Activation of Alternative Bilirubin Clearance Pathways Partially Reduces Hyperbilirubinemia in a Mouse Model Lacking Functional Ugt1a1 Activity
Abstract
1. Introduction
2. Results
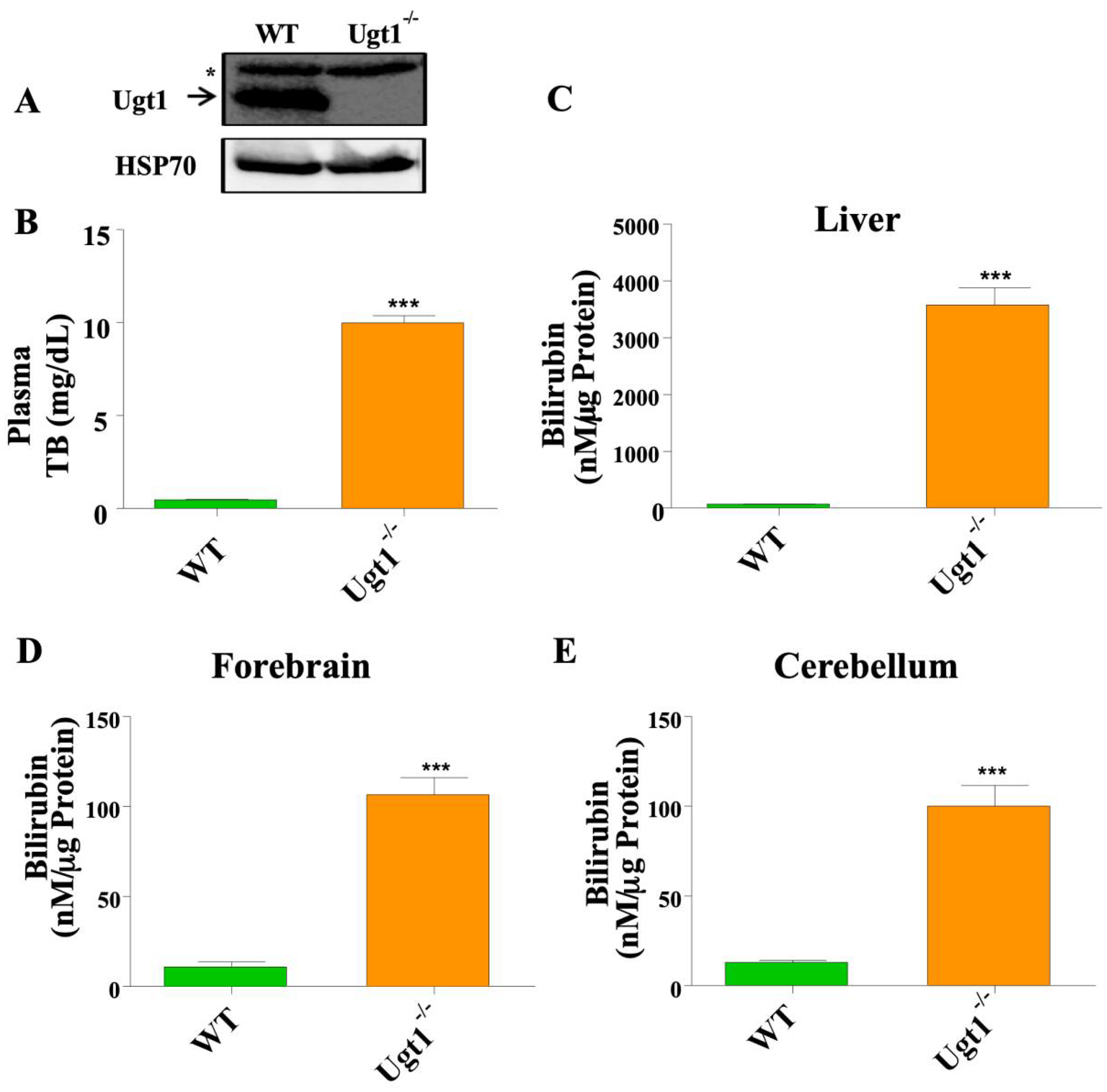
2.1. Adult Ugt1 KO Animals Have High Plasma and Brain Bilirubin Level
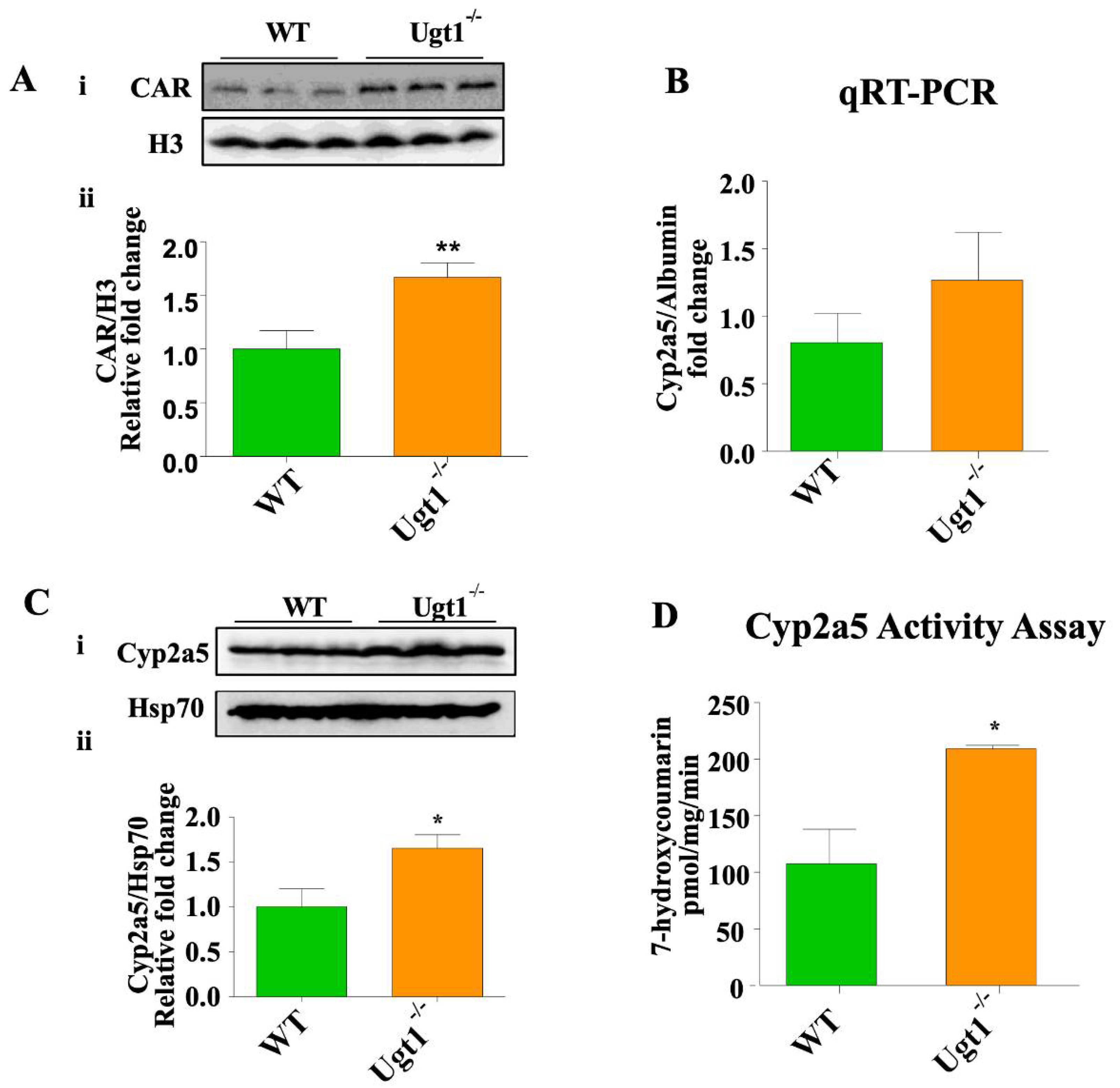
2.2. CAR and Cyp2a5 Are Up-Regulated in Untreated Ugt1 Mutant Animals Compared to Wild Type Animals
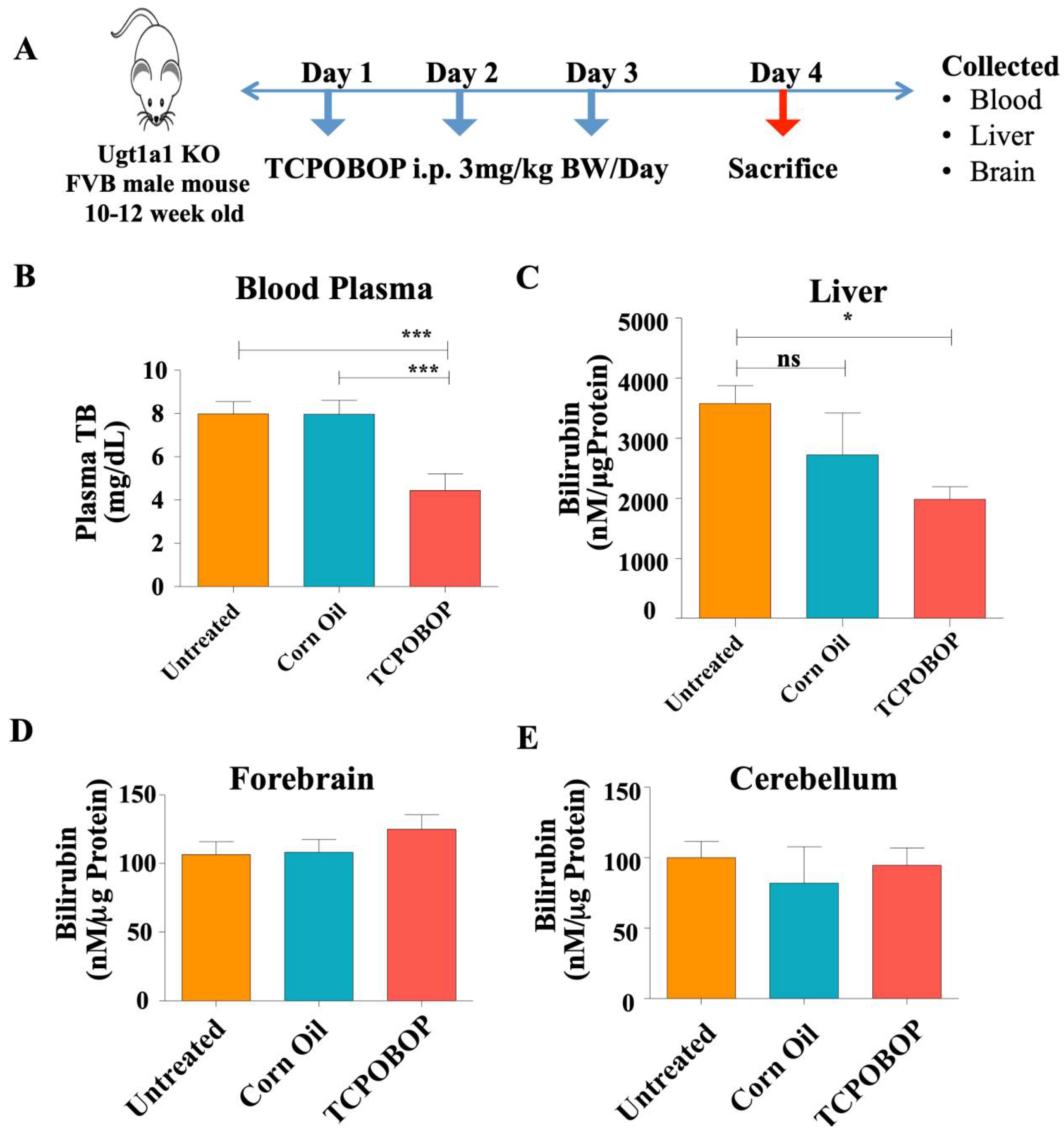
2.3. CAR Activation Decreases Plasma Bilirubin Level in Ugt1−/− Mice
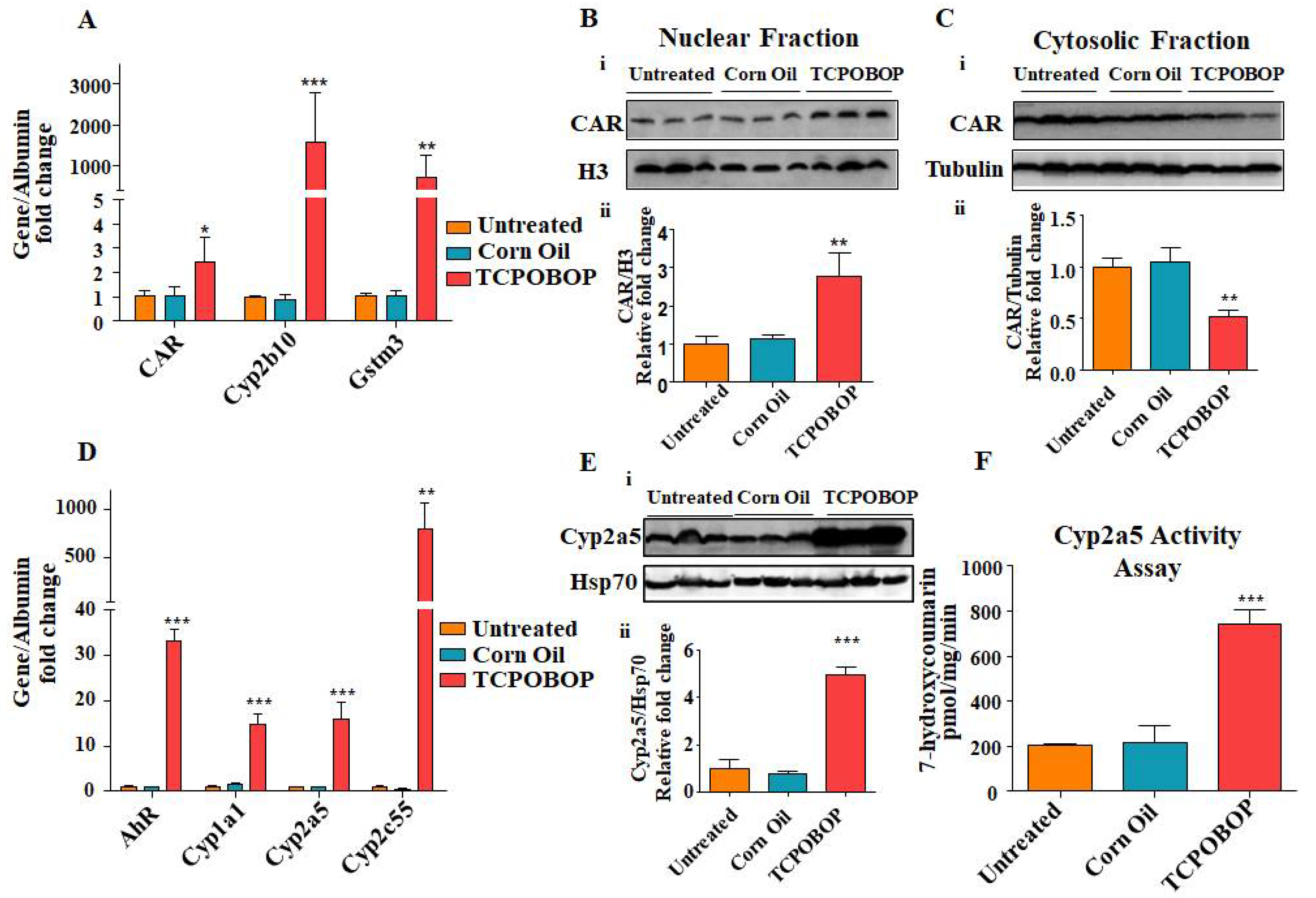
2.4. TCPOBOP Treatment Activated CAR and Induced CAR-Target Genes
2.5. TCPOBOP Treatment Up-Regulated Transcription of Bilirubin Transporter Genes
2.6. TCPOBOP Treatment Activated Nrf2-Related Genes
2.7. Liver Microsomes from TCPOBOP Treated Ugt1a1 KO Mice Show Increased Ability to Degrade Bilirubin In Vitro
3. Discussion
4. Materials and Methods
4.1. Animals and Drug Treatment
4.2. Cell Culture and Treatment
4.3. Biochemical Analysis of Plasma Samples
4.4. mRNA Extraction and qRT-PCR Analysis
4.5. Protein Extraction and Western Blot Analysis
4.6. Tissue Bilirubin Measurement
4.7. Microsome Preparation
4.8. Bilirubin Disappearance Assay
4.9. Coumarin 7-Hydroxylase Activity Assay
4.10. Reactive Oxygen Species (ROS) Quantification in Liver Tissue
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hinds, T.D., Jr.; Stec, D.E. Bilirubin, a Cardiometabolic Signaling Molecule. Hypertension 2018, 72, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Ngashangva, L.; Bachu, V.; Goswami, P. Development of new methods for determination of bilirubin. J. Pharm. Biomed. Anal. 2019, 162, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Bortolussi, G.; Muro, A.F. Advances in understanding disease mechanisms and potential treatments for Crigler–Najjar syndrome. Expert Opin. Orphan Drugs 2018, 6, 425–439. [Google Scholar] [CrossRef]
- Kalakonda, A.; Jenkins, B.A.; John, S. Physiology, Bilirubin. In StatPearls© 2022; StatPearls Publishing LLC: Treasure Island, FL, USA, 2022. [Google Scholar]
- Bosma, P.J.; Seppen, J.; Goldhoorn, B.; Bakker, C.; Oude Elferink, R.P.; Chowdhury, J.R.; Chowdhury, N.R.; Jansen, P.L. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J. Biol. Chem. 1994, 269, 17960–17964. [Google Scholar] [CrossRef]
- Čvorović, J.; Passamonti, S. Membrane Transporters for Bilirubin and Its Conjugates: A Systematic Review. Front. Pharmacol. 2017, 8, 887. [Google Scholar] [CrossRef]
