ZNF281 Promotes Colon Fibroblast Activation in TGFβ1-Induced Gut Fibrosis
Abstract
:1. Introduction
2. Results
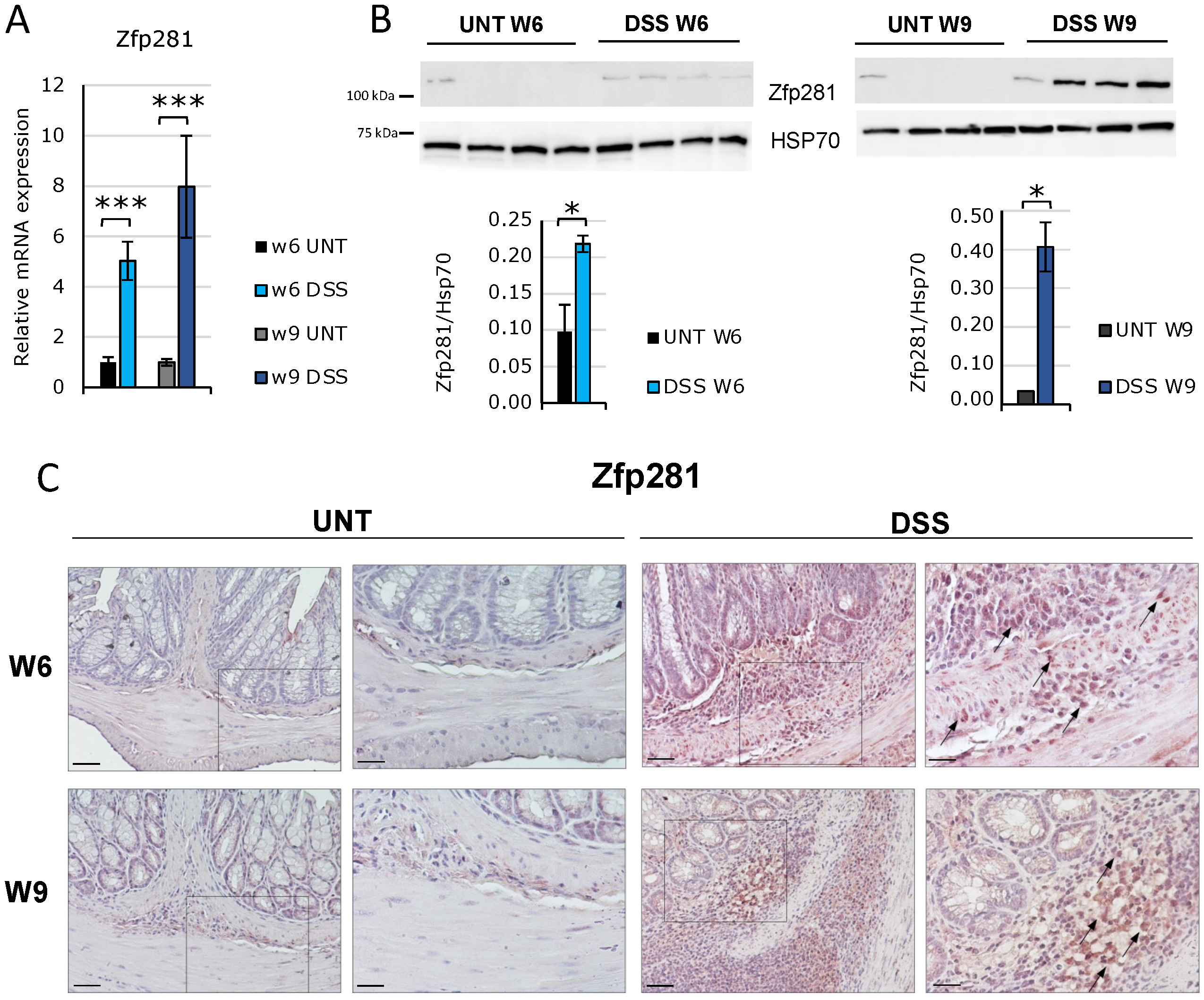
2.1. Zfp281/ZNF281 mRNA and Protein Expressions Are Significantly Increased from the Onset of Intestinal Fibrosis and Continue to Increase According to Its Progression
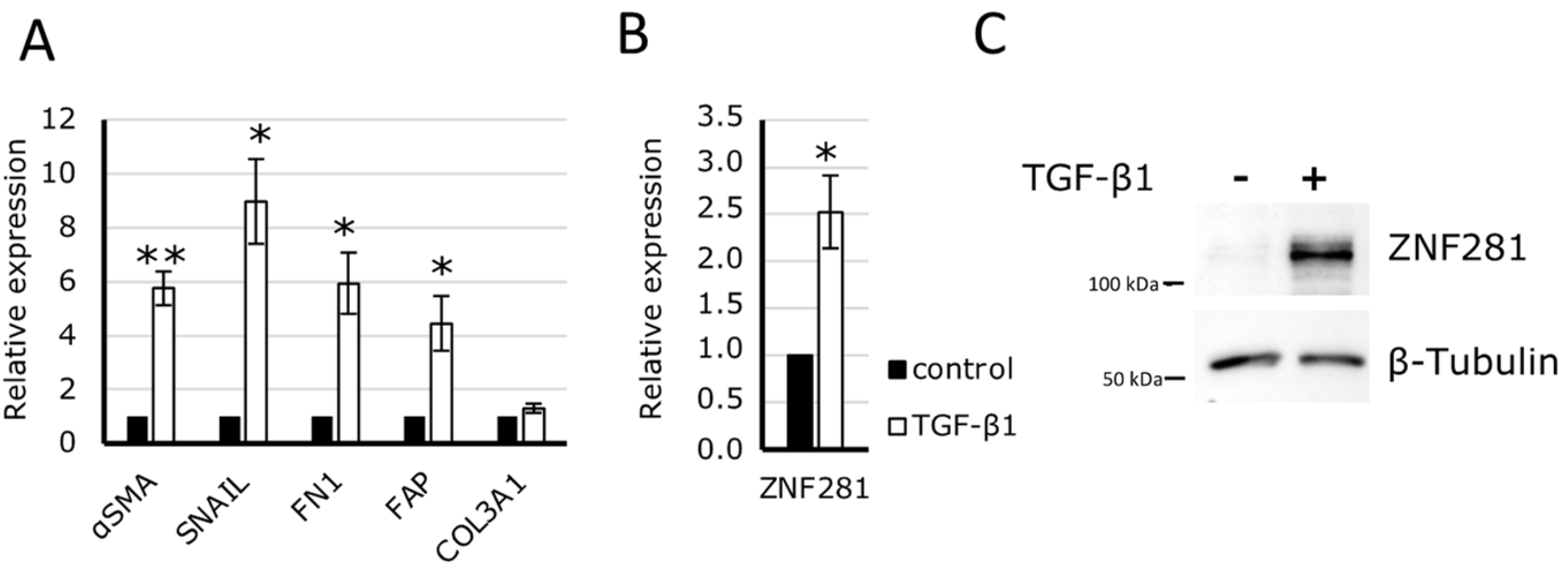
2.2. ZNF281 Silencing Significantly Decreases Fibrotic Genes αSMA, SNAIL, and FN1 in Human Colon Fibroblasts
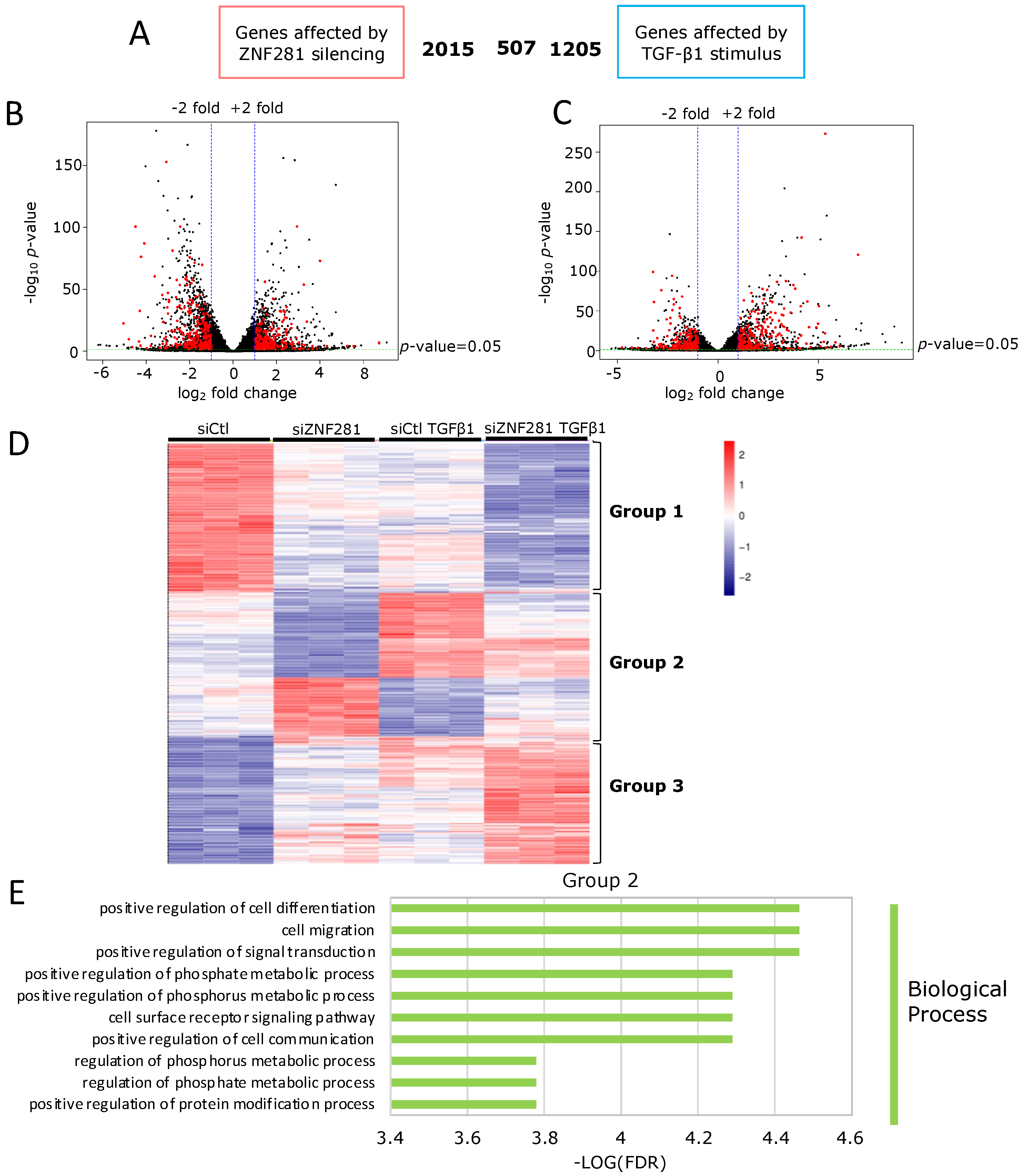
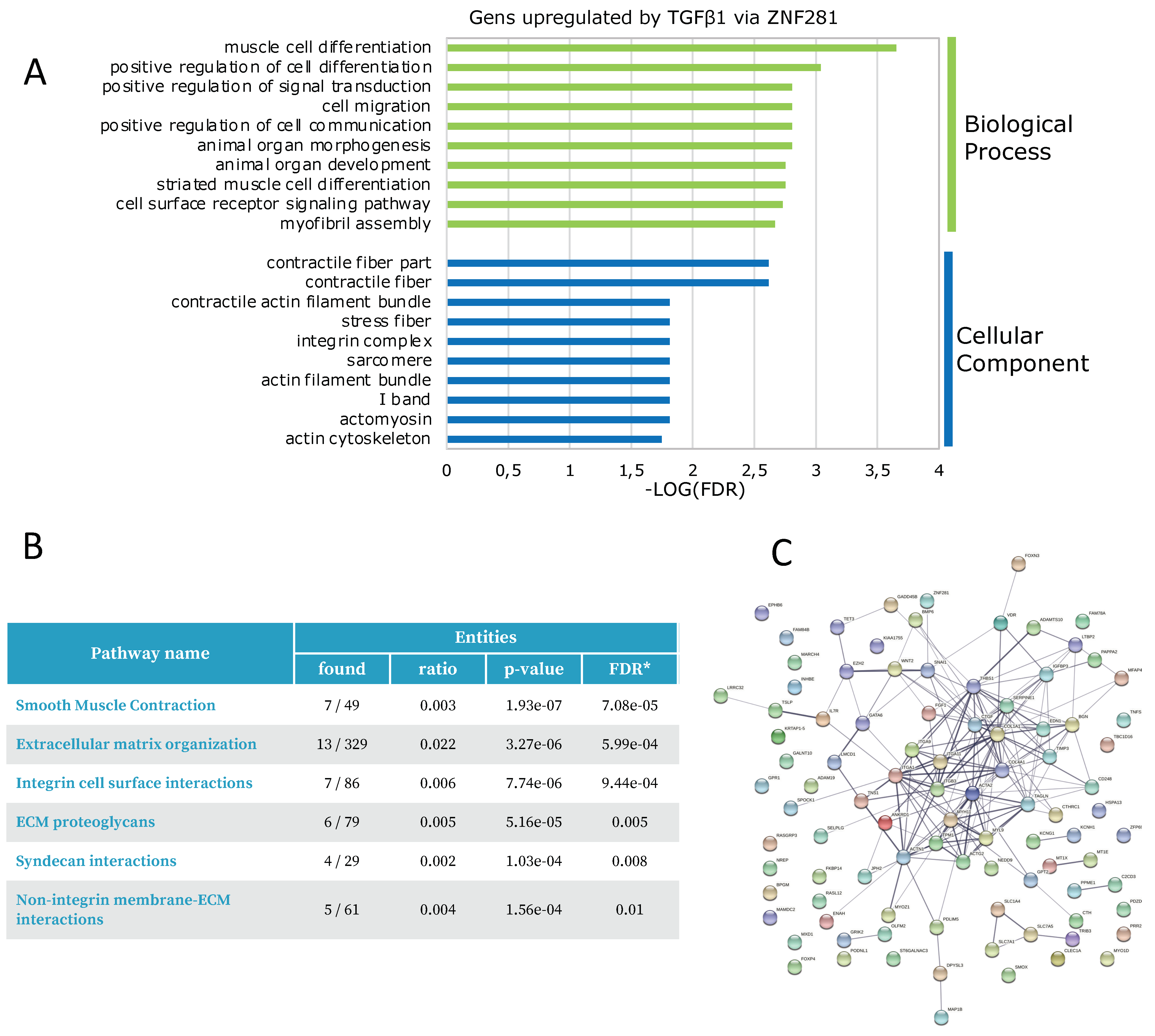
2.3. ZNF281 Controls the Expression of Genes Involved in Cell Migration, Differentiation, and Signal Transduction in Activated Colon Fibroblasts
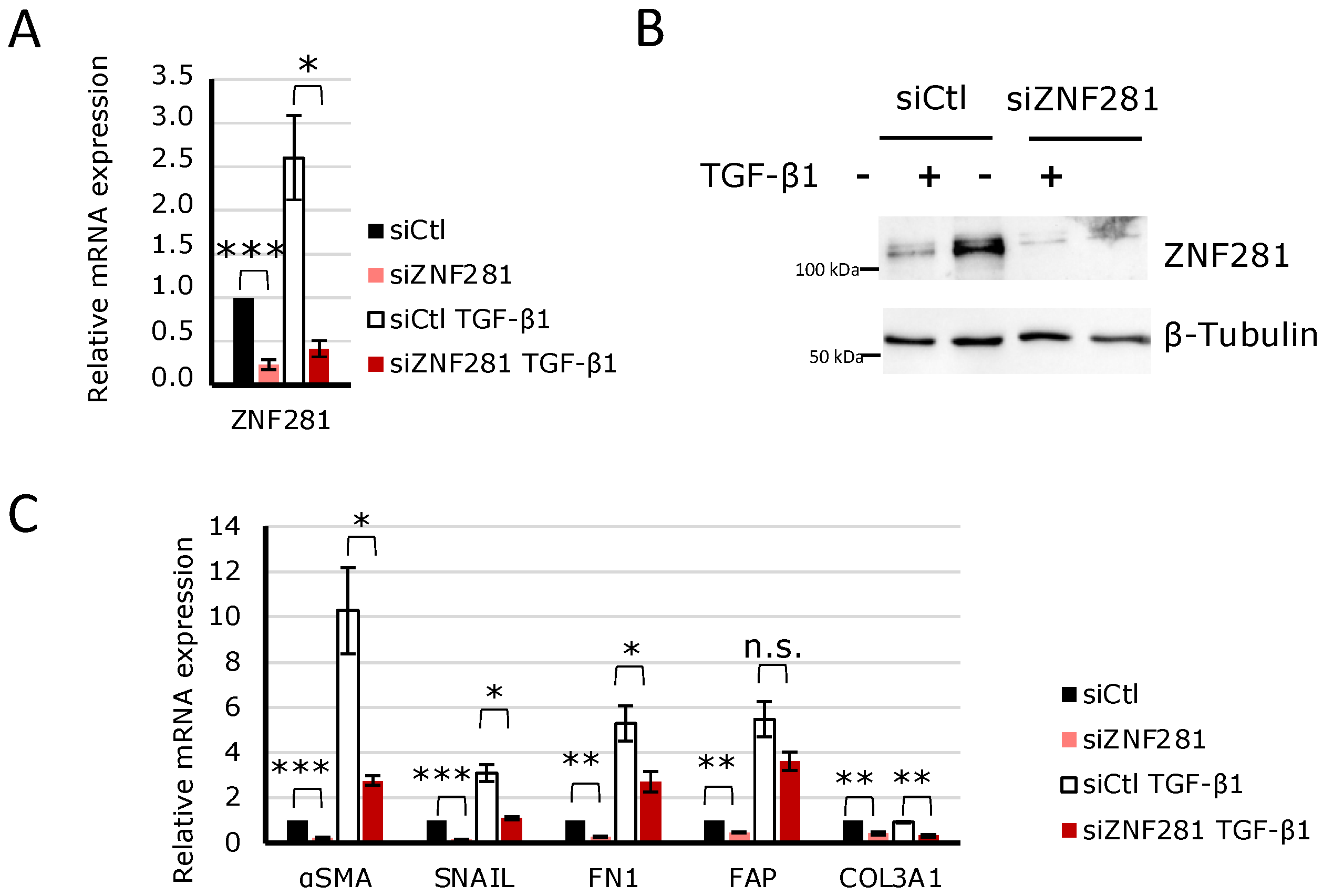
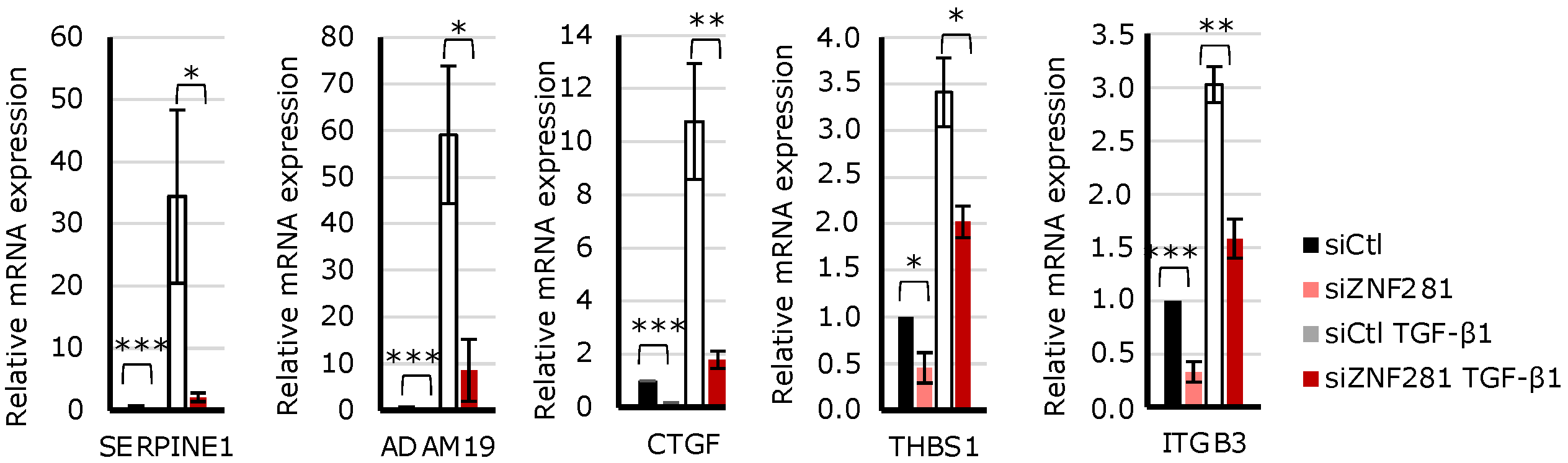
2.4. TGFβ1 Regulates the Expression of Fibrosis-Related Genes via ZNF281
2.5. ZNF281 Acts as a Transcriptional Activator of Pro-Fibrotic Genes Involved in Myofibroblast Differentiation
3. Discussion
4. Methods
4.1. Ethic Statement
4.2. Chronic DSS-Induced Colitis Model
4.3. Immunohistochemistry
4.4. Cell Culture, Treatment, and Transfection
4.5. RNA Isolation and RT-qPCR
4.6. Protein Isolation and Western Blot
4.7. RNA-Sequencing
4.8. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Souza, H.S.P.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, K.; Wu, Y.; Yang, Y.; Tso, P.; Wu, Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front. Immunol. 2017, 8, 942. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Fiocchi, C. Intestinal fibrosis in IBD—A dynamic, multifactorial process. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Fiocchi, C.; Rogler, G. Mechanisms, Management, and Treatment of Fibrosis in Patients With Inflammatory Bowel Diseases. Gastroenterology 2017, 152, 340–350.e6. [Google Scholar] [CrossRef]
- Lawrance, I.C.; Rogler, G.; Bamias, G.; Breynaert, C.; Florholmen, J.; Pellino, G.; Reif, S.; Speca, S.; Latella, G. Cellular and Molecular Mediators of Intestinal Fibrosis. J. Crohns Colitis 2017, 11, 1491–1503. [Google Scholar] [CrossRef]
- Bettenworth, D.; Rieder, F. Pathogenesis of Intestinal Fibrosis in Inflammatory Bowel Disease and Perspectives for Therapeutic Implication. Dig. Dis. 2017, 35, 25–31. [Google Scholar] [CrossRef]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef]
- D’Alessio, S.; Ungaro, F.; Noviello, D.; Lovisa, S.; Peyrin-Biroulet, L.; Danese, S. Revisiting fibrosis in inflammatory bowel disease: The gut thickens. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 169–184. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, B.; Jin, T.; Ocansey, D.K.W.; Jiang, J.; Mao, F. Intestinal Fibrosis in Inflammatory Bowel Disease and the Prospects of Mesenchymal Stem Cell Therapy. Front. Immunol. 2022, 13, 835005. [Google Scholar] [CrossRef]
- Yue, X.; Shan, B.; Lasky, J.A. TGF-β: Titan of Lung Fibrogenesis. Curr. Enzyme Inhib. 2010, 6, 10067. [Google Scholar] [CrossRef]
- Xu, F.; Liu, C.; Zhou, D.; Zhang, L. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2016, 64, 157–167. [Google Scholar] [CrossRef]
