Figure 1.
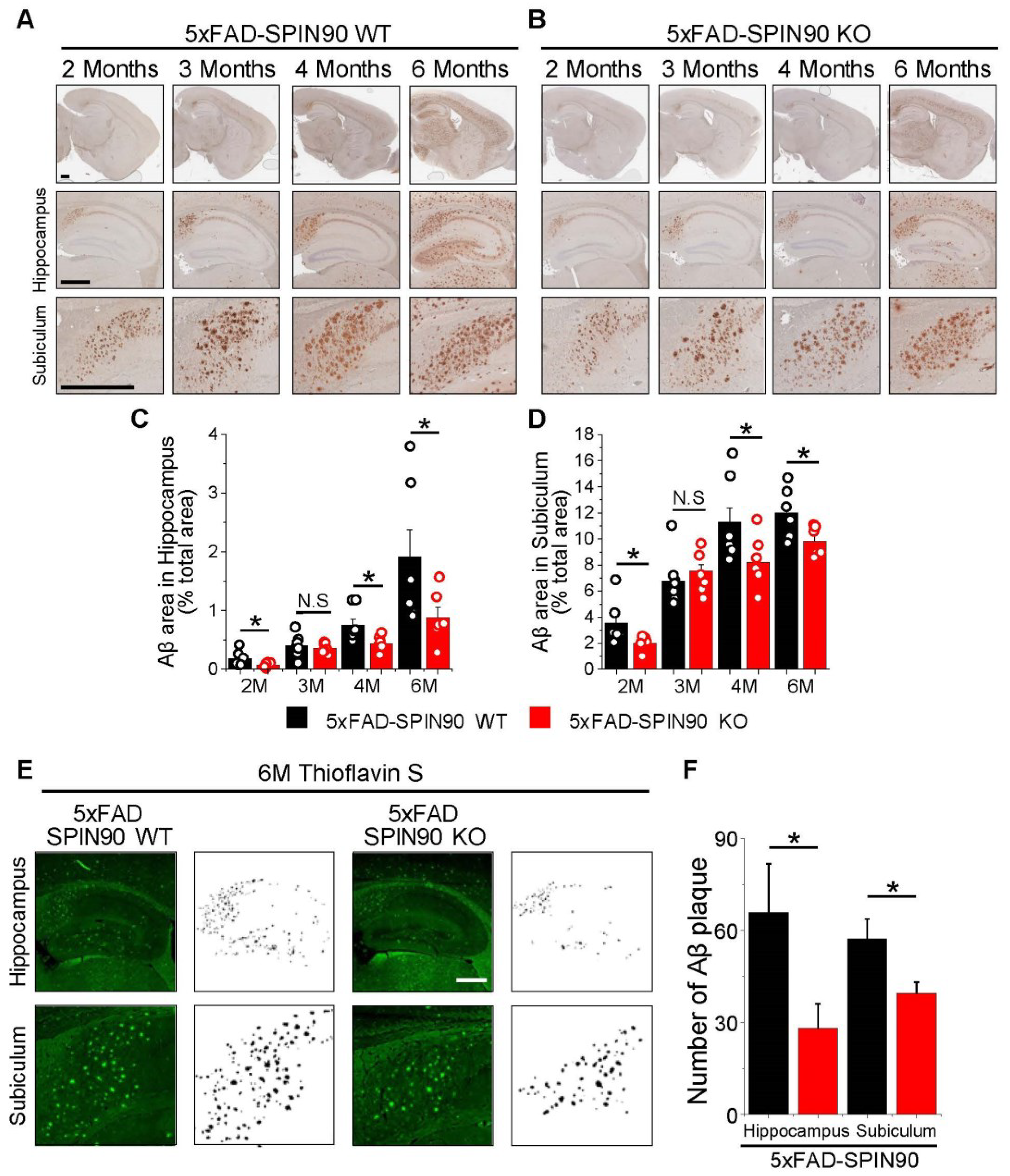
SPIN90 knockout reduces amyloid β (Aβ) deposition in AD model mice. (A,B) Representative images of Aβ plaques in the brains of 5xFAD-SPIN90 WT (A) and 5xFAD-SPIN90 KO mice (B), as shown by Dab reaction (brown) and hematoxylin straining (blue). Brains of mice aged 2, 3, 4, and 6 months were fixed and stained with anti-Aβ antibody. (Top) The entire area of a hemisphere; (middle) magnified images of the hippocampus; (bottom) magnified images of the subiculum. Scale bars, 500 μm. (C,D) Mean areas of Aβ deposition in the hippocampus (C) and subiculum (D) of brains from 2-, 3-, 4-, and 6-month old 5xFAD-SPIN90 WT (Black) and 5xFAD-SPIN90 KO (Red) mice. Total areas of Aβ plaque were quantified and normalized relative to areas of the hippocampus or subiculum. Hippocampus: [2M; Aβ plaque]5xFAD-SPIN90WT = 0.18 ± 0.05%, n = 18 (6 mice), [2M; Aβ plaque]5xFAD-SPIN90KO = 0.08 ± 0.01%, n = 18 (6 mice), [3M; Aβ plaque]5xFAD-SPIN90WT = 0.4 ± 0.07%, n = 21 (7 mice), [3M; Aβ plaque]5xFAD-SPIN90KO = 0.36 ± 0.03%, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90WT = 0.75 ± 0.1%, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90KO = 0.44 ± 0.04%, n = 21 (7 mice), [6M; Aβ plaque]5xFAD-SPIN90WT = 1.9 ± 0.47%, n = 18 (6 mice), [6M; Aβ plaque]5xFAD-SPIN90KO = 0.89 ± 0.17, n = 18 (6 mice). Subiculum: [2M; Aβ plaque]5xFAD-SPIN90WT = 3.58 ± 0.66, n = 18 (6 mice), [2M; Aβ plaque]5xFAD-SPIN90KO = 2.04 ± 0.2, n = 18 (6 mice), [3M; Aβ plaque]5xFAD-SPIN90WT = 6.79 ± 0.7, n = 21 (7 mice), [3M; Aβ plaque]5xFAD-SPIN90KO = 7.54 ± 0.5, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90WT = 11.29 ± 1.09, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90KO = 8.22 ± 0.67, n = 21 (7 mice), [6M; Aβ plaque]5xFAD-SPIN90WT = 12.02 ± 0.73, n = 18 (6 mice), [6M; Aβ plaque]5xFAD-SPIN90KO = 9.85 ± 0.43, n = 18 (6 mice). (E) Representative images of thioflavin S- stained Aβ plaques in the hippocampus and subiculum in the brains of 6-month-old 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) mice. Images are shown inverted to clearly display Aβ plaques. Scale bars, 500 μm. (F) Quantification of Aβ aggregates in the hippocampus and subiculum of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO mice. [Hippocampus: Aβ aggregates]5xFAD-SPIN90WT = 65.83 ± 15.88 (n = 6), [Hippocampus: Aβ aggregates]5xFAD-SPIN90KO = 28.17 ± 7.94 (n = 6), [Subiculum: Aβ aggregates]5xFAD-SPIN90WT = 57.33 ± 6.32 (n = 6), [Subiculum: Aβ aggregates]5xFAD-SPIN90KO = 39.5 ± 3.61 (n = 6). * p < 0.05. N.S.; not significant.