- Keppler, D. The roles of MRP2, MRP3, OATP1B1, and OATP1B3 in conjugated hyperbilirubinemia. Drug Metab. Dispos. Biol. Fate Chem. 2014, 42, 561–565. [Google Scholar] [CrossRef]
- Bosma, P.J. Inherited disorders of bilirubin metabolism. J. Hepatol. 2003, 38, 107–117. [Google Scholar] [CrossRef]
- Ostrow, J.D.; Tiribelli, C. Bilirubin, a curse and a boon. Gut 2003, 52, 1668–1670. [Google Scholar] [CrossRef]
- Petrova, A.; Mehta, R.; Birchwood, G.; Ostfeld, B.; Hegyi, T. Management of neonatal hyperbilirubinemia: Pediatricians’ practices and educational needs. BMC Pediatr. 2006, 6, 6. [Google Scholar] [CrossRef]
- Watchko, J.F. Kernicterus and the molecular mechanisms of bilirubin-induced CNS injury in newborns. Neuromolecular Med. 2006, 8, 513–529. [Google Scholar] [CrossRef]
- Burgos, A.E.; Flaherman, V.J.; Newman, T.B. Screening and Follow-Up for Neonatal Hyperbilirubinemia: A Review. Clin. Pediatr. 2012, 51, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Hyperbilirubinemia, S.O. Management of hyperbilirubinemia in the newborn infant 35 or more weeks of gestation. Pediatrics 2004, 114, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Abu-Bakar, A.; Arthur, D.M.; Wikman, A.S.; Rahnasto, M.; Juvonen, R.O.; Vepsäläinen, J.; Raunio, H.; Ng, J.C.; Lang, M.A. Metabolism of bilirubin by human cytochrome P450 2A6. Toxicol. Appl. Pharmacol. 2012, 261, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.W.; Köhle, C. Contributions of the Ah receptor to bilirubin homeostasis and its antioxidative and atheroprotective functions. Biol. Chem. 2010, 391, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Kapitulnik, J.; Gonzalez, F.J. Marked endogenous activation of the CYP1A1 and CYP1A2 genes in the congenitally jaundiced Gunn rat. Mol. Pharmacol. 1993, 43, 722–725. [Google Scholar]
- Kapitulnik, J.; Ostrow, J.D. Stimulation of bilirubin catabolism in jaundiced Gunn rats by an induced of microsomal mixed-function monooxygenases. Proc. Natl. Acad. Sci. USA 1978, 75, 682–685. [Google Scholar] [CrossRef]
- Kaul, R.; Kaul, H.K.; Murti, C.R. An alternate pathway for bilirubin catabolism. FEBS Lett. 1980, 111, 240–242. [Google Scholar] [CrossRef][Green Version]
- Fujiwara, R.; Haag, M.; Schaeffeler, E.; Nies, A.T.; Zanger, U.M.; Schwab, M. Systemic regulation of bilirubin homeostasis: Potential benefits of hyperbilirubinemia. Hepatology 2018, 67, 1609–1619. [Google Scholar] [CrossRef]