- Marinelli Busilacchi, E.; Costantini, A.; Mancini, G.; Tossetta, G.; Olivieri, J.; Poloni, A.; Viola, N.; Butini, L.; Campanati, A.; Goteri, G.; et al. Nilotinib Treatment of Patients Affected by Chronic Graft-versus-Host Disease Reduces Collagen Production and Skin Fibrosis by Downmodulating the TGF-β and p-SMAD Pathway. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2020, 26, 823–834. [Google Scholar] [CrossRef]
- Ahmed, H.; Umar, M.I.; Imran, S.; Javaid, F.; Syed, S.K.; Riaz, R.; Hassan, W. TGF-β1 signaling can worsen NAFLD with liver fibrosis backdrop. Exp. Mol. Pathol. 2022, 124, 104733. [Google Scholar] [CrossRef]
- Ong, C.H.; Tham, C.L.; Harith, H.H.; Firdaus, N.; Israf, D.A. TGF-β-induced fibrosis: A review on the underlying mechanism and potential therapeutic strategies. Eur. J. Pharmacol. 2021, 911, 174510. [Google Scholar] [CrossRef]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, Q.; Zhu, G.; Xing, Y.; Li, S.; Ren, N.; Liu, T.; Zhu, A.; Bai, Y.; Piao, D. GSK-3β phosphorylation-dependent degradation of ZNF281 by β-TrCP2 suppresses colorectal cancer progression. Oncotarget 2017, 8, 88599–88612. [Google Scholar] [CrossRef]
- Lisowsky, T.; Polosa, P.L.; Sagliano, A.; Roberti, M.; Gadaleta, M.N.; Cantatore, P. Identification of human GC-box-binding zinc finger protein, a new Krüppel-like zinc finger protein, by the yeast one-hybrid screening with a GC-rich target sequence. FEBS Lett. 1999, 453, 369–374. [Google Scholar] [CrossRef]
- Fidalgo, M.; Shekar, P.C.; Ang, Y.-S.; Fujiwara, Y.; Orkin, S.H.; Wang, J. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells 2011, 29, 1705–1716. [Google Scholar] [CrossRef]