Figure 1.
SPIN90 knockout reduces amyloid β (Aβ) deposition in AD model mice. (A,B) Representative images of Aβ plaques in the brains of 5xFAD-SPIN90 WT (A) and 5xFAD-SPIN90 KO mice (B), as shown by Dab reaction (brown) and hematoxylin straining (blue). Brains of mice aged 2, 3, 4, and 6 months were fixed and stained with anti-Aβ antibody. (Top) The entire area of a hemisphere; (middle) magnified images of the hippocampus; (bottom) magnified images of the subiculum. Scale bars, 500 μm. (C,D) Mean areas of Aβ deposition in the hippocampus (C) and subiculum (D) of brains from 2-, 3-, 4-, and 6-month old 5xFAD-SPIN90 WT (Black) and 5xFAD-SPIN90 KO (Red) mice. Total areas of Aβ plaque were quantified and normalized relative to areas of the hippocampus or subiculum. Hippocampus: [2M; Aβ plaque]5xFAD-SPIN90WT = 0.18 ± 0.05%, n = 18 (6 mice), [2M; Aβ plaque]5xFAD-SPIN90KO = 0.08 ± 0.01%, n = 18 (6 mice), [3M; Aβ plaque]5xFAD-SPIN90WT = 0.4 ± 0.07%, n = 21 (7 mice), [3M; Aβ plaque]5xFAD-SPIN90KO = 0.36 ± 0.03%, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90WT = 0.75 ± 0.1%, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90KO = 0.44 ± 0.04%, n = 21 (7 mice), [6M; Aβ plaque]5xFAD-SPIN90WT = 1.9 ± 0.47%, n = 18 (6 mice), [6M; Aβ plaque]5xFAD-SPIN90KO = 0.89 ± 0.17, n = 18 (6 mice). Subiculum: [2M; Aβ plaque]5xFAD-SPIN90WT = 3.58 ± 0.66, n = 18 (6 mice), [2M; Aβ plaque]5xFAD-SPIN90KO = 2.04 ± 0.2, n = 18 (6 mice), [3M; Aβ plaque]5xFAD-SPIN90WT = 6.79 ± 0.7, n = 21 (7 mice), [3M; Aβ plaque]5xFAD-SPIN90KO = 7.54 ± 0.5, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90WT = 11.29 ± 1.09, n = 21 (7 mice), [4M; Aβ plaque]5xFAD-SPIN90KO = 8.22 ± 0.67, n = 21 (7 mice), [6M; Aβ plaque]5xFAD-SPIN90WT = 12.02 ± 0.73, n = 18 (6 mice), [6M; Aβ plaque]5xFAD-SPIN90KO = 9.85 ± 0.43, n = 18 (6 mice). (E) Representative images of thioflavin S- stained Aβ plaques in the hippocampus and subiculum in the brains of 6-month-old 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) mice. Images are shown inverted to clearly display Aβ plaques. Scale bars, 500 μm. (F) Quantification of Aβ aggregates in the hippocampus and subiculum of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO mice. [Hippocampus: Aβ aggregates]5xFAD-SPIN90WT = 65.83 ± 15.88 (n = 6), [Hippocampus: Aβ aggregates]5xFAD-SPIN90KO = 28.17 ± 7.94 (n = 6), [Subiculum: Aβ aggregates]5xFAD-SPIN90WT = 57.33 ± 6.32 (n = 6), [Subiculum: Aβ aggregates]5xFAD-SPIN90KO = 39.5 ± 3.61 (n = 6). * p < 0.05. N.S.; not significant.
![Ijms 23 10563 g001]()
Figure 2.
Deficiency of SPIN90 alters surface fraction of APP in axons and dendrites of 5xFAD neurons. (A) Schematic diagram of pHluorin-conjugated APP (pH-APP). (B) Illustration of surface pH-APP (left) and endosomal pH-APP (right). (C) Representative images of pH-APP in axons of 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons at rest (top), after acid quenching (middle), and after NH4Cl treatment (bottom). Scale bar: 5 μm. (D) Representative traces of pH-APP in response to acid quenching in axons of 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (E) Mean surface fraction of APP in axons of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [Surface fraction: axon]5xFAD-SPIN90WT = 5.85 ± 0.42% (n = 9 cells), [Surface fraction: axon]5xFAD-SPIN90KO = 2.66 ± 0.38% (n = 13 cells). (F) Representative images of pH-APP in dendrites of 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons at rest (top), after acid quenching (middle), and after NH4Cl treatment (bottom). Scale bar, 5 μm. (G) Representative traces of pH-APP in response to acid quenching in dendrites of 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (H) Mean surface fraction of APP in dendrites of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [surface fraction: dendrite]5xFAD-SPIN90WT = 11.56 ± 1.63% (n = 15 cells), [surface fraction: dendrite]5xFAD-SPIN90KO = 23.26 ± 2.23% (n = 19 cells). *** p < 0.001.
Figure 2.
Deficiency of SPIN90 alters surface fraction of APP in axons and dendrites of 5xFAD neurons. (A) Schematic diagram of pHluorin-conjugated APP (pH-APP). (B) Illustration of surface pH-APP (left) and endosomal pH-APP (right). (C) Representative images of pH-APP in axons of 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons at rest (top), after acid quenching (middle), and after NH4Cl treatment (bottom). Scale bar: 5 μm. (D) Representative traces of pH-APP in response to acid quenching in axons of 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (E) Mean surface fraction of APP in axons of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [Surface fraction: axon]5xFAD-SPIN90WT = 5.85 ± 0.42% (n = 9 cells), [Surface fraction: axon]5xFAD-SPIN90KO = 2.66 ± 0.38% (n = 13 cells). (F) Representative images of pH-APP in dendrites of 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons at rest (top), after acid quenching (middle), and after NH4Cl treatment (bottom). Scale bar, 5 μm. (G) Representative traces of pH-APP in response to acid quenching in dendrites of 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (H) Mean surface fraction of APP in dendrites of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [surface fraction: dendrite]5xFAD-SPIN90WT = 11.56 ± 1.63% (n = 15 cells), [surface fraction: dendrite]5xFAD-SPIN90KO = 23.26 ± 2.23% (n = 19 cells). *** p < 0.001.