- Nishioka, T.; Hafkamp, A.M.; Havinga, R.; vn Lierop, P.P.; Velvis, H.; Verkade, H.J. Orlistat treatment increases fecal bilirubin excretion and decreases plasma bilirubin concentrations in hyperbilirubinemic Gunn rats. J. Pediatr. 2003, 143, 327–334. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, J.; Moore, D.D. A traditional herbal medicine enhances bilirubin clearance by activating the nuclear receptor CAR. J. Clin. Investig. 2004, 113, 137–143. [Google Scholar] [CrossRef]
- Saini, S.P.; Mu, Y.; Gong, H.; Toma, D.; Uppal, H.; Ren, S.; Li, S.; Poloyac, S.M.; Xie, W. Dual role of orphan nuclear receptor pregnane X receptor in bilirubin detoxification in mice. Hepatology 2005, 41, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, L.; Wu, J.; Tang, B.; Zhang, M.; Zhu, D.; Lin, X. Constitutive androstane receptor activation promotes bilirubin clearance in a murine model of alcoholic liver disease. Mol. Med. Rep. 2017, 15, 3459–3466. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van der Schoor, L.W.E.; Verkade, H.J.; Bertolini, A.; de Wit, S.; Mennillo, E.; Rettenmeier, E.; Weber, A.A.; Havinga, R.; Valášková, P.; Jašprová, J.; et al. Potential of therapeutic bile acids in the treatment of neonatal Hyperbilirubinemia. Sci. Rep. 2021, 11, 11107. [Google Scholar] [CrossRef]
- Fujiwara, R.; Mitsugi, R.; Uemura, A.; Itoh, T.; Tukey, R.H. Severe Neonatal Hyperbilirubinemia in Crigler-Najjar Syndrome Model Mice Can Be Reversed With Zinc Protoporphyrin. Hepatol. Commun. 2017, 1, 792–802. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, J.; Chua, S.S.; Qatanani, M.; Han, Y.; Granata, R.; Moore, D.D. Induction of bilirubin clearance by the constitutive androstane receptor (CAR). Proc. Natl. Acad. Sci. USA 2003, 100, 4156–4161. [Google Scholar] [CrossRef]
- Wagner, M.; Halilbasic, E.; Marschall, H.U.; Zollner, G.; Fickert, P.; Langner, C.; Zatloukal, K.; Denk, H.; Trauner, M. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology 2005, 42, 420–430. [Google Scholar] [CrossRef]
- Yoda, E.; Paszek, M.; Konopnicki, C.; Fujiwara, R.; Chen, S.; Tukey, R.H. Isothiocyanates induce UGT1A1 in humanized UGT1 mice in a CAR dependent fashion that is highly dependent upon oxidative stress. Sci. Rep. 2017, 7, 46489. [Google Scholar] [CrossRef]
- Bortolussi, G.; Baj, G.; Vodret, S.; Viviani, G.; Bittolo, T.; Muro, A.F. Age-dependent pattern of cerebellar susceptibility to bilirubin neurotoxicity in vivo in mice. Dis. Models Mech. 2014, 7, 1057–1068. [Google Scholar] [CrossRef]