- Pieraccioli, M.; Nicolai, S.; Antonov, A.; Somers, J.; Malewicz, M.; Melino, G.; Raschellà, G. ZNF281 contributes to the DNA damage response by controlling the expression of XRCC2 and XRCC4. Oncogene 2016, 35, 2592–2601. [Google Scholar] [CrossRef]
- Hahn, S.; Hermeking, H. ZNF281/ZBP-99: A new player in epithelial-mesenchymal transition, stemness, and cancer. J. Mol. Med. Berl. Ger. 2014, 92, 571–581. [Google Scholar] [CrossRef]
- Pierdomenico, M.; Palone, F.; Cesi, V.; Vitali, R.; Mancuso, A.B.; Cucchiara, S.; Oliva, S.; Aloi, M.; Stronati, L. Transcription Factor ZNF281: A Novel Player in Intestinal Inflammation and Fibrosis. Front. Immunol. 2018, 9, 2907. [Google Scholar] [CrossRef] [PubMed]
- Holvoet, T.; Devriese, S.; Castermans, K.; Boland, S.; Leysen, D.; Vandewynckel, Y.-P.; Devisscher, L.; Van den Bossche, L.; Van Welden, S.; Dullaers, M.; et al. Treatment of Intestinal Fibrosis in Experimental Inflammatory Bowel Disease by the Pleiotropic Actions of a Local Rho Kinase Inhibitor. Gastroenterology 2017, 153, 1054–1067. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Imai, J.; Yahata, T.; Ichikawa, H.; Ibrahim, A.A.; Yazawa, M.; Sumiyoshi, H.; Inagaki, Y.; Matsushima, M.; Suzuki, T.; Mine, T.; et al. Inhibition of plasminogen activator inhibitor-1 attenuates against intestinal fibrosis in mice. Intest. Res. 2020, 18, 219–228. [Google Scholar] [CrossRef]
