Glycyrrhizic Acid Derivatives Bearing Amino Acid Residues in the Carbohydrate Part as Dengue Virus E Protein Inhibitors: Synthesis and Antiviral Activity
Abstract
:1. Introduction
2. Results
2.1. Synthesis of GL Conjugates with Amino Acids and Their Methyl/Ethyl Esters
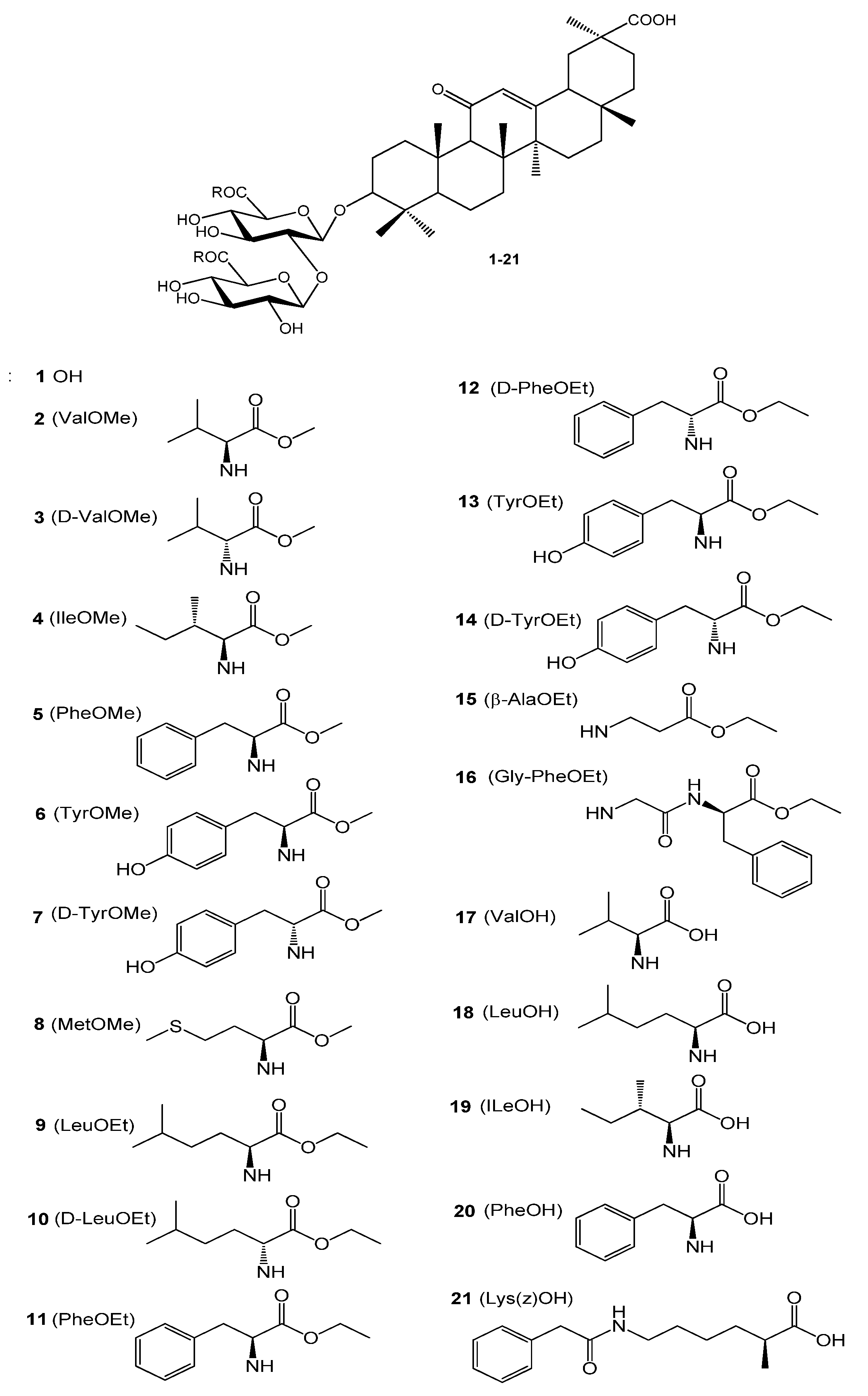
2.2. Antiviral Screening of GL Derivatives in CPE Assay
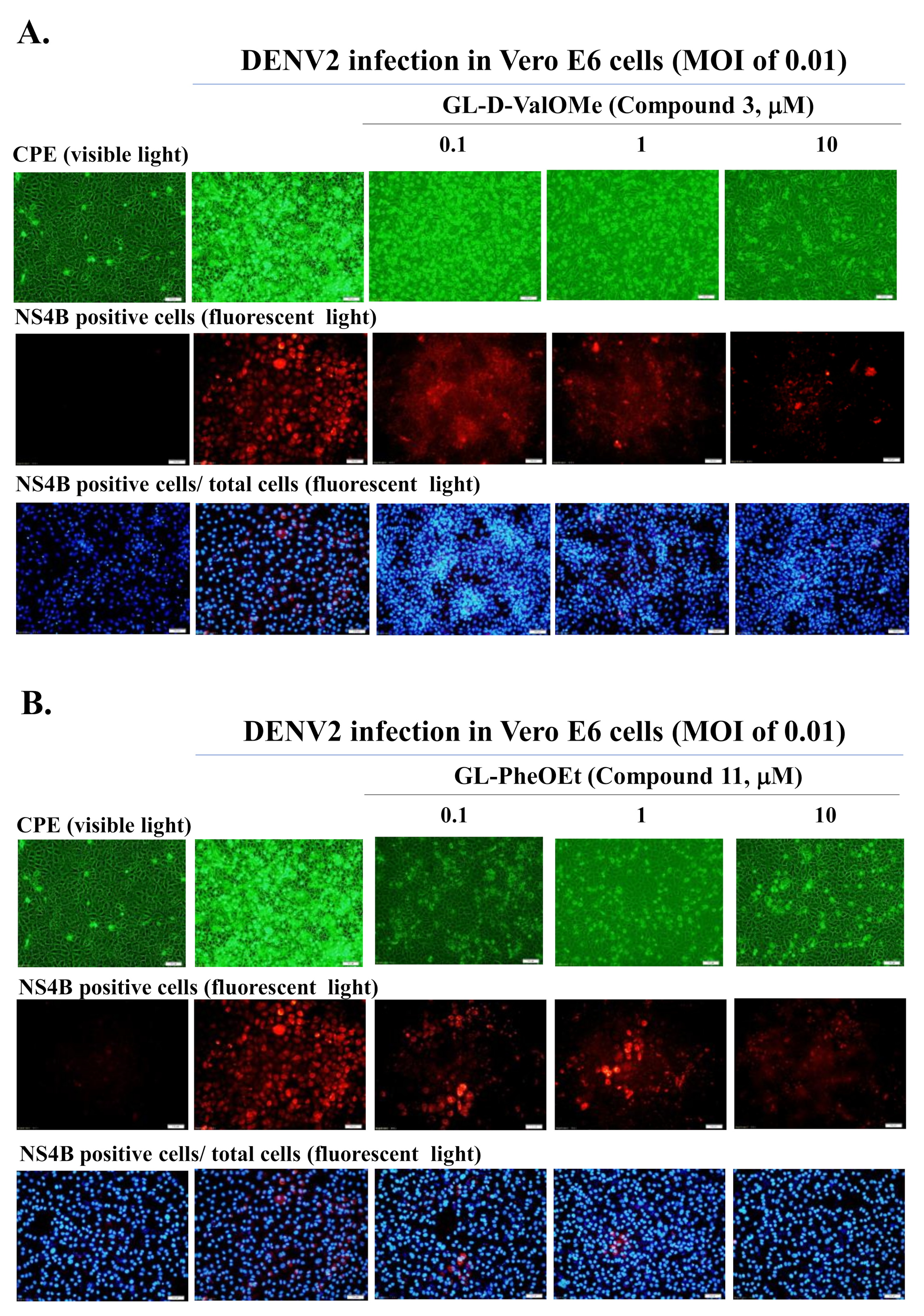
2.3. Antiviral Activity of GL Derivatives in DENV Infectivity Assay
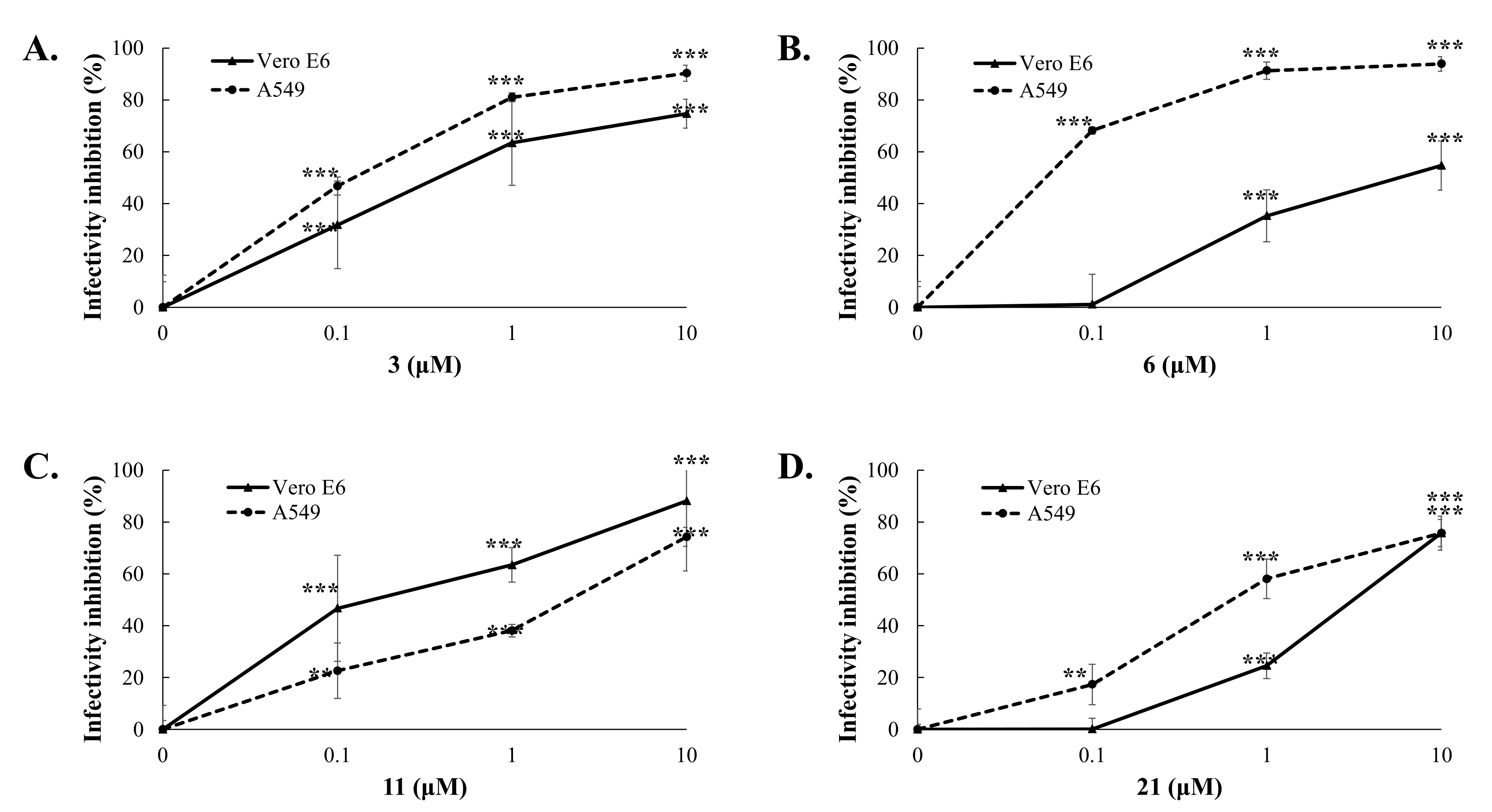
2.4. Cytotoxicity and Antiviral Potency of Active GL Conjugates
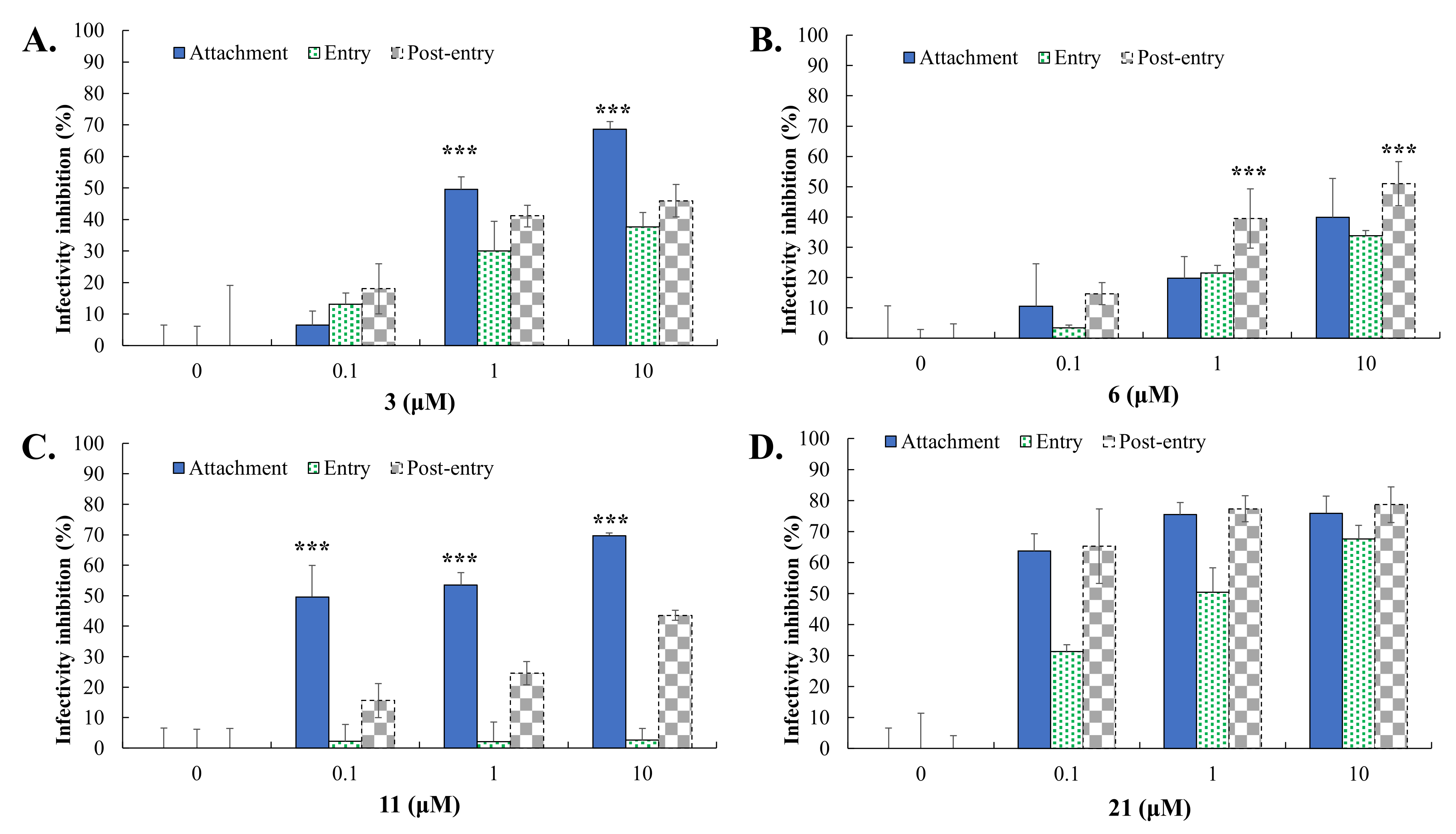
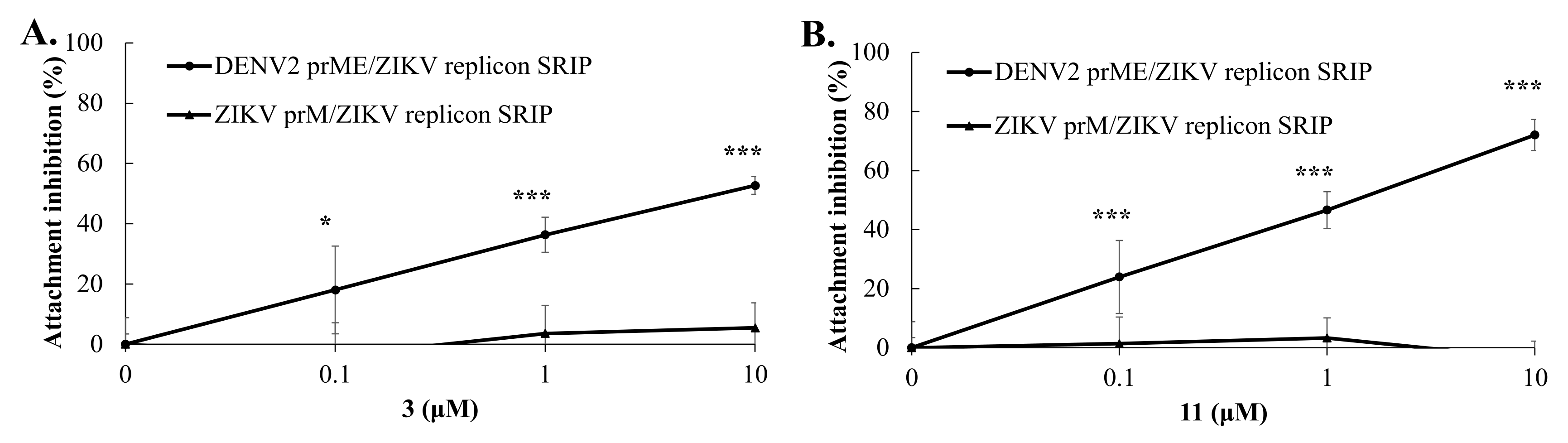
2.5. Virus Yield Reduction and Attachment Inhibition by Active GL Derivatives
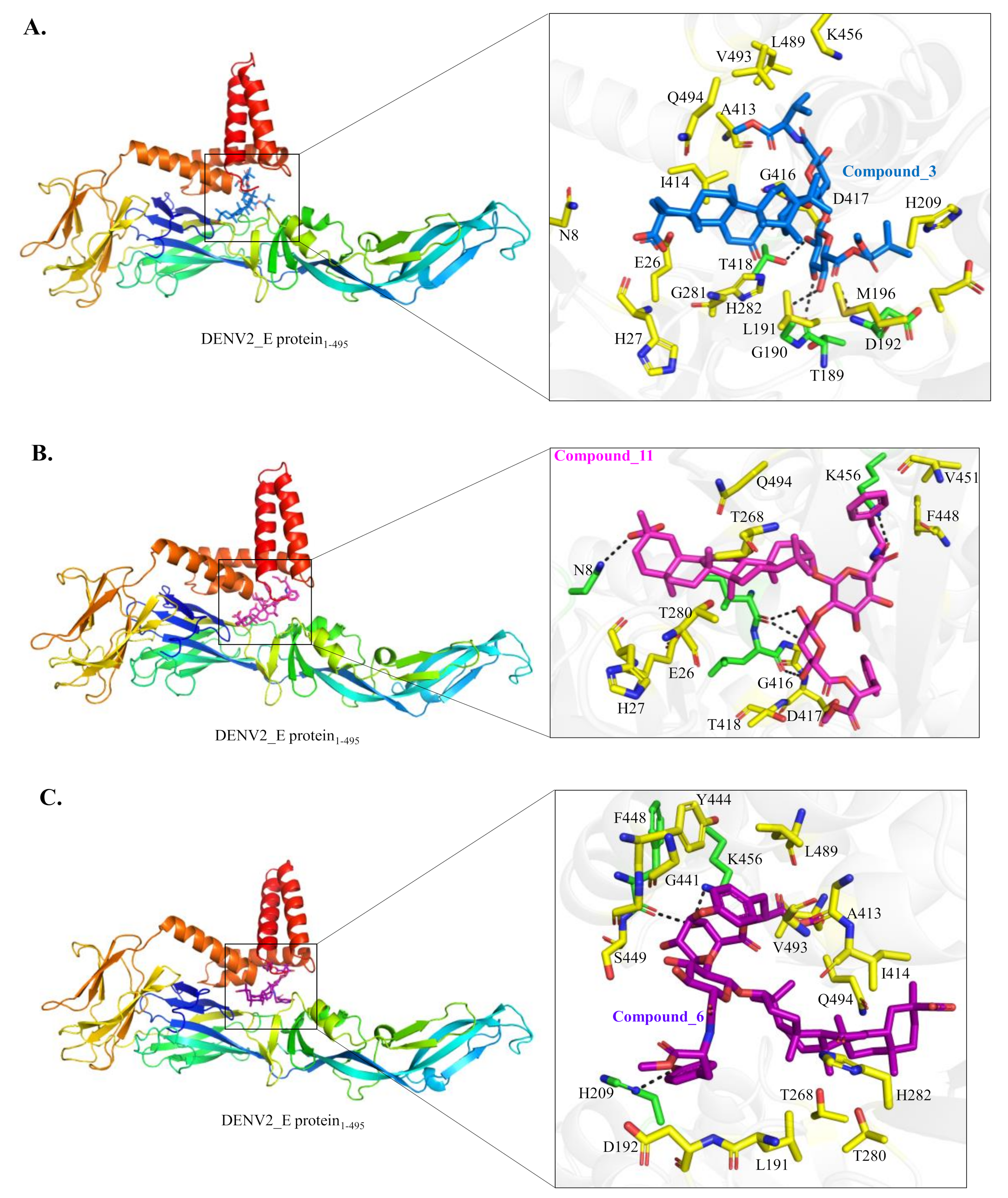
2.6. Target Identification of Active GL Derivative Hits 3 and 11 against DENV
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures for Chemical Synthesis
4.2. Chemicals
4.3. A General Procedure to Synthesize Compounds 2–5
4.4. A General Procedure to Synthesize Compounds 6–16
4.5. A General Procedure to Synthesize Compounds 17–21
4.6. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-valine methyl ester]-N-(β-D-glucopyranosyluronoyl)-L-valine methyl ester}-(3β, 20β)-11-oxo-30-norolean-12-ene-30-oic Acid (2) [42]
4.7. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-D-valine methyl ester]-N-(β-D-glucopyranosyluronoyl)-D-valine methyl ester}-(3β, 20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (3) [43]
4.8. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-isoleucine methyl ester]-N-(β-D-glucopyranosyluronoyl)-L-isoleucine methyl ester}-(3β, 20β)-11-oxo-30-norolean-12-ene-30-oic Acid (4)