- Bortolussi, G.; Zentilin, L.; Baj, G.; Giraudi, P.; Bellarosa, C.; Giacca, M.; Tiribelli, C.; Muro, A.F. Rescue of bilirubin-induced neonatal lethality in a mouse model of Crigler-Najjar syndrome type I by AAV9-mediated gene transfer. FASEB J. 2012, 26, 1052–1063. [Google Scholar] [CrossRef]
- Baskin-Bey, E.S.; Huang, W.; Ishimura, N.; Isomoto, H.; Bronk, S.F.; Braley, K.; Craig, R.W.; Moore, D.D.; Gores, G.J. Constitutive androstane receptor (CAR) ligand, TCPOBOP, attenuates Fas-induced murine liver injury by altering Bcl-2 proteins. Hepatology 2006, 44, 252–262. [Google Scholar] [CrossRef]
- Abu-Bakar, A.; Arthur, D.M.; Aganovic, S.; Ng, J.C.; Lang, M.A. Inducible bilirubin oxidase: A novel function for the mouse cytochrome P450 2A5. Toxicol. Appl. Pharmacol. 2011, 257, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.D.; Antenos, M.; Squires, E.J.; Kirby, G.M. Cytochrome P450 2A5 and bilirubin: Mechanisms of gene regulation and cytoprotection. Toxicol. Appl. Pharmacol. 2013, 270, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Kirby, G.M.; Nichols, K.D.; Antenos, M. CYP2A5 induction and hepatocellular stress: An adaptive response to perturbations of heme homeostasis. Curr. Drug Metab. 2011, 12, 186–197. [Google Scholar] [CrossRef]
- Wieneke, N.; Hirsch-Ernst, K.I.; Kuna, M.; Kersten, S.; Püschel, G.P. PPARalpha-dependent induction of the energy homeostasis-regulating nuclear receptor NR1i3 (CAR) in rat hepatocytes: Potential role in starvation adaptation. FEBS Lett. 2007, 581, 5617–5626. [Google Scholar] [CrossRef]
- Hinds, T.D., Jr.; Sodhi, K.; Meadows, C.; Fedorova, L.; Puri, N.; Kim, D.H.; Peterson, S.J.; Shapiro, J.; Abraham, N.G.; Kappas, A. Increased HO-1 levels ameliorate fatty liver development through a reduction of heme and recruitment of FGF21. Obesity 2014, 22, 705–712. [Google Scholar] [CrossRef]
- Stec, D.E.; John, K.; Trabbic, C.J.; Luniwal, A.; Hankins, M.W.; Baum, J.; Hinds, T.D., Jr. Bilirubin Binding to PPARα Inhibits Lipid Accumulation. PLoS ONE 2016, 11, e0153427. [Google Scholar] [CrossRef]
- Cuperus, F.J.; Hafkamp, A.M.; Hulzebos, C.V.; Verkade, H.J. Pharmacological therapies for unconjugated hyperbilirubinemia. Curr. Pharm. Des. 2009, 15, 2927–2938. [Google Scholar] [CrossRef]