- Hugo, C.; Shankland, S.J.; Pichler, R.H.; Couser, W.G.; Johnson, R.J. Thrombospondin 1 precedes and predicts the development of tubulointerstitial fibrosis in glomerular disease in the rat. Kidney Int. 1998, 53, 302–311. [Google Scholar] [CrossRef]
- Yanagihara, T.; Tsubouchi, K.; Gholiof, M.; Chong, S.G.; Lipson, K.E.; Zhou, Q.; Scallan, C.; Upagupta, C.; Tikkanen, J.; Keshavjee, S.; et al. Connective-Tissue Growth Factor Contributes to TGF-β1-induced Lung Fibrosis. Am. J. Respir. Cell Mol. Biol. 2022, 66, 260–270. [Google Scholar] [CrossRef]
- Tsai, H.; Karube, S.; Kondou, K.; Yamaguchi, N.; Ishii, F.; Otani, Y. Clear variation of spin splitting by changing electron distribution at non-magnetic metal/Bi2O3 interfaces. Sci. Rep. 2018, 8, 5564. [Google Scholar] [CrossRef]
- Qi, B.; Newcomer, R.G.; Sang, Q.-X.A. ADAM19/adamalysin 19 structure, function, and role as a putative target in tumors and inflammatory diseases. Curr. Pharm. Des. 2009, 15, 2336–2348. [Google Scholar] [CrossRef]
- Li, S.; Jiang, S.; Zhang, Q.; Jin, B.; Lv, D.; Li, W.; Zhao, M.; Jiang, C.; Dai, C.; Liu, Z. Integrin β3 Induction Promotes Tubular Cell Senescence and Kidney Fibrosis. Front. Cell Dev. Biol. 2021, 9, 733831. [Google Scholar] [CrossRef]
- Burke, J.P.; Watson, R.W.G.; Murphy, M.; Docherty, N.G.; Coffey, J.C.; O’Connell, P.R. Simvastatin impairs smad-3 phosphorylation and modulates transforming growth factor beta1-mediated activation of intestinal fibroblasts. Br. J. Surg. 2009, 96, 541–551. [Google Scholar] [CrossRef]
- Li, G.; Ren, J.; Hu, Q.; Deng, Y.; Chen, G.; Guo, K.; Li, R.; Li, Y.; Wu, L.; Wang, G.; et al. Oral pirfenidone protects against fibrosis by inhibiting fibroblast proliferation and TGF-β signaling in a murine colitis model. Biochem. Pharmacol. 2016, 117, 57–67. [Google Scholar] [CrossRef]