![Ijms 23 10563 g002]()
Figure 3.
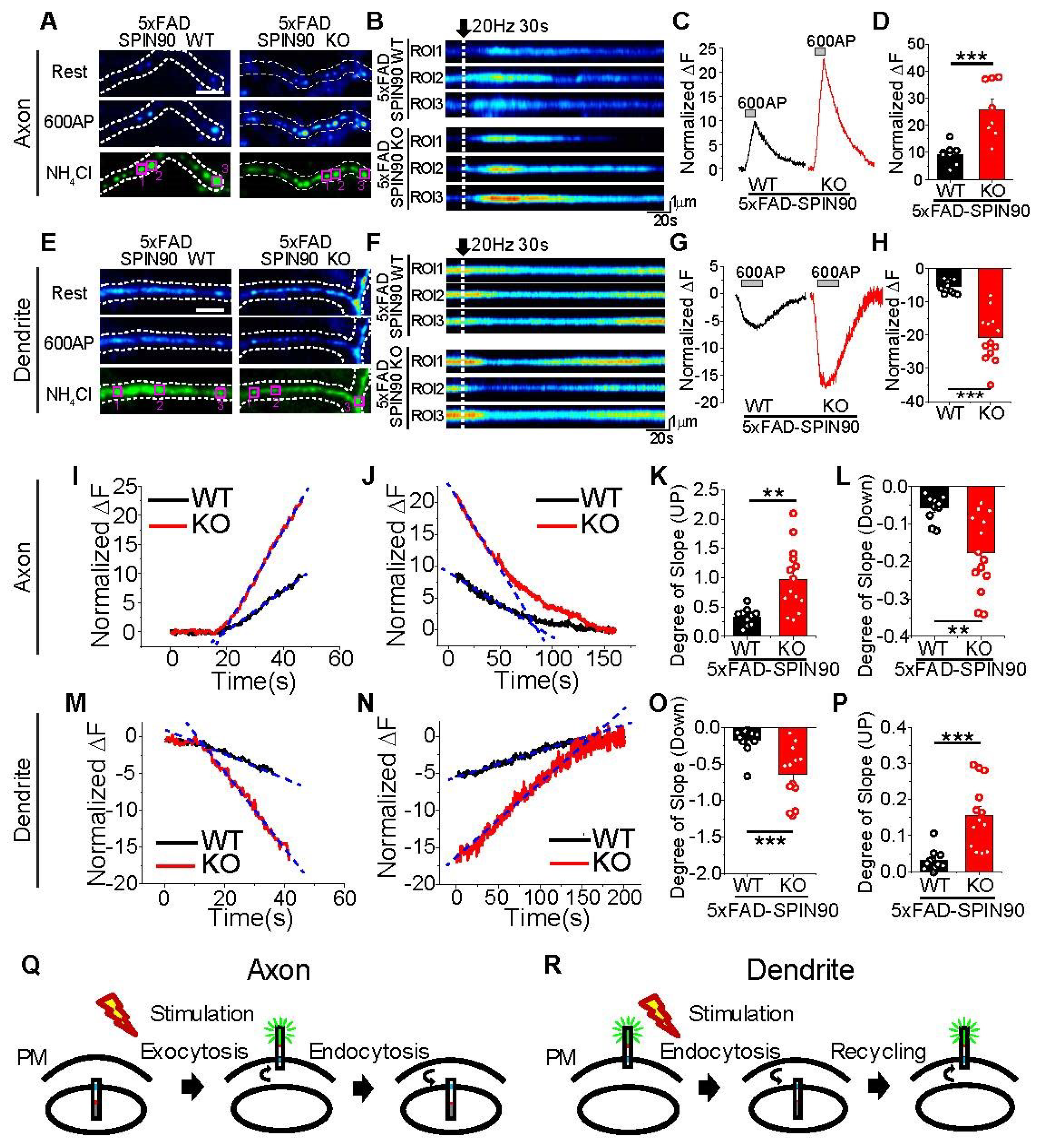
Activity-driven recycling of APP in axons and dendrites is enhanced in 5xFAD-SPIN90 KO neurons. (A,E) Representative images of pH-APP in axons (A) and dendrites (E) of 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons at rest (top), in the presence of an action potential (AP) of 600 (middle), and after NH4Cl treatment (bottom). Scale bar: 5 μm. (B,F) Kymographs of pH-APP in corresponding areas of axons (B) and dendrites (F) in 5xFAD-SPIN90 WT (top) and 5xFAD-SPIN90 KO (bottom) neurons in response to 600AP. (C,G) Representative traces of pH-APP in axons (C) and dendrites (G) of 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons in response to 600AP. (D,H) Mean peak amplitudes in axons (D) and dendrites (H) of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons in response to 600AP. [600AP: axon]5xFAD-SPIN90WT = 9.30 ± 1.31% (n = 8 cells), [600AP: axon]5xFAD-SPIN90KO = 25.87 ± 3.69% (n = 8 cells); [600AP: dendrite]5xFAD-SPIN90WT = −5.48 ± 0.62% (n = 10 cells), [600AP: dendrite]5xFAD-SPIN90KO = −20.80 ± 2.05% (n = 13 cells). Representative traces of pH-APP with linear fit-line during stimulation of (I) axons and (M) dendrites and after stimulation of (J) axons and (N) dendrites in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (K,O) Mean slopes as determined by linear fitting during stimulation of axons (K) and dendrites (O) in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [Degree of slope: axon]5xFAD-SPIN90WT = 0.33 ± 0.05 (n = 10 cells); [Degree of slope: axon]5xFAD-SPIN90KO = 0.97 ± 0.15 (n = 14 cells); [Degree of slope: dendrite]5xFAD-SPIN90WT = −0.17 ± 0.05 (n = 11 cells); [Degree of slope: dendrite]5xFAD-SPIN90KO = −0.65 ± 0.11 (n = 13 cells). (L,P) Mean slopes as determined by linear fitting after stimulation of axons (L) and dendrites (P) in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [Degree of slope: axon]5xFAD-SPIN90WT = −0.057 ± 0.01 (n = 10 cells); [Degree of slope: axon]5xFAD-SPIN90KO = −0.18 ± 0.03 (n = 14 cells); [Degree of slope: dendrite]5xFAD-SPIN90WT = 0.032 ± 0.009 (n = 11 cells); [Degree of slope: dendrite]5xFAD-SPIN90KO = 0.16 ± 0.03 (n = 13 cells). (Q) Schematic illustration of activity-driven pH-APP recycling in axons (endosome-surface-endosome). (R) Schematic illustration of activity-driven pH-APP recycling in dendrites (surface-endosome-surface). ** p < 0.01, *** p < 0.001.