4.9. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-phenylalanine methyl ester]-N-(β-D-glucopyranosyluronoyl)-L-phenylalanine methyl ester}-(3β, 20β)-11-oxo-30-norolean-12-ene-30-oic Acid (5) [42]
4.10. 3-O-{2-O-[N-(β-D-glucopyranosyluronoyl)-L-tyrosine methyl ester]-N-(β-D-glucopyranosyluronoyl)-L-tyrosine methyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic acid (6) [42,46]
4.11. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-D-tyrosine methyl ester]-N-(β-D-glucopyranosyluronoyl)-D-tyrosine methyl ester}-(3β, 20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (7)
4.12. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-methionine methyl ester]-N-(β-D-glucopyranosyluronoyl)-L-methionine methyl ester}-(3β, 20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (8) [50]
4.13. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-leucine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-L-leucine ethyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (9) [46]
4.14. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-D-leucine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-D-leucine ethyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (10)
4.15. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-phenylalanine ethyl ester]-N-(β-D-Glucopyranosyluronoyl)-L-phenylalanine ethyl ester}-(3β, 20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (11) [46]
4.16. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-D-phenylalanine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-D-phenylalanine ethyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (12)
4.17. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-tyrosine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-L-tyrosine ethyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (13)
4.18. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-D-tyrosine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-D-tyrosine ethyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-acid (14)
4.19. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-β-alanine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-β-alanine ethyl ester}- (3β, 20β)-11-oxo-30-noroleane-12-ene-30-acid (15)
4.20. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-glycyl-L-phenylalanine ethyl ester]-N-(β-D-glucopyranosyluronoyl)-glycyl-L-phenylalanine ethyl ester}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-acid (16) [46]
4.21. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-valine]-N-(β-D-glucopyranosyluronoyl)-L-valine}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (17) [47]
4.22. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-leucine]-N-(β-D-glucopyranosyluronoyl)-L-leucine}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (18) [46,47]
4.23. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-isoleucine]-N-(β-D-glucopyranosyluronoyl)-L-isoleucine}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (19) [47,49]
4.24. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-phenylalanine]-N-(β-D-glucopyranosyluronoyl)-L-phenylalanine}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (20) [47,49]