- Guan, Y.; Tan, Y.; Liu, W.; Yang, J.; Wang, D.; Pan, D.; Sun, Y.; Zheng, C. NF-E2-Related Factor 2 Suppresses Intestinal Fibrosis by Inhibiting Reactive Oxygen Species-Dependent TGF-β1/SMADs Pathway. Dig. Dis. Sci. 2018, 63, 366–380. [Google Scholar] [CrossRef]
- Nicolai, S.; Pieraccioli, M.; Smirnov, A.; Pitolli, C.; Anemona, L.; Mauriello, A.; Candi, E.; Annicchiarico-Petruzzelli, M.; Shi, Y.; Wang, Y.; et al. ZNF281/Zfp281 is a target of miR-1 and counteracts muscle differentiation. Mol. Oncol. 2020, 14, 294–308. [Google Scholar] [CrossRef]
- Hahn, S.; Jackstadt, R.; Siemens, H.; Hünten, S.; Hermeking, H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013, 32, 3079–3095. [Google Scholar] [CrossRef]
- Qin, C.-J.; Bu, P.-L.; Zhang, Q.; Chen, J.-T.; Li, Q.-Y.; Liu, J.-T.; Dong, H.-C.; Ren, X.-Q. ZNF281 Regulates Cell Proliferation, Migration and Invasion in Colorectal Cancer through Wnt/β-Catenin Signaling. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2019, 52, 1503–1516. [Google Scholar] [CrossRef]
- Qian, Y.; Li, J.; Xia, S. ZNF281 Promotes Growth and Invasion of Pancreatic Cancer Cells by Activating Wnt/β-Catenin Signaling. Dig. Dis. Sci. 2017, 62, 2011–2020. [Google Scholar] [CrossRef]
- Oliviero, G.; Brien, G.L.; Waston, A.; Streubel, G.; Jerman, E.; Andrews, D.; Doyle, B.; Munawar, N.; Wynne, K.; Crean, J.; et al. Dynamic Protein Interactions of the Polycomb Repressive Complex 2 during Differentiation of Pluripotent Cells. Mol. Cell. Proteom. MCP 2016, 15, 3450–3460. [Google Scholar] [CrossRef]