Figure 3.
Activity-driven recycling of APP in axons and dendrites is enhanced in 5xFAD-SPIN90 KO neurons. (A,E) Representative images of pH-APP in axons (A) and dendrites (E) of 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons at rest (top), in the presence of an action potential (AP) of 600 (middle), and after NH4Cl treatment (bottom). Scale bar: 5 μm. (B,F) Kymographs of pH-APP in corresponding areas of axons (B) and dendrites (F) in 5xFAD-SPIN90 WT (top) and 5xFAD-SPIN90 KO (bottom) neurons in response to 600AP. (C,G) Representative traces of pH-APP in axons (C) and dendrites (G) of 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons in response to 600AP. (D,H) Mean peak amplitudes in axons (D) and dendrites (H) of 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons in response to 600AP. [600AP: axon]5xFAD-SPIN90WT = 9.30 ± 1.31% (n = 8 cells), [600AP: axon]5xFAD-SPIN90KO = 25.87 ± 3.69% (n = 8 cells); [600AP: dendrite]5xFAD-SPIN90WT = −5.48 ± 0.62% (n = 10 cells), [600AP: dendrite]5xFAD-SPIN90KO = −20.80 ± 2.05% (n = 13 cells). Representative traces of pH-APP with linear fit-line during stimulation of (I) axons and (M) dendrites and after stimulation of (J) axons and (N) dendrites in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (K,O) Mean slopes as determined by linear fitting during stimulation of axons (K) and dendrites (O) in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [Degree of slope: axon]5xFAD-SPIN90WT = 0.33 ± 0.05 (n = 10 cells); [Degree of slope: axon]5xFAD-SPIN90KO = 0.97 ± 0.15 (n = 14 cells); [Degree of slope: dendrite]5xFAD-SPIN90WT = −0.17 ± 0.05 (n = 11 cells); [Degree of slope: dendrite]5xFAD-SPIN90KO = −0.65 ± 0.11 (n = 13 cells). (L,P) Mean slopes as determined by linear fitting after stimulation of axons (L) and dendrites (P) in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [Degree of slope: axon]5xFAD-SPIN90WT = −0.057 ± 0.01 (n = 10 cells); [Degree of slope: axon]5xFAD-SPIN90KO = −0.18 ± 0.03 (n = 14 cells); [Degree of slope: dendrite]5xFAD-SPIN90WT = 0.032 ± 0.009 (n = 11 cells); [Degree of slope: dendrite]5xFAD-SPIN90KO = 0.16 ± 0.03 (n = 13 cells). (Q) Schematic illustration of activity-driven pH-APP recycling in axons (endosome-surface-endosome). (R) Schematic illustration of activity-driven pH-APP recycling in dendrites (surface-endosome-surface). ** p < 0.01, *** p < 0.001.
![Ijms 23 10563 g003]()
Figure 4.
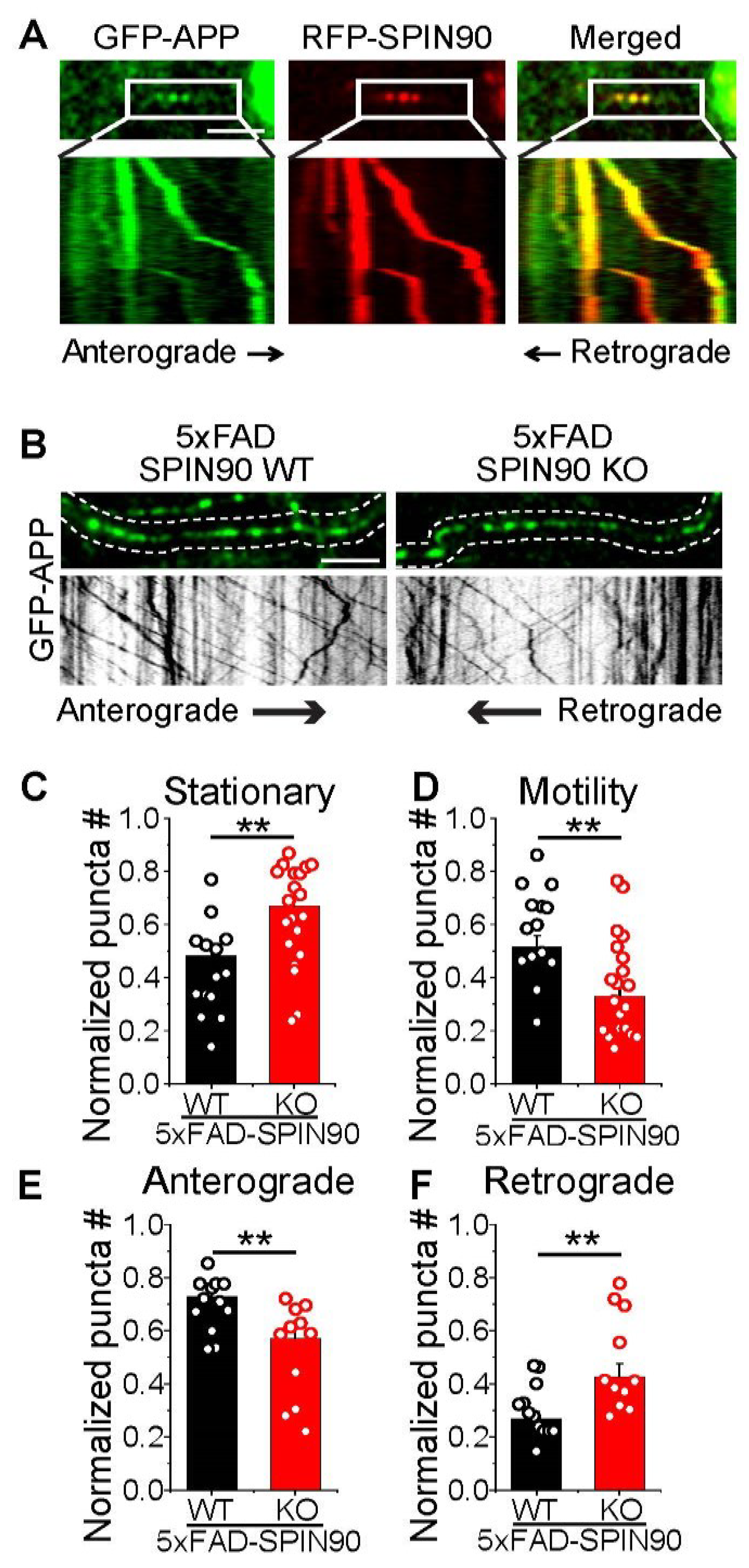
SPIN90 modulates axonal motility of APP. (A) Representative images of GFP-APP and RFP-SPIN90 in rat hippocampal neurons. Selected images from the boxed area in top and kymographs (bottom) of GFP-APP and RFP-SPIN90. Scale bar: 5 μm. (B) (Top) Representative images of GFP-APP in 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons. (Bottom) Kymographs of GFP-APP in 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons. Scale bar: 10 μm. (C) Mean stationary APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [stationary APP]5xFAD-SPIN90WT = 0.48 ± 0.04 (n = 14 cells); [stationary APP]5xFAD-SPIN90KO = 0.67 ± 0.04 (n = 20 cells). (D) Mean motile APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [motility APP]5xFAD-SPIN90WT = 0.52 ± 0.04 (n = 14 cells); [motility APP]5xFAD-SPIN90KO = 0.33 ± 0.04 (n = 20 cells). (E) Mean anterograde transport of APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [anterograde transport of APP]5xFAD-SPIN90WT = 0.73 ± 0.03 (n = 12 cells); [anterograde transport of APP]5xFAD-SPIN90KO = 0.57 ± 0.05 (n = 11 cells). (F) Mean retrograde transport of APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [retrograde transport of APP]5xFAD-SPIN90WT = 0.27 ± 0.03 (n = 12 cells); [retrograde transport of APP]5xFAD-SPIN90KO = 0.43 ± 0.05 (n = 11 cells). ** p < 0.01. #; number.
Figure 4.