4.25. 3-O-{2-O-[N-(β-D-Glucopyranosyluronoyl)-L-lysine(ϵ-carbobenzoxy)]-N-(β-D-glucopyranosyluronoyl)-L-lysine(ϵ-carbobenzoxy)}-(3β,20β)-11-oxo-30-noroleane-12-ene-30-oic Acid (21) [46,51]
4.26. Viruses and Cells
4.27. Cytotoxicity Assay
4.28. Antiviral Activity Assays
4.29. DENV2 Yield Reduction Assay
4.30. Time-of-Addition/Compound Removal Assay
4.31. Targeting Viral E Protein-Mediated Attachment Assay with Chimeric DENV2 CprME/JEV and Chimeric DENV2 prME/ZIKV Single-Round Infectious Particles (SRIPs)
4.32. Docking Studies to DENV-2 E Protein
4.33. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GL | Glycyrrhizic acid |
| DENV | Dengue virus |
| ZIKV | Zika virus |
| JEV | Japanese Encephalitis Virus |
| WNV | West Nile virus |
| RdRp | RNA-dependent RNA polymerase |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl)carbodiimid hydrochloride |
| DMF | N,N′-Dimethylformamide |
| HOSu | N-hydroxysuccinimide |
| DCC | N,N′-dicyclohexylcarbodiimide |
| IR | Infrared |
| NMR | Nuclear magnetic resonance |
| Hz | Hertz |
| J | Coupling constants |
| s | Singlet |
| d | Doublet |
| m | Multiplet |
| DMSO | Dimethyl sulfoxide |
| EtOH | Ethanol |
| MeOH | Methanol |
| Me | Methyl |
| Z | Carbobenzoxy |
| CC | Column chromatography |
| TLC | Thin-layer chromatography |
| HPLC | High-performance liquid chromatography |
| τ | Retention time |
| CPE | Cytopathic effect |
| IFA | Immunofluorescence assay |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide |
| IC50 | 50% inhibitory concentration |
| CC50 | 50% cytotoxic concentration |
| SI | Selectivity index |
| TCID50 | 50%Tissue culture infectious dose |