- Jansen, P.L. Diagnosis and management of Crigler-Najjar syndrome. Eur. J. Pediatr. 1999, 158 (Suppl. S2), S89–S94. [Google Scholar] [CrossRef]
- Mutoh, S.; Sobhany, M.; Moore, R.; Perera, L.; Pedersen, L.; Sueyoshi, T.; Negishi, M. Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci. Signal. 2013, 6, ra31. [Google Scholar] [CrossRef]
- Kranc, K.R.; Pyne, G.J.; Tao, L.; Claridge, T.D.W.; Harris, D.A.; Cadoux-Hudson, T.A.D.; Turnbull, J.J.; Schofield, C.J.; Clark, J.F. Oxidative degradation of bilirubin produces vasoactive compounds. Eur. J. Biochem. 2000, 267, 7094–7101. [Google Scholar] [CrossRef]
- Bock, K.W. Regulation of bilirubin clearance by ligand-activated transcription factors of the endo- and xenobiotic metabolism system. Front. Pharmacol. 2011, 2, 82. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.L.; Bammler, T.K.; Cui, J.Y. RNA Sequencing Reveals Age and Species Differences of Constitutive Androstane Receptor-Targeted Drug-Processing Genes in the Liver. Drug Metab. Dispos. Biol. Fate Chem. 2017, 45, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, B.; Stoops, J.W.; Mars, W.M.; Orr, A.; Bowen, W.C.; Paranjpe, S.; Michalopoulos, G.K. TCPOBOP-Induced Hepatomegaly and Hepatocyte Proliferation are Attenuated by Combined Disruption of MET and EGFR Signaling. Hepatology 2019, 69, 1702–1718. [Google Scholar] [CrossRef]
- Maglich, J.M.; Lobe, D.C.; Moore, J.T. The nuclear receptor CAR (NR1I3) regulates serum triglyceride levels under conditions of metabolic stress. J. Lipid Res. 2009, 50, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Honkakoski, P.; Sueyoshi, T.; Negishi, M. Drug-activated nuclear receptors CAR and PXR. Ann. Med. 2003, 35, 172–182. [Google Scholar] [CrossRef]
- Forman, B.M.; Tzameli, I.; Choi, H.S.; Chen, J.; Simha, D.; Seol, W.; Evans, R.M.; Moore, D.D. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature 1998, 395, 612–615. [Google Scholar] [CrossRef]
- Qatanani, M.; Moore, D.D. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr. Drug Metab. 2005, 6, 329–339. [Google Scholar] [CrossRef]
- Kim, S.D.; Morgan, L.; Hargreaves, E.; Zhang, X.; Jiang, Z.; Antenos, M.; Li, B.; Kirby, G.M. Regulation of Cytochrome P450 2a5 by Artemisia capillaris and 6,7-Dimethylesculetin in Mouse Hepatocytes. Front. Pharmacol. 2021, 12, 3390. [Google Scholar] [CrossRef]
- Mutoh, S.; Osabe, M.; Inoue, K.; Moore, R.; Pedersen, L.; Perera, L.; Rebolloso, Y.; Sueyoshi, T.; Negishi, M. Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3). J. Biol. Chem. 2009, 284, 34785–34792. [Google Scholar] [CrossRef]