SPIN90 modulates axonal motility of APP. (A) Representative images of GFP-APP and RFP-SPIN90 in rat hippocampal neurons. Selected images from the boxed area in top and kymographs (bottom) of GFP-APP and RFP-SPIN90. Scale bar: 5 μm. (B) (Top) Representative images of GFP-APP in 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons. (Bottom) Kymographs of GFP-APP in 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons. Scale bar: 10 μm. (C) Mean stationary APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [stationary APP]5xFAD-SPIN90WT = 0.48 ± 0.04 (n = 14 cells); [stationary APP]5xFAD-SPIN90KO = 0.67 ± 0.04 (n = 20 cells). (D) Mean motile APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [motility APP]5xFAD-SPIN90WT = 0.52 ± 0.04 (n = 14 cells); [motility APP]5xFAD-SPIN90KO = 0.33 ± 0.04 (n = 20 cells). (E) Mean anterograde transport of APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [anterograde transport of APP]5xFAD-SPIN90WT = 0.73 ± 0.03 (n = 12 cells); [anterograde transport of APP]5xFAD-SPIN90KO = 0.57 ± 0.05 (n = 11 cells). (F) Mean retrograde transport of APP in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [retrograde transport of APP]5xFAD-SPIN90WT = 0.27 ± 0.03 (n = 12 cells); [retrograde transport of APP]5xFAD-SPIN90KO = 0.43 ± 0.05 (n = 11 cells). ** p < 0.01. #; number.
![Ijms 23 10563 g004]()
Figure 5.
Depletion of SPIN90 regulates internal accumulation of APP in 5xFAD neurons. (A) Representative images of pH-APP accumulation in 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons. Cells transfected with pH-APP were fixed at DIV 14~17. Scale bar: 20 μm. (B) Magnified images of the boxed area in (A) at rest (top), during acid quenching (middle), and during NH4Cl application (bottom). Scale bar: 5 μm. (C) Pie chart showing the percentages of cells with/without APP endosomal accumulation. [Cell without APP endosomal accumulation]5xFAD-SPIN90WT = 9.09% (n = 11 cells); [Cell without APP endosomal accumulation]5xFAD-SPIN90KO = 43.75% (n = 16 cells); [Cell with APP endosomal accumulation]5xFAD-SPIN90WT = 90.91% (n = 11 cells); [Cell with APP endosomal accumulation]5xFAD-SPIN90KO = 56.25% (n = 16 cells). (D) Mean pH-APP accumulation in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [pH-APP accumulation/Cell]5xFAD-SPIN90WT = 3.18 ± 0.6 (n = 11 cells); [pH-APP accumulation/Cell]5xFAD-SPIN90KO = 0.88 ± 0.26 (n = 16 cells). (E) Cumulative pH-APP accumulation in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (F) Mean sizes of endosomal pH-APP accumulation in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [size of pH-APP accumulation]5xFAD-SPIN90WT = 179.34 ± 17.03 pixel (n = 35 puncta); [size of pH-APP accumulation]5xFAD-SPIN90KO = 105 ± 7.63 pixel (n = 14 puncta). (G) Cumulative sizes of endosomal pH-APP accumulation in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. ** p < 0.01, *** p < 0.001.
Figure 5.
Depletion of SPIN90 regulates internal accumulation of APP in 5xFAD neurons. (A) Representative images of pH-APP accumulation in 5xFAD-SPIN90 WT (left) and 5xFAD-SPIN90 KO (right) neurons. Cells transfected with pH-APP were fixed at DIV 14~17. Scale bar: 20 μm. (B) Magnified images of the boxed area in (A) at rest (top), during acid quenching (middle), and during NH4Cl application (bottom). Scale bar: 5 μm. (C) Pie chart showing the percentages of cells with/without APP endosomal accumulation. [Cell without APP endosomal accumulation]5xFAD-SPIN90WT = 9.09% (n = 11 cells); [Cell without APP endosomal accumulation]5xFAD-SPIN90KO = 43.75% (n = 16 cells); [Cell with APP endosomal accumulation]5xFAD-SPIN90WT = 90.91% (n = 11 cells); [Cell with APP endosomal accumulation]5xFAD-SPIN90KO = 56.25% (n = 16 cells). (D) Mean pH-APP accumulation in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [pH-APP accumulation/Cell]5xFAD-SPIN90WT = 3.18 ± 0.6 (n = 11 cells); [pH-APP accumulation/Cell]5xFAD-SPIN90KO = 0.88 ± 0.26 (n = 16 cells). (E) Cumulative pH-APP accumulation in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. (F) Mean sizes of endosomal pH-APP accumulation in 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [size of pH-APP accumulation]5xFAD-SPIN90WT = 179.34 ± 17.03 pixel (n = 35 puncta); [size of pH-APP accumulation]5xFAD-SPIN90KO = 105 ± 7.63 pixel (n = 14 puncta). (G) Cumulative sizes of endosomal pH-APP accumulation in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons. ** p < 0.01, *** p < 0.001.