| MOI | multiplicity of infection |
| DAPI | 4′,6-diamidino-2-phenylindole |
| PBS | Phosphate-buffered saline |
| C-prM-E | capsid-pr-membrane-envelope protein |
| SRIP | single-round infectious particle |
| BHK-21 | Baby Hamster Kidney-21 |
| CMV | Cytomegalovirus |
| PCR | polymerase chain reaction |
References
- Daep, C.A.; Muñoz-Jordán, J.L.; Eugenin, E.A. Flaviviruses, an expanding threat in public health: Focus on dengue, West Nile, and Japanese encephalitis virus. J. Neurovirol. 2014, 20, 539–560. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Charlier, C.; Vasilakis, N.; Lecuit, M. Zika, Chikungunya, and other emerging vector-borne viral diseases. Annu. Rev. Med. 2018, 69, 395–408. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.-M.; Musso, D. Emerging arboviruses in the Pacific. Lancet 2014, 384, 1571–1572. [Google Scholar] [CrossRef]
- Gubler, D.J. Dengue, urbanization and globalization: The unholy trinity of the 21st century. Trop. Med. Health 2011, 39, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.P.; Farrar, J.J.; Chau, N.; Wills, B. Dengue. N. Engl. J. Med. 2012, 366, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Wilder-Smith, A.; Ooi, E.-E.; Vasudevan, S.G.; Gubler, D.J. Update on Dengue: Epidemiology, Virus Evolution, Antiviral Drugs, and Vaccine Development. Curr. Infect. Dis. Rep. 2010, 12, 157–164. [Google Scholar] [CrossRef]
- WHO/PAHO Epidemiological Update Dengue and other Arboviruses. Available online: https://reliefweb.int/report/argentina/epidemiological-update-dengue-and-other-arboviruses-10-june-2020 (accessed on 28 July 2022).
