119640. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Zhou, X.; Chen, Y.; Bao, J. Comparative phosphoproteomic analysis of the developing seeds in two indica rice (Oryza sativa L.) cultivars with different starch quality. J. Agric. Food Chem. 2018, 66, 3030–3037. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bao, J. Underlying Mechanisms of zymographic diversity in starch synthase I and pullulanase in rice-developing endosperm. J. Agric. Food Chem. 2016, 64, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pang, Y.; Bao, J. Expression profiles and protein complexes of starch biosynthetic enzymes from white-core and waxy mutants induced from high amylose indica rice. Rice Sci. 2020, 27, 152–161. [Google Scholar]
- Qu, J.; Xu, S.; Tian, X.; Li, T.; Wang, L.; Zhong, Y.; Xue, J.; Guo, D. Comparative transcriptomics reveals the difference in early endosperm development between maize with different amylose contents. PeerJ 2019, 7, e7528. [Google Scholar] [CrossRef]
- Waduge, R.N.; Xu, S.; Seetharaman, K. Iodine absorption properties and its effect on the crystallinity of developing wheat starch granules. Carbohydr. Polym. 2010, 82, 786–794. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, J.; Chen, Y.; Zhou, W.; Xu, B.; Wang, Y.; Chen, J. Physicochemical properties and development of wheat large and small starch granules during endosperm development. Acta Physiol. Plant. 2010, 32, 905–916. [Google Scholar] [CrossRef]
- Källman, A.; Bertoft, E.; Koch, K.; Sun, C.; Åman, P.; Andersson, R. Starch structure in developing barley endosperm. Int. J. Biol. Macromol. 2015, 81, 730–735. [Google Scholar] [CrossRef]
- Li, L.; Blanco, M.; Jane, J.-L. Physicochemical properties of endosperm and pericarp starches during maize development. Carbohydr. Polym. 2007, 67, 630–639. [Google Scholar] [CrossRef]
- Pang, Y.; Hu, Y.; Bao, J. Comparative phosphoproteomic analysis reveals the response of starch metabolism to high-temperature stress in rice endosperm. Int. J. Mol. Sci. 2021, 22, 10546. [Google Scholar] [CrossRef]
- Cuesta-Seijo, J.A.; Nielsen, M.M.; Ruzanski, C.; Krucewicz, K.; Beeren, S.R.; Rydhal, M.G.; Yoshimura, Y.; Striebeck, A.; Motawia, M.S.; Willats, W.G.; et al. In vitro biochemical characterization of all barley endosperm starch synthases. Front. Plant Sci. 2015, 6, 1265. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, Y.; Li, X.; Yan, Z.; Xie, Y.; Xiong, H.; Zhao, L.; Gu, J.; Zhao, S.; Liu, L. Novel mutant alleles of the starch synthesis gene TaSSIVb-D result in the reduction of starch granule number per chloroplast in wheat. BMC Genom. 2017, 18, 358. [Google Scholar] [CrossRef] [PubMed]
- Malinova, I.; Qasim, H.M.; Brust, H.; Fettke, J. Parameters of starch granule genesis in chloroplasts of Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Seung, D.; Smith, A.M. Starch granule initiation and morphogenesis-progress in Arabidopsis and cereals. J. Exp. Bot. 2019, 70, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Toyosawa, Y.; Kawagoe, Y.; Matsushima, R.; Crofts, N.; Ogawa, M.; Fukuda, M.; Kumamaru, T.; Okazaki, Y.; Kusano, M.; Saito, K.; et al. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 2016, 170, 1255–1270. [Google Scholar] [CrossRef]