![Ijms 23 10563 g005]()
Figure 6.
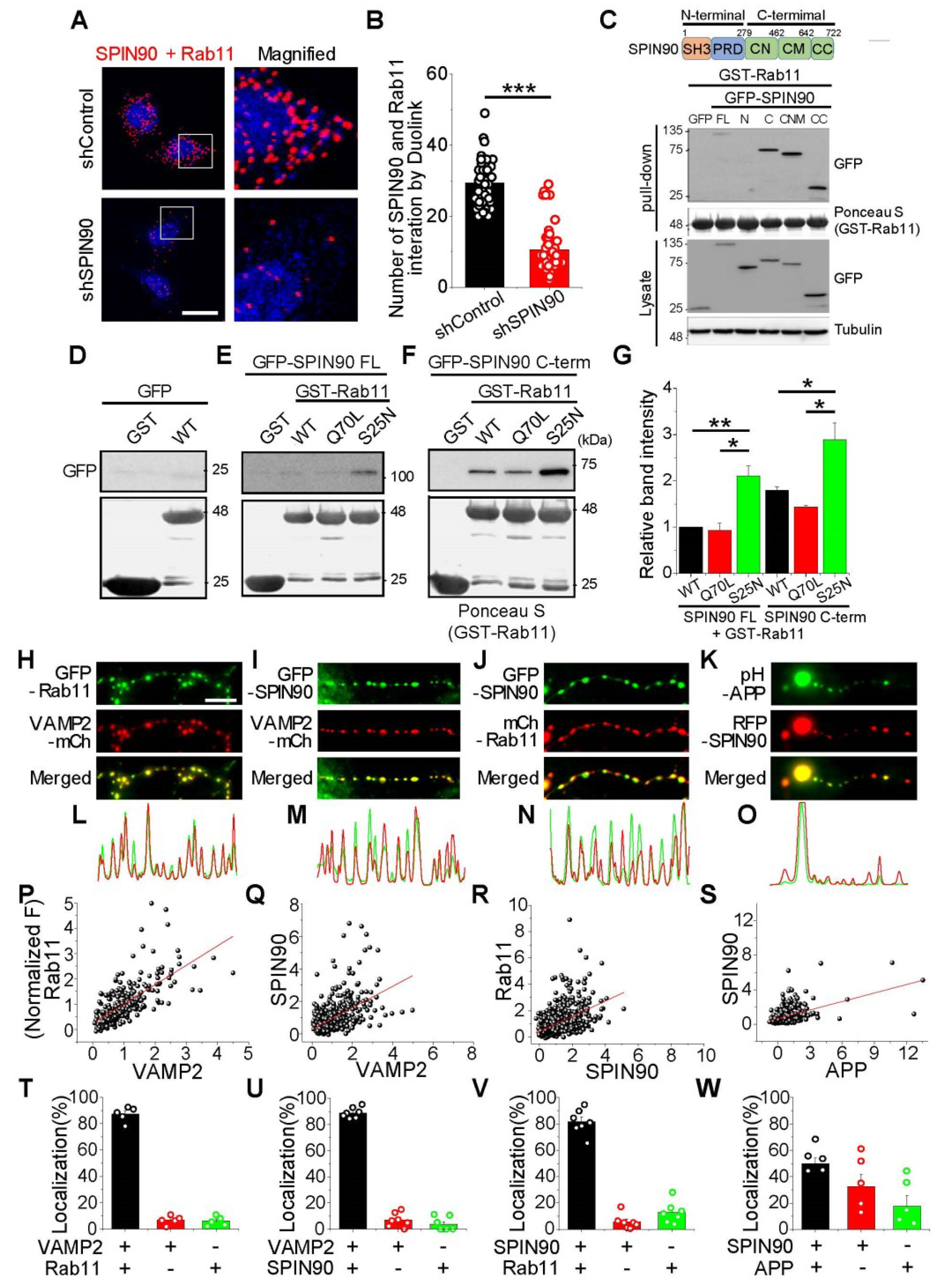
SPIN90 interacts with inactive Rab11. (A) Left: Representative images of SPIN90 and Rab11 interaction by Duolink in HeLa cells bearing shControl (top) and shSPIN90 (bottom). Right: Magnified image of the box areas. Red puncta indicate in situ interactions between SPIN90 and Rab11. (B) Mean numbers of puncta in shControl and shSPIN90-HeLa cells. [Puncta #]shControl = 20.25 ± 0.05 (564 cells, n = 4 experiments), [Puncta #]shSPIN90 = 9.92 ± 0.58 (272 cells, n = 4 experiments). scale bar: 20 μm. (C) Representative immunoblot of SPIN90 and Rab11 interactions, as shown by GST-binding assays. Cells transfected with GFP-SPIN90 full-length (FL), N-term (N), C-term (C), CN+CM (CNM), and CC domain were lysed and the lysates subsequently incubated with GST-Rab11 protein. GST-Rab11 was pull-downed and interactions were verified by immunoblotting with anti-GFP antibody. (D–F) Representative immunoblots of the interactions of SPIN90 and Rab11. Interactions of the active/inactive forms of Rab11 with full-length SPIN90 (E), or with its C-terminus (F). (G) Quantification of relative band intensities in (E,F). The intensities, normalized to the band intensities of GST-Rab11 WT and SPIN90 FL. [GST-Rab11 Q70L]SPIN90 FL = 0.93 ± 0.16; [GST-Rab11 S25N]SPIN90 FL = 2.11 ± 0.22; [GST-Rab11 WT]SPIN90 C-term = 1.8 ± 0.07; [GST-Rab11 Q70L]SPIN90 C-term = 1.43 ± 035; [GST-Rab11 S25N]SPIN90 C-term = 2.89 ± 0.36 (n = 3). (H–K) Representative images of Rab11, SPIN90, APP, and VAMP2 in hippocampal neurons. Neurons were cotransfected with GFP-Rab11/VAMP2-mCh (H), GFP-SPIN90/VAMP2-mCh (I), GFP-SPIN90/mCh-Rab11 (J) and pH-APP/RFP-SPIN90 (K) at DIV 8 and fixed at DIV 14–16. (L–O) Line scans of each image. (P–S) Distribution and correlation of expression levels of GFP-Rab11/VAMP2-mCh (P: n = 277 synapses), GFP-SPIN90/VAMP2-mCh (Q: n = 470 synapses), GFP-SPIN90/mCh-Rab11 (R: n = 887 synapses) and pH-APP/RFP-SPIN90 (S: n = 330 synapses). (T–W) Quantification of colocalization of corresponding proteins (T: GFP-Rab11/VAMP2-mCh, U: GFP-SPIN90/VAMP2-mCh, V: GFP-SPIN90/mCh-Rab11, and W: pH-APP/RFP-SPIN90). * p < 0.05, ** p < 0.01 *** p < 0.001.