- World Health Organization. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control; World Health Organization: Geneva, Switzerland, 2009. [Google Scholar]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 507–594. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Mohammed, A.T.; Vu, T.T.; Khattab, M.; Dohein, M.F.; Mohamed, A.A.; Abdelhamed, M.M.; Shamandy, B.E.; Dawod, M.T.; Alesaei, W.A.; et al. Prevelance and burden of dengue infection in Europe: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 30, e2093. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.J.; Gahan, M.E.; Mahalingam, S.; Keller, P.A. The medicinal chemistry of Dengue Fever. J. Med. Chem. 2009, 52, 7911–7926. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Pierson, T.C. Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell 2015, 162, 488–492. [Google Scholar] [CrossRef] [Green Version]
- Zonetti, L.F.C.; Coutinho, M.C.; De Araujo, A.S. Molecular aspects of Denge virus infection process: A review. Protein Pept. Lett. 2018, 25, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Murrell, S.; Wu, S.-C.; Butler, M. Review of dengue virus and development of a vaccine. Biotechnol. Adv. 2011, 29, 239–247. [Google Scholar] [CrossRef]
- Kim, K.-S. Current challenges in the development of vaccines and drugs against emerging vector-borne diseases. Curr. Med. Chem. 2019, 26, 2974–2986. [Google Scholar] [CrossRef]
- Abdullah, A.A.; Lee, Y.K.; Chin, S.P.; Lim, S.K.; Lee, V.S.; Othman, R.; Othman, S.; Rahman, N.A.; Yusof, R.; Heh, C.H. Discovery of Dengure virus inhibitors. Curr. Med. Chem. 2020, 27, 4945–5036. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Dimopoulos, G. Antiviral Compounds for Blocking Arboviral Transmission in Mosquitoes. Viruses 2021, 13, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Chen, Y.-L.; Dong, H.; Gu, F.; Lim, S.P.; Schul, W.; Wang, Q.-Y.; Shi, P.-Y. Strategies for development of dengue virus inhibitors. Antiviral Res. 2010, 85, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Sampath, A.; Padmanabhan, R. Molecular targets for flavivirus drug discovery. Antivir. Res. 2009, 81, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Jadav, S.S.; Kaptein, S.; Timiri, A.; De Burghgrave, T.; Badavath, V.N.; Ganesan, R.; Sinha, B.N.; Neyts, J.; Leyssen, P.; Jayaprakash, V. Design, synthesis optimization and antiviral activity of a class of hybrid dengue virus E protein inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Lescar, J.; Luo, D.; Xu, T.; Sampath, A.; Lim, S.P.; Canard, B.; Vasudevan, S.G. Towards the design of antiviral inhibitors against flaviviruses: The case for the multifunctional NS3 protein from Dengue virus as a target. Antivir. Res. 2008, 80, 94–101. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, B.; Shi, P.-Y. Flavirus methyltransferase: A novel antiviral target. Antivir. Res. 2008, 80, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Tomlinson, S.M.; Malmstrom, R.D.; Russo, A.; Mueller, N.; Pang, Y.-P.; Watowich, S.J. Structure-based discovery of dengue virus protease inhibitors. Antivir. Res. 2009, 82, 110–114. [Google Scholar] [CrossRef]
- De Clercq, E.; Li, G. Approved Antiviral Drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef]
- Martinez, J.P.; Sasse, F.; Bronstrup, M.; Diez, J.; Meyerhans, A. Antiviral drug discovery: Broad-spectrum drugs from nature. Nat. Prod. Rep. 2015, 32, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Tahs, M.M.E.; Makeen, H.A.; Alhazmi, H.A.; Al Bratty, M.; Sultana, S.; Ahsan, W.; Najmi, A.; Khalid, A. Bioactive Natural Antivirals: An Updated Review of the Available Plants isolated Molecules. Molecules 2020, 25, 4878–4913. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, R.; Yuan, B.; Liu, Y.; Liu, C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm. Sin. B. 2015, 5, 310–315. [Google Scholar] [CrossRef]
- Tolstikov, G.A.; Baltina, L.A.; Grankina, V.P.; Kondratenko, R.M.; Tolstikova, T.G. Licorice: Biodiversity, Chemistry, and Application in Medicine; Academic Publishing House “Geo”: Novosibirsk, Russia, 2007; pp. 147–186. (In Russian) [Google Scholar]
- Musayeva, S.; Valiyeva, M.; Madatli, F.; Mehraliyeva, S.; Khalilov, R.; Vahedi, P.; Eftekhari, A. Prospects for the development of drugs with antiviral activity based on licorice. Eurasian Chem. Comm. 2021, 3, 301–309. [Google Scholar] [CrossRef]
- Pompei, R.; Laconi, S.; Ingianni, A. Antiviral properties of Glycyrrhizic acid and its semisynthetic derivatives. Mini-Rev. Med. Chem. 2009, 9, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.-G.; Zhao, T.-T.; Lu, N.; Yang, Y.-A.; Zhu, H.-L. Research progress of Glycyrrhizic acid on antiviral activity. Mini-Rev. Med. Chem. 2019, 19, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Baltina, L.A.; Kondratenko, R.M.; Baltina, L.A., Jr.; Plyasunova, O.A.; Pokrovskyi, A.G.; Tolstikov, G.A. Prospects for the creation of new antiviral drugs based on Glycyrrhizic acid and its derivatives. Pharm. Chem. J. 2009, 43, 539–548. [Google Scholar] [CrossRef]
- Baltina, L.; Kondratenko, R. Glycyrrhizic Acid Derivatives as New Antiviral and Immune Modulating Agents (a review). Curr. Bioact. Compds. 2021, 17, 41–58. [Google Scholar] [CrossRef]