- Kawamoto, T.; Sueyoshi, T.; Zelko, I.; Moore, R.; Washburn, K.; Negishi, M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 1999, 19, 6318–6322. [Google Scholar] [CrossRef]
- Sugatani, J.; Kojima, H.; Ueda, A.; Kakizaki, S.; Yoshinari, K.; Gong, Q.H.; Owens, I.S.; Negishi, M.; Sueyoshi, T. The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology 2001, 33, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, S.J.; Levy, G.; Matsuzawa, T.; Baliah, T. Enhancement of Glucuronide-Conjugating Capacity in a Hyperbilirubinemic Infant Due to Apparent Enzyme Induction by Phenobarbital. N. Engl. J. Med. 1966, 275, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Tzameli, I.; Pissios, P.; Schuetz, E.G.; Moore, D.D. The Xenobiotic Compound 1,4-Bis[2-(3,5-Dichloropyridyloxy)]Benzene Is an Agonist Ligand for the Nuclear Receptor CAR. Mol. Cell. Biol. 2000, 20, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Elferink, R.O. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 2002, 71, 537–592. [Google Scholar] [CrossRef]
- Xiong, H.; Yoshinari, K.; Brouwer, K.L.; Negishi, M. Role of constitutive androstane receptor in the in vivo induction of Mrp3 and CYP2B1/2 by phenobarbital. Drug Metab. Dispos. Biol. Fate Chem. 2002, 30, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Bockor, L.; Bortolussi, G.; Vodret, S.; Iaconcig, A.; Jašprová, J.; Zelenka, J.; Vitek, L.; Tiribelli, C.; Muro, A.F. Modulation of bilirubin neurotoxicity by the Abcb1 transporter in the Ugt1-/- lethal mouse model of neonatal hyperbilirubinemia. Hum. Mol. Genet. 2017, 26, 145–157. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Thimmulappa, R.K.; Mai, K.H.; Srisuma, S.; Kensler, T.W.; Yamamoto, M.; Biswal, S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002, 62, 5196–5203. [Google Scholar]
- Martin, D.; Rojo, A.I.; Salinas, M.; Diaz, R.; Gallardo, G.; Alam, J.; de Galarreta, C.M.R.; Cuadrado, A. Regulation of Heme Oxygenase-1 Expression through the Phosphatidylinositol 3-Kinase/Akt Pathway and the Nrf2 Transcription Factor in Response to the Antioxidant Phytochemical Carnosol. J. Biol. Chem. 2004, 279, 8919–8929. [Google Scholar] [CrossRef]
- Sugatani, J.; Sueyoshi, T.; Negishi, M.; Miwa, M. Regulation of the human UGT1A1 gene by nuclear receptors constitutive active/androstane receptor, pregnane X receptor, and glucocorticoid receptor. Methods Enzymol. 2005, 400, 92–104. [Google Scholar] [CrossRef]