Figure 6.
SPIN90 interacts with inactive Rab11. (A) Left: Representative images of SPIN90 and Rab11 interaction by Duolink in HeLa cells bearing shControl (top) and shSPIN90 (bottom). Right: Magnified image of the box areas. Red puncta indicate in situ interactions between SPIN90 and Rab11. (B) Mean numbers of puncta in shControl and shSPIN90-HeLa cells. [Puncta #]shControl = 20.25 ± 0.05 (564 cells, n = 4 experiments), [Puncta #]shSPIN90 = 9.92 ± 0.58 (272 cells, n = 4 experiments). scale bar: 20 μm. (C) Representative immunoblot of SPIN90 and Rab11 interactions, as shown by GST-binding assays. Cells transfected with GFP-SPIN90 full-length (FL), N-term (N), C-term (C), CN+CM (CNM), and CC domain were lysed and the lysates subsequently incubated with GST-Rab11 protein. GST-Rab11 was pull-downed and interactions were verified by immunoblotting with anti-GFP antibody. (D–F) Representative immunoblots of the interactions of SPIN90 and Rab11. Interactions of the active/inactive forms of Rab11 with full-length SPIN90 (E), or with its C-terminus (F). (G) Quantification of relative band intensities in (E,F). The intensities, normalized to the band intensities of GST-Rab11 WT and SPIN90 FL. [GST-Rab11 Q70L]SPIN90 FL = 0.93 ± 0.16; [GST-Rab11 S25N]SPIN90 FL = 2.11 ± 0.22; [GST-Rab11 WT]SPIN90 C-term = 1.8 ± 0.07; [GST-Rab11 Q70L]SPIN90 C-term = 1.43 ± 035; [GST-Rab11 S25N]SPIN90 C-term = 2.89 ± 0.36 (n = 3). (H–K) Representative images of Rab11, SPIN90, APP, and VAMP2 in hippocampal neurons. Neurons were cotransfected with GFP-Rab11/VAMP2-mCh (H), GFP-SPIN90/VAMP2-mCh (I), GFP-SPIN90/mCh-Rab11 (J) and pH-APP/RFP-SPIN90 (K) at DIV 8 and fixed at DIV 14–16. (L–O) Line scans of each image. (P–S) Distribution and correlation of expression levels of GFP-Rab11/VAMP2-mCh (P: n = 277 synapses), GFP-SPIN90/VAMP2-mCh (Q: n = 470 synapses), GFP-SPIN90/mCh-Rab11 (R: n = 887 synapses) and pH-APP/RFP-SPIN90 (S: n = 330 synapses). (T–W) Quantification of colocalization of corresponding proteins (T: GFP-Rab11/VAMP2-mCh, U: GFP-SPIN90/VAMP2-mCh, V: GFP-SPIN90/mCh-Rab11, and W: pH-APP/RFP-SPIN90). * p < 0.05, ** p < 0.01 *** p < 0.001.
![Ijms 23 10563 g006]()
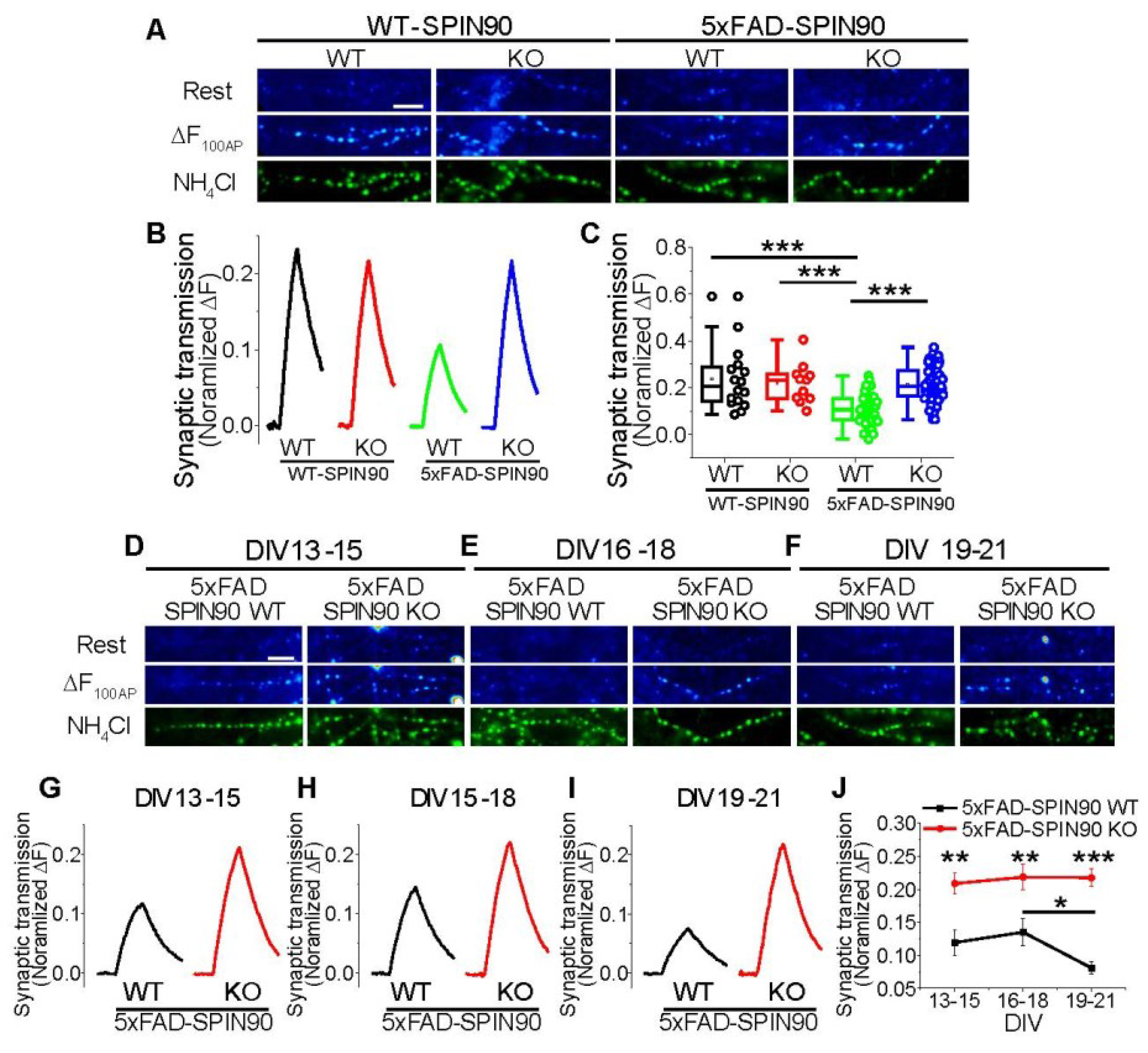
Figure 7.