- He, Y.; Cheng, J.; He, Y.; Yang, B.; Cheng, Y.; Yang, C.; Zhang, H.; Wang, Z. Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice. Plant Biotechnol. J. 2019, 17, 322–337. [Google Scholar] [CrossRef]
- Tappiban, P.; Ying, Y.; Xu, F.; Bao, J. Proteomics and Post-Translational Modifications of Starch Biosynthesis-Related Proteins in Developing Seeds of Rice. Int. J. Mol. Sci. 2021, 22, 5901. [Google Scholar] [CrossRef]
- Itoh, Y.; Crofts, N.; Abe, M.; Hosaka, Y.; Fujita, N. Characterization of the endosperm starch and the pleiotropic effects of biosynthetic enzymes on their properties in novel mutant rice lines with high resistant starch and amylose content. Plant Sci. 2017, 258, 52–60. [Google Scholar] [CrossRef]
- Seung, D.; Schreier, T.B.; Bürgy, L.; Eicke, S.; Zeeman, S.C. Two plastidial coiled-coil proteins are essential for normal starch granule initiation in Arabidopsis. Plant Cell 2018, 30, 1523–1542. [Google Scholar] [CrossRef]
- Vandromme, C.; Spriet, C.; Dauvillée, D.; Courseaux, A.; Putaux, J.L.; Wychowski, A.; Krzewinski, F.; Facon, M.; D’Hulst, C.; Wattebled, F. PII1: A protein involved in starch initiation that determines granule number and size in Arabidopsis chloroplast. New Phytol. 2019, 221, 356–370. [Google Scholar] [CrossRef]
- Zhang, L.; Li, N.; Zhang, J.; Zhao, L.; Qiu, J.; Wei, C. The CBM48 domain-containing protein FLO6 regulates starch synthesis by interacting with SSIVb and GBSS in rice. Plant Mol. Biol. 2022, 108, 343–361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tappiban, P.; Ying, Y.; Hu, Y.; Bao, J. Functional interactions between enzymes involved in amylose and amylopectin biosynthesis in rice based on mathematical models. Biomacromolecules 2022, 23, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Chastain, C.J.; Failing, C.J.; Manandhar, L.; Zimmerman, M.A.; Lakner, M.; Nguyen, T. Functional evolution of C(4) pyruvate, orthophosphate dikinase. J. Exp. Bot. 2011, 62, 3083–3091. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef]
- Subasinghe, R.M.; Liu, F.; Polack, U.C.; Lee, E.A.; Emes, M.J.; Tetlow, I.J. Multimeric states of starch phosphorylase determine protein-protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 2014, 83, 168–179. [Google Scholar] [CrossRef]
- Prabhakar, S.; Asuthkar, S.; Tsung, A.J.; Velpula, K.K. Dual-specificity Protein Phosphatases. In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer: New York, NY, USA, 2017; pp. 1–9. [Google Scholar]
- Zhong, Y.; Blennow, A.; Kofoed-Enevoldsen, O.; Jiang, D.; Hebelstrup, K.H. Protein Targeting to Starch 1 is essential for starchy endosperm development in barley. J. Exp. Bot. 2019, 70, 485–496. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Ross, F.A.; Thomas, E.J.; Hardie, D.G. Differential regulation by AMP and ADP of AMPK complexes containing different γ subunit isoforms. Biochem. J. 2016, 473, 189–199. [Google Scholar] [CrossRef]
- Xu, F.; Jia, M.; Li, X.; Tang, Y.; Jiang, K.; Bao, J.; Gu, Y. Exportin-4 coordinates nuclear shuttling of TOPLESS family transcription corepressors to regulate plant immunity. Plant Cell 2021, 33, 697–713. [Google Scholar] [CrossRef]