- Yueh, M.-F.; Tukey, R.H. Nrf2-Keap1 Signaling Pathway Regulates Human UGT1A1 Expression in Vitro and in Transgenic UGT1 Mice. J. Biol. Chem. 2007, 282, 8749–8758. [Google Scholar] [CrossRef] [PubMed]
- Gow, P.J.; Ghabrial, H.; Smallwood, R.A.; Morgan, D.J.; Ching, M.S. Neonatal Hepatic Drug Elimination. Pharmacol. Toxicol. 2001, 88, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, P.; Nagini, S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Sabrina, E.; Gambaro, R.M.; Tiribelli, C.; Gazzin, S. Brain Cytochrome p450 enzymes: A possible Therapeutic Targets for Neurological Diseases. Ther. Targets Neurol. Dis. 2015, 2, 1–6. [Google Scholar] [CrossRef]
- Raghunath, A.; Sundarraj, K.; Arfuso, F.; Sethi, G.; Perumal, E. Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers 2018, 10, 481. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Moore, R.; Goldsworthy, T.L.; Negishi, M.; Maronpot, R.R. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004, 64, 7197–7200. [Google Scholar] [CrossRef]
- Dimauro, I.; Pearson, T.; Caporossi, D.; Jackson, M.J. A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res. Notes 2012, 5, 513. [Google Scholar] [CrossRef]
- Kumagai, A.; Ando, R.; Miyatake, H.; Greimel, P.; Kobayashi, T.; Hirabayashi, Y.; Shimogori, T.; Miyawaki, A. A bilirubin-inducible fluorescent protein from eel muscle. Cell 2013, 153, 1602–1611. [Google Scholar] [CrossRef]
- Adeosun, S.O.; Moore, K.H.; Lang, D.M.; Nwaneri, A.C.; Hinds, T.D., Jr.; Stec, D.E. A Novel Fluorescence-Based Assay for the Measurement of Biliverdin Reductase Activity. React. Oxyg. Species (Apex) 2018, 5, 35–45. [Google Scholar] [CrossRef][Green Version]
- Aitio, A. A simple and sensitive assay of 7-ethoxycoumarin deethylation. Anal. Biochem. 1978, 85, 488–491. [Google Scholar] [CrossRef]
- Gabbia, D.; Pozzo, L.; Zigiotto, G.; Roverso, M.; Sacchi, D.; Dalla Pozza, A.; Carrara, M.; Bogialli, S.; Floreani, A.; Guido, M.; et al. Dexamethasone counteracts hepatic inflammation and oxidative stress in cholestatic rats via CAR activation. PLoS ONE 2018, 13, e0204336. [Google Scholar] [CrossRef] [PubMed]

| Forward (5′-3′) | Reverse (5′-3′) | |
|---|---|---|
| Albumin | GCATGAAGTTGCCAGAAGAC | TCTGCATACTGGAGCACTTC |
| Cyp2a5 | TGCGCTATGGCTTTCTGTTG | GAAACTTGGTGTCCTTGGTG |
| Cyp1a1 | TCATTCCTGTCCTCCGTTAC | ACATTGGCATTCTCGTCCAG |
| HO 1 | ACAGGGTGACAGAAGAGGCTAAGAC | ATTTTCCTCGGGGCGTCTCT |