SPIN90 deficiency restores synaptic function in 5xFAD neurons. (A) Representative synapse images of vG-pH at rest (top) and differences from 100 AP-treated (ΔF100AP, middle), and NH4Cl-treated (bottom) neurons from WT-SPIN90 WT, WT-SPIN90 KO, 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO mice. Scale bar: 5 μm. (B) Representative traces of vG-pH in response to 100AP in WT-SPIN90 WT (black), WT-SPIN90 KO (red), 5xFAD-SPIN90 WT (green) and 5xFAD-SPIN90 KO (blue) neurons. (C) Mean synaptic transmission in WT-SPIN90 WT, WT-SPIN90 KO, 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [synaptic transmission]WT-SPIN90WT = 0.24 ± 0.03 (n = 16 cells); [synaptic transmission]WT-SPIN90KO = 0.22 ± 0.026 (n = 11 cells); [synaptic transmission]5xFAD-SPIN90WT = 0.11 ± 0.01 (n = 46 cells); [synaptic transmission]5xFAD-SPIN90KO = 0.21 ± 0.01 (n = 63 cells). (D–F) Representative synapse images of vG-pH at rest (top) and differences from 100 AP treated (ΔF100AP, middle), and NH4Cl treated (bottom) neurons from 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO mice at DIVs (D) 13–15, (E) 16–18, and (F) 19–21. Scale bar: 5 μm. (G–I) Representative traces of vG-pH in response to 100AP in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons at DIVs (G) 13–15, (H) 16–18, and (I) 19–21. (J) The mean synaptic transmission in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons in various time window at DIVs 13–15, 16–18 and 19–21. [synaptic transmission: DIV 13–15]5xFAD-SPIN90WT = 0.12 ± 0.02 (n = 15 cells), [synaptic transmission: DIV 16–18]5xFAD-SPIN90WT = 0.13 ± 0.02 (n = 12 cells), [synaptic transmission: DIV 19–21]5xFAD-SPIN90WT = 0.08 ± 0.01 (n = 17 cells), [synaptic transmission: DIV 13–15]5xFAD-SPIN90KO = 0.21 ± 0.02 (n = 22 cells), [synaptic transmission: DIV 16–18]5xFAD-SPIN90KO = 0.22 ± 0.02 (n = 19 cells), [synaptic transmission: DIV 19-21]5xFAD-SPIN90KO = 0.22 ± 0.01 (n = 22 cells). * p < 0.05, ** p < 0.01, *** p < 0.001.
Figure 7.
SPIN90 deficiency restores synaptic function in 5xFAD neurons. (A) Representative synapse images of vG-pH at rest (top) and differences from 100 AP-treated (ΔF100AP, middle), and NH4Cl-treated (bottom) neurons from WT-SPIN90 WT, WT-SPIN90 KO, 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO mice. Scale bar: 5 μm. (B) Representative traces of vG-pH in response to 100AP in WT-SPIN90 WT (black), WT-SPIN90 KO (red), 5xFAD-SPIN90 WT (green) and 5xFAD-SPIN90 KO (blue) neurons. (C) Mean synaptic transmission in WT-SPIN90 WT, WT-SPIN90 KO, 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO neurons. [synaptic transmission]WT-SPIN90WT = 0.24 ± 0.03 (n = 16 cells); [synaptic transmission]WT-SPIN90KO = 0.22 ± 0.026 (n = 11 cells); [synaptic transmission]5xFAD-SPIN90WT = 0.11 ± 0.01 (n = 46 cells); [synaptic transmission]5xFAD-SPIN90KO = 0.21 ± 0.01 (n = 63 cells). (D–F) Representative synapse images of vG-pH at rest (top) and differences from 100 AP treated (ΔF100AP, middle), and NH4Cl treated (bottom) neurons from 5xFAD-SPIN90 WT and 5xFAD-SPIN90 KO mice at DIVs (D) 13–15, (E) 16–18, and (F) 19–21. Scale bar: 5 μm. (G–I) Representative traces of vG-pH in response to 100AP in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons at DIVs (G) 13–15, (H) 16–18, and (I) 19–21. (J) The mean synaptic transmission in 5xFAD-SPIN90 WT (black) and 5xFAD-SPIN90 KO (red) neurons in various time window at DIVs 13–15, 16–18 and 19–21. [synaptic transmission: DIV 13–15]5xFAD-SPIN90WT = 0.12 ± 0.02 (n = 15 cells), [synaptic transmission: DIV 16–18]5xFAD-SPIN90WT = 0.13 ± 0.02 (n = 12 cells), [synaptic transmission: DIV 19–21]5xFAD-SPIN90WT = 0.08 ± 0.01 (n = 17 cells), [synaptic transmission: DIV 13–15]5xFAD-SPIN90KO = 0.21 ± 0.02 (n = 22 cells), [synaptic transmission: DIV 16–18]5xFAD-SPIN90KO = 0.22 ± 0.02 (n = 19 cells), [synaptic transmission: DIV 19-21]5xFAD-SPIN90KO = 0.22 ± 0.01 (n = 22 cells). * p < 0.05, ** p < 0.01, *** p < 0.001.
![Ijms 23 10563 g007]()