- Crofts, N.; Abe, K.; Aihara, S.; Itoh, R.; Nakamura, Y.; Itoh, K.; Fujita, N. Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. Plant Sci. 2012, 193–194, 62–69. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and Characterization of Starch Synthase I Using Mutants in Rice. Plant Physiol. 2006, 140, 1070–1084. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Takeichi, T.; Kawaguchi, K.; Yamanouchi, H. Purification of two forms of starch branching enzyme (Q-enzyme) from developing rice endosperm. Physiol. Plant. 1992, 84, 329–335. [Google Scholar] [CrossRef]
- Nakamura, Y.; Umemoto, T.; Ogata, N.; Kuboki, Y.; Yano, M.; Sasaki, T. Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: Purification, cDNA and chromosomal localization of the gene. Planta 1996, 199, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]

| Sample | Reciprocal | One Sided a | |||
|---|---|---|---|---|---|
| Strong Signal b | Weak Signal b | Strong Signal b | Weak Signal b | ||
| 5 DAF | WT | BEIIb–SSI BEIIb–SSIIa BEIIa–SSIVb BEIIb–SSIVb BEIIb–BEI | BEI–SSI BEI–SSIIa BEI–Pho1 | SSIIa–SSI BEIIa–SSI BEIIa–SSIIa SSIVb–SSIIIa SSI–SSIVb BEI–SSIVb BEIIa–BEI BEIIa–BEIIb BEIIb–Pho1 | |
| be2b | BEIIa–SSIVb | SSIIa–SSI BEIIa–SSI BEIIa–SSIIa | BEI–SSIIa BEI–SSIIIa BEIIa–SSIIIa | ||
| 10 DAF | WT | BEIIb–SSI BEIIb–SSIIa SSIVb–SSIIIa BEIIb–BEI | SSIIa–SSI BEIIb–SSIIIa BEIIa–SSIVb BEIIb–BEIIa | BEI–SSI BEIIa–SSI BEI–SSIIa SSIIa–SSIIIa SSI–SSIVb BEIIa–BEI SSI–PUL BEI–PUL BEIIb–PUL BEI–Pho1 | BEIIa–SSIIa SSI–SSIIIa BEIIa–SSIIIa BEI–SSIVb BEIIb–SSIVb SSIIa–PUL SSIIIa –PUL BEIIa–PUL BEIIb–Pho1 |
| be2b | SSIVb–SSIIIa | BEIIa–SSIVb BEIIa–BEI | BEI–SSI BEI–SSIVb BEI–Pho1 | BEIIa–SSIIa SSI–SSIIIa SSIIa–SSIIIa | |
| 15 DAF | WT | SSIIa–SSI BEI–SSI BEIIb–SSI BEIIb–SSIIa | BEI–SSIIa SSIVb–SSIIIa BEIIb–BEI | BEIIa–SSI BEIIa–SSIIa BEIIb–SSIIIa BEIIb–BEIIa SSI–PUL BEI–PUL | BEI–SSIIIa BEIIa–BEI SSIIa–PUL BEIIb–PUL BEIIb–ISA1 |
| be2b | BEI–SSI | SSIVb–SSIIIa SSI–PUL | BEIIa–SSIIa SSI–SSIIIa BEI–SSIIIa SSIIa–BEI BEIIa–BEI SSIIa–PUL | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, Y.; Xu, F.; Zhang, Z.; Tappiban, P.; Bao, J. Dynamic Change in Starch Biosynthetic Enzymes Complexes during Grain-Filling Stages in BEIIb Active and Deficient Rice. Int. J. Mol. Sci. 2022, 23, 10714. https://doi.org/10.3390/ijms231810714
Ying Y, Xu F, Zhang Z, Tappiban P, Bao J. Dynamic Change in Starch Biosynthetic Enzymes Complexes during Grain-Filling Stages in BEIIb Active and Deficient Rice. International Journal of Molecular Sciences. 2022; 23(18):10714. https://doi.org/10.3390/ijms231810714
Chicago/Turabian StyleYing, Yining, Feifei Xu, Zhongwei Zhang, Piengtawan Tappiban, and Jinsong Bao. 2022. "Dynamic Change in Starch Biosynthetic Enzymes Complexes during Grain-Filling Stages in BEIIb Active and Deficient Rice" International Journal of Molecular Sciences 23, no. 18: 10714. https://doi.org/10.3390/ijms231810714
APA StyleYing, Y., Xu, F., Zhang, Z., Tappiban, P., & Bao, J. (2022). Dynamic Change in Starch Biosynthetic Enzymes Complexes during Grain-Filling Stages in BEIIb Active and Deficient Rice. International Journal of Molecular Sciences, 23(18), 10714. https://doi.org/10.3390/ijms231810714