- Ishiuchi, T.; Ohishi, H.; Sato, T.; Kamimura, S.; Yorino, M.; Abe, S.; Suzuki, A.; Wakayama, T.; Suyama, M.; Sasaki, H. Zfp281 Shapes the Transcriptome of Trophoblast Stem Cells and Is Essential for Placental Development. Cell Rep. 2019, 27, 1742–1754. [Google Scholar] [CrossRef] [Green Version]
- Fidalgo, M.; Huang, X.; Guallar, D.; Sanchez-Priego, C.; Valdes, V.J.; Saunders, A.; Ding, J.; Wu, W.-S.; Clavel, C.; Wang, J. Zfp281 Coordinates Opposing Functions of Tet1 and Tet2 in Pluripotent States. Cell Stem Cell 2016, 19, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.-M.; Kim, S.-H.; Kim, E.-H. The Molecular Mechanism of Transforming Growth Factor-β Signaling for Intestinal Fibrosis: A Mini-Review. Front. Pharmacol. 2019, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Finnson, K.W.; Almadani, Y.; Philip, A. Non-canonical (non-SMAD2/3) TGF-β signaling in fibrosis: Mechanisms and targets. Semin. Cell Dev. Biol. 2020, 101, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Walton, K.L.W.; Blue, R.E.; McNaughton, K.K.; Macnaughton, K.; Magness, S.T.; Lund, P.K. Mucosal healing and fibrosis after acute or chronic inflammation in wild type FVB-N mice and C57BL6 procollagen α1(I)-promoter-GFP reporter mice. PLoS ONE 2012, 7, e42568. [Google Scholar] [CrossRef]
- Babraham Bioinformatics—FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 13 July 2022).