- Sato, H.; Goto, W.; Yamamura, J.; Kurokava, M.; Kageyama, S.; Takahara, T.; Watanabe, A.; Shiraki, K. Therapeutic basis of glycyrrhizin in chronic hepatitis B. Antivir. Res. 1996, 30, 171–177. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Matsuura, T.; Aoyagi, H.; Matsuda, M.; Hmwe, S.S.; Date, T.; Watanabe, N.; Watashi, K.; Suzuki, R.; Ichinose, S.; et al. Antiviral activity of Glycyrrhizin against hepatitis C virus in vitro. PLoS ONE 2013, 8, e68992. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, U.A.; Masoud, M.S.; Nawaz, Z.; Riazuddin, S. Glycyrrhizin as antiviral agent against Hepatitis C Virus. J. Transl. Med. 2011, 9, 112. [Google Scholar] [CrossRef]
- Ito, M.; Sato, A.; Hirabayashi, K.; Tanabe, F.; Shigeta, S.; Baba, M.; De Clerq, E.; Nakashima, H.; Yamamoto, N. Mechanism of inhibitory effect of glycyrrhizin on replication of human immunodeficiency virus (HIV). Antivir. Res. 1988, 10, 289–298. [Google Scholar] [CrossRef]
- Ikegami, N.; Kinoshita, S.; Kanesaki, T.; Uno, K.; Akatani, K.; Kisida, T. Evalluation of long term treatment with glycyrrhizin and of combination therapy with glycyrrhizin and AZT or DDI on HIV-1 carries. Antivir. Res. 1996, 30, A33. [Google Scholar] [CrossRef]
- Baltina, L.A. Chemical Modification of Glycyrrhizic Acid As A Route to New Bioactive Compounds for Medicine. Curr. Med. Chem. 2003, 10, 155–171. [Google Scholar] [CrossRef]
- Hoever, G.; Baltina, L.A.; Michaelis, M.; Kondratenko, R.; Baltina, L.A., Jr.; Tolstikov, G.A. Antiviral activity of Glycyrrhizic acid derivatives against SARS-Coronavirus. J. Med. Chem. 2005, 48, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Cherng, J.M.; Hung, M.S.; Baltina, L.A.; Baltina, L.A.; Kondratenko, R.M. Inhibitory effects of some derivatives of Glycyrrhizic acid against Epstein-Barr virus infection: Structure-activity relationships. Antivir. Res. 2008, 79, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Baltina, L.A.; Zarubaev, V.V.; Baltina, L.A.; Orshanskaya, I.A.; Fairushina, A.I.; Kiselev, O.I.; Yunusov, M.S. Glycyrrhizic acid derivatives as influenza A/H1N1 virus inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 1742–1746. [Google Scholar] [CrossRef]
- Fairushina, A.I.; Baltina, L.A., Jr.; Baltina, L.A.; Konovalova, N.I.; Petrova, P.A.; Eropkin, M.Y. Synthesis and antiviral activity of novel Glycyrrhizic acid conjugates with D-amino acid esters. Russ. J. Bioorg. Chem. 2017, 43, 456–462. [Google Scholar] [CrossRef]
- Crance, J.M.; Scaramozzino, N.; Jouan, A.; Garin, D. Interferon, Ribavirin, 6-Azauridine and Glycyrrhizin: Antiviral Compounds Active against Pathogenic Flaviviruses. Antivir. Res. 2003, 58, 73–77. [Google Scholar] [CrossRef]
- Baltina, L.A.; Tasi, Y.-T.; Huang, S.-H.; Lai, H.-C.; Baltina, L.A.; Petrova, S.F.; Yunusov, M.S.; Lin, C.W. Glycyrrhizic acid derivatives as Dengue virus inhibitors. Bioorg. Med. Chem. Lett. 2019, 29, 126645. [Google Scholar] [CrossRef] [PubMed]
- Baltina, L.A.; Hour, M.-J.; Liu, Y.-C.; Chang, Y.-S.; Huang, S.-H.; Lai, H.-C.; Kondratenko, R.M.; Petrova, S.F.; Yunusov, M.S.; Lin, C.-W. Antiviral activity of glycyrrhizic acid conjugates with amino acid esters against Zika virus. Virus Res. 2021, 294, 198290. [Google Scholar] [CrossRef]
- Baltina, L.A.; Sapozhnikova, T.A.; Gabdrakhmanova, S.F.; Makara, N.S.; Khisamutdinova, R.Y.; Baltina, L.A., Jr.; Petrova, S.F.; Saifullina, D.R.; Kondratenko, R.M. Hypoglycemic activity of Glycyrrhizic acid and some of its derivatives in the alloxan diabetes model in rats. Pharm. Chem. J. 2021, 55, 340–344. [Google Scholar] [CrossRef]
- Baltina, L.A.; Baltina, L.A., Jr.; Kondratenko, R.M.; Petrova, S.F. 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide in the synthesis of glycyrrhizic acid amino-acid conjugates. Chem. Nat. Compd. 2020, 56, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Baltina, L.A., Jr.; Kondratenko, R.M.; Baltina, L.A.; Baschenko, N.Z.; Plyasunova, O.A. Synthesis and biological activity of new Glycyrrhizic acid conjugates with amino acids and dipeptides. Russ. J. Bioorg. Chem. 2009, 35, 510–517. [Google Scholar] [CrossRef]
- Baltina, L.A., Jr.; Kondratenko, R.M.; Baltina, L.A.; Plyasunova, O.A.; Galin, F.Z.; Tolstikov, G.A. Synthesis of Glycyrrhizic acid conjugates containing L-lysine. Chem. Nat. Compd. 2006, 42, 543–548. [Google Scholar] [CrossRef]
- Kondratenko, R.M.; Baltina, L.A.; Vasil’eva, E.V.; Nasyrov, K.M.; Kireeva, R.M.; Baschenko, N.Z.; Fridman, S.M.; Baltina, L.A., Jr.; Tolstikov, G.A. Synthesis and immunomodulating activity of new amino acid derivatives of Glycyrrhizic acid and its methyl ester. Russ. J. Bioorg. Chem. 2004, 30, 148–153. [Google Scholar] [CrossRef]
- Smit, J.; Moesker, B.; Rodenhuis-Zybert, I.; Wilschut, J. Flavivirus cell entry and membrane fusion. Viruses 2011, 3, 160–171. [Google Scholar] [CrossRef]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. A ligand-binding pocket in dengue virus ebvelope glycoprotein. Proc. Natl. Acad. Sci. USA 2003, 100, 6986–6991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boldescu, V.; Behnam, M.A.M.; Vasilakis, N.; Klein, C.D. Broad-spectrum agents for flaviviral infrctions: Dengue, Zika and beyond. Nat. Rev. Drug Discov. 2017, 16, 565–586. [Google Scholar] [CrossRef]