| Nqo1 | TTTAGGGTCGTCTTGGCAAC | GTCTTCTCTGAATGGGCCAG |
| CAR | TGCAGGTTGCAGAAGTGTCTA | GGACCAGTTCTTTCTGCTGC |
| Cyp2b10 | GTTCCACACAGCATAACCAC | CCTTGTGGGCTTCTTGTTAC |
| Cyp2c55 | CGCATTCAAGAGGAAGCATC | ACAGATCACATTGGAGGGAG |
| Gstm3 | TCCGTGTGGATACTTTGGAG | ATGGCCTTCAAGAACTCTGG |
| Nrf2 | GGCAGAGACATTCCCATTTG | AAACTTGCTCCATGTCCTGC |
| Abcc1 | TGATACAGCTTGAACGGAGG | AGCACAAGGGTGAAGTACAG |
| Abcc2 | TGGTTCCTGTCCATGTTCTG | TCTGGAAACCATAGGAGACG |
| Abcc3 | TTACCTGAGACACCATCAGC | CCAAGGGTGTGACAAAGAAG |
| Abcc4 | TTCTTCTGCCTCTGCAAAGC | ACGATTTCTCCCACGCATAC |
| Abcc5 | TGAATGAAGTTGGGCCAGAC | TGTACTCCAAGAGGTGCTTC |
| Abcc6 | TCCTAGCTGTGCTGATGTTC | TGGACTTTCTGGAACCACTG |
| Slco1b2 | GAAACAAGCTCCTGCCAACC | GTCTGACCAAACTGCTGCTC |
| Abcb1 | GATAGGCTGGTTTGATGTGC | TCACAAGGGTTAGCTTCCAG |
| Gsta1 | AGCCCGTGCTTCACTACTTC | TCTTCAAACTCCACCCCT GC |
| Gsta2 | AATCAGCAGCCTCCCCAAT | TCCATCAATGCAGCCACACT |
| Ugt1a1 | TCTGGCTGATGAGAAGTGACT | GAAAACAACGATGCCATGCT |
| AhR | GGCAGCTTATTCTGGGCTATAC | ATCAAGCGTGCATTGGACTG |
| PXR | GACGCTCAGATGCAAACCTT | TCTTCTCCGCGCAGCTGCA |
| Gapdh | ATGGTGAAGGTCGGTGTGAA | GTTGATGGCAACAATCTCCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, B.; Olajide, O.J.; Bortolussi, G.; Muro, A.F. Activation of Alternative Bilirubin Clearance Pathways Partially Reduces Hyperbilirubinemia in a Mouse Model Lacking Functional Ugt1a1 Activity. Int. J. Mol. Sci. 2022, 23, 10703. https://doi.org/10.3390/ijms231810703
Banerjee B, Olajide OJ, Bortolussi G, Muro AF. Activation of Alternative Bilirubin Clearance Pathways Partially Reduces Hyperbilirubinemia in a Mouse Model Lacking Functional Ugt1a1 Activity. International Journal of Molecular Sciences. 2022; 23(18):10703. https://doi.org/10.3390/ijms231810703
Chicago/Turabian StyleBanerjee, Bhaswati, Olayemi Joseph Olajide, Giulia Bortolussi, and Andrés F. Muro. 2022. "Activation of Alternative Bilirubin Clearance Pathways Partially Reduces Hyperbilirubinemia in a Mouse Model Lacking Functional Ugt1a1 Activity" International Journal of Molecular Sciences 23, no. 18: 10703. https://doi.org/10.3390/ijms231810703
APA StyleBanerjee, B., Olajide, O. J., Bortolussi, G., & Muro, A. F. (2022). Activation of Alternative Bilirubin Clearance Pathways Partially Reduces Hyperbilirubinemia in a Mouse Model Lacking Functional Ugt1a1 Activity. International Journal of Molecular Sciences, 23(18), 10703. https://doi.org/10.3390/ijms231810703