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinform. Oxf. Engl. 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://cran.microsoft.com/snapshot/2018-06-22/web/packages/pheatmap/pheatmap.pdf (accessed on 13 July 2022).
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laudadio, I.; Bastianelli, A.; Fulci, V.; Carissimi, C.; Colantoni, E.; Palone, F.; Vitali, R.; Lorefice, E.; Cucchiara, S.; Negroni, A.; et al. ZNF281 Promotes Colon Fibroblast Activation in TGFβ1-Induced Gut Fibrosis. Int. J. Mol. Sci. 2022, 23, 10261. https://doi.org/10.3390/ijms231810261
Laudadio I, Bastianelli A, Fulci V, Carissimi C, Colantoni E, Palone F, Vitali R, Lorefice E, Cucchiara S, Negroni A, et al. ZNF281 Promotes Colon Fibroblast Activation in TGFβ1-Induced Gut Fibrosis. International Journal of Molecular Sciences. 2022; 23(18):10261. https://doi.org/10.3390/ijms231810261
Chicago/Turabian StyleLaudadio, Ilaria, Alex Bastianelli, Valerio Fulci, Claudia Carissimi, Eleonora Colantoni, Francesca Palone, Roberta Vitali, Elisa Lorefice, Salvatore Cucchiara, Anna Negroni, and et al. 2022. "ZNF281 Promotes Colon Fibroblast Activation in TGFβ1-Induced Gut Fibrosis" International Journal of Molecular Sciences 23, no. 18: 10261. https://doi.org/10.3390/ijms231810261
APA StyleLaudadio, I., Bastianelli, A., Fulci, V., Carissimi, C., Colantoni, E., Palone, F., Vitali, R., Lorefice, E., Cucchiara, S., Negroni, A., & Stronati, L. (2022). ZNF281 Promotes Colon Fibroblast Activation in TGFβ1-Induced Gut Fibrosis. International Journal of Molecular Sciences, 23(18), 10261. https://doi.org/10.3390/ijms231810261