- Zhou, Z.; Khaliq, M.; Suk, J.-E.; Patkar, C.; Li, L.; Kuhn, R.J.; Post, C.B. Antiviral compounds discovered by virtual screening of small-molecule libraries against Dengue virus E Protein. Ach. Chem. Biol. 2008, 3, 765–775. [Google Scholar] [CrossRef]
- Kampmann, T.; Yennamalli, R.; Campbell, P.; Stoermer, M.J.; Fairlie, D.P.; Kobe, B.; Young, P.R. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antivir. Res. 2009, 84, 234–241. [Google Scholar] [CrossRef]
- Cuardia, C.; Stephens, D.H.; Dang, H.T.; Quijada, M.; Larionov, O.V.; Lleonart, R. Antiviral activity of novel quinolone derivatives against Dengue virus serotype 2. Molecules 2018, 23, 672. [Google Scholar] [CrossRef]
- Kondratenko, R.M.; Baltina, L.A.; Mustafina, S.R.; Makarova, N.V.; Nasyrov, K.M.; Tolstikov, G.A. Crystalline Glycyrrhizic acid synthesized from commercial Glycyrrham. Immunomodulant properties of high-purity Glycyrrhizic acid. Pharm. Chem. J. 2001, 35, 101–104. [Google Scholar] [CrossRef]
- Baltina, L.A.; Lai, H.C.; Liu, Y.C.; Huang, S.H.; Hour, M.J.; Baltina, L.A.; Nugumanov, T.R.; Borisevich, S.S.; Khalilov, L.M.; Petrova, S.F.; et al. Glycyrrhetinic acid derivatives as Zika virus inhibitors: Synthesis and antiviral activity in vitro. Bioorg. Med. Chem. 2021, 41, 116204. [Google Scholar] [CrossRef] [PubMed]
- Tsypysheva, I.P.; Lai, H.C.; Kiu, Y.T.; Koval’skaya, A.V.; Tsypyshev, D.O.; Huang, S.H.; Lin, C.W. Synthesis and antiviral evaluation of cytisine derivatives against dengue virus types 1 and 2. Bioorg. Med. Chem. Lett. 2021, 54, 128437. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85–86. [Google Scholar] [CrossRef]
- Lu, C.Y.; Hour, M.J.; Wang, C.Y.; Huang, S.H.; Mu, W.X.; Chang, Y.C.; Lin, C.W. Single-Round Infectious Particle Antiviral Screening Assays for the Japanese Encephalitis Virus. Viruses 2017, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.Y.; Lin, C.S.; Lai, H.C.; Yu, Y.W.; Liao, C.Y.; Su, W.C.; Ko, B.H.; Chang, Y.S.; Huang, S.H.; Lin, C.W. The Rescue and Characterization of Recombinant, Microcephaly-Associated Zika Viruses as Single-Round Infectious Particles. Viruses 2019, 11, 1005. [Google Scholar] [CrossRef] [Green Version]
- Hardy, J.M.; Newton, N.D.; Modhiran, N.; Scott, C.; Venugopal, H.; Vet, L.J.; Young, P.R.; Hall, R.A.; Hobson-Peters, J.; Coulibaly, F.; et al. A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses. Nat. Commun. 2021, 12, 3266. [Google Scholar] [CrossRef] [PubMed]


| Compounds | Vero E6 Cells | A549 Cells | ||||||
|---|---|---|---|---|---|---|---|---|
| CPE Reduction at 10 μM a | Inhibition (%) at 10 μM b | Cytotoxicity (CC50, μM) c | Inhibition of DENV2 Infectivity (IC50, μM) e | Selectivity Index (CC50/ IC50) | Cytotoxicity (CC50, μM) c | Inhibition of DENV2 Infectivity (IC50, μM) e | Selectivity Index (CC50/ IC50) | |
| 1 | +++ | 70.5 | >100 μM | 8.1 ± 0.21 | >12.3 | ND | ND | ND |
| 2 | − | ND d | ND | ND | ND | ND | ND | ND |
| 3 | ++++ | 90.7 | >100 μM | 0.50 ± 0.17 | >201.6 | >100 μM | 0.12 ± 0.028 | >833 |
| 4 | − | ND | ND | ND | ND | ND | ND | ND |
| 6 | ++++ | 94.0 | >100 μM | 5.98 ± 0.41 | >16.7 | >100 μM | <0.1 | >1000 |
| 7 | ++ | 51.7 | ND | ND | ND | ND | ND | ND |
| 8 | ++ | 51.6 | ND | ND | ND | ND | ND | ND |
| 9 | +++ | 68.9 | ND | ND | ND | ND | ND | ND |
| 11 | ++++ | 91.3 | >100 μM | 0.18 ± 0.01 | >552 | >100 μM | 1.56 ± 0.49 | >64 |
| 15 | ++ | 53.6 | ND | ND | ND | ND | ND | ND |
| 16 | +++ | 63.8 | ND | ND | ND | ND | ND | ND |
| 18 | +++ | 63.1 | ND | ND | ND | ND | ND | ND |
| 21 | ++++ | 92.8 | >100 μM | 2.68 ± 0.54 | >37.3 | >100 μM | 0.97 ± 0.34 | >103 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hour, M.-J.; Chen, Y.; Lin, C.-S.; Baltina, L.A.; Kan, J.-Y.; Tsai, Y.-T.; Kiu, Y.-T.; Lai, H.-C.; Baltina, L.A.; Petrova, S.F.; et al. Glycyrrhizic Acid Derivatives Bearing Amino Acid Residues in the Carbohydrate Part as Dengue Virus E Protein Inhibitors: Synthesis and Antiviral Activity. Int. J. Mol. Sci. 2022, 23, 10309. https://doi.org/10.3390/ijms231810309
Hour M-J, Chen Y, Lin C-S, Baltina LA, Kan J-Y, Tsai Y-T, Kiu Y-T, Lai H-C, Baltina LA, Petrova SF, et al. Glycyrrhizic Acid Derivatives Bearing Amino Acid Residues in the Carbohydrate Part as Dengue Virus E Protein Inhibitors: Synthesis and Antiviral Activity. International Journal of Molecular Sciences. 2022; 23(18):10309. https://doi.org/10.3390/ijms231810309
Chicago/Turabian StyleHour, Mann-Jen, Yeh Chen, Chen-Sheng Lin, Lidia A. Baltina, Ju-Ying Kan, Yan-Ting Tsai, Yan-Tung Kiu, Hsueh-Chou Lai, Lia A. Baltina, Svetlana F. Petrova, and et al. 2022. "Glycyrrhizic Acid Derivatives Bearing Amino Acid Residues in the Carbohydrate Part as Dengue Virus E Protein Inhibitors: Synthesis and Antiviral Activity" International Journal of Molecular Sciences 23, no. 18: 10309. https://doi.org/10.3390/ijms231810309
APA StyleHour, M.-J., Chen, Y., Lin, C.-S., Baltina, L. A., Kan, J.-Y., Tsai, Y.-T., Kiu, Y.-T., Lai, H.-C., Baltina, L. A., Petrova, S. F., & Lin, C.-W. (2022). Glycyrrhizic Acid Derivatives Bearing Amino Acid Residues in the Carbohydrate Part as Dengue Virus E Protein Inhibitors: Synthesis and Antiviral Activity. International Journal of Molecular Sciences, 23(18), 10309. https://doi.org/10.3390/ijms231810309







