Ectopic Expression of CsSUN in Tomato Results in Elongated Fruit Shape via Regulation of Longitudinal Cell Division
Abstract
:1. Introduction
2. Results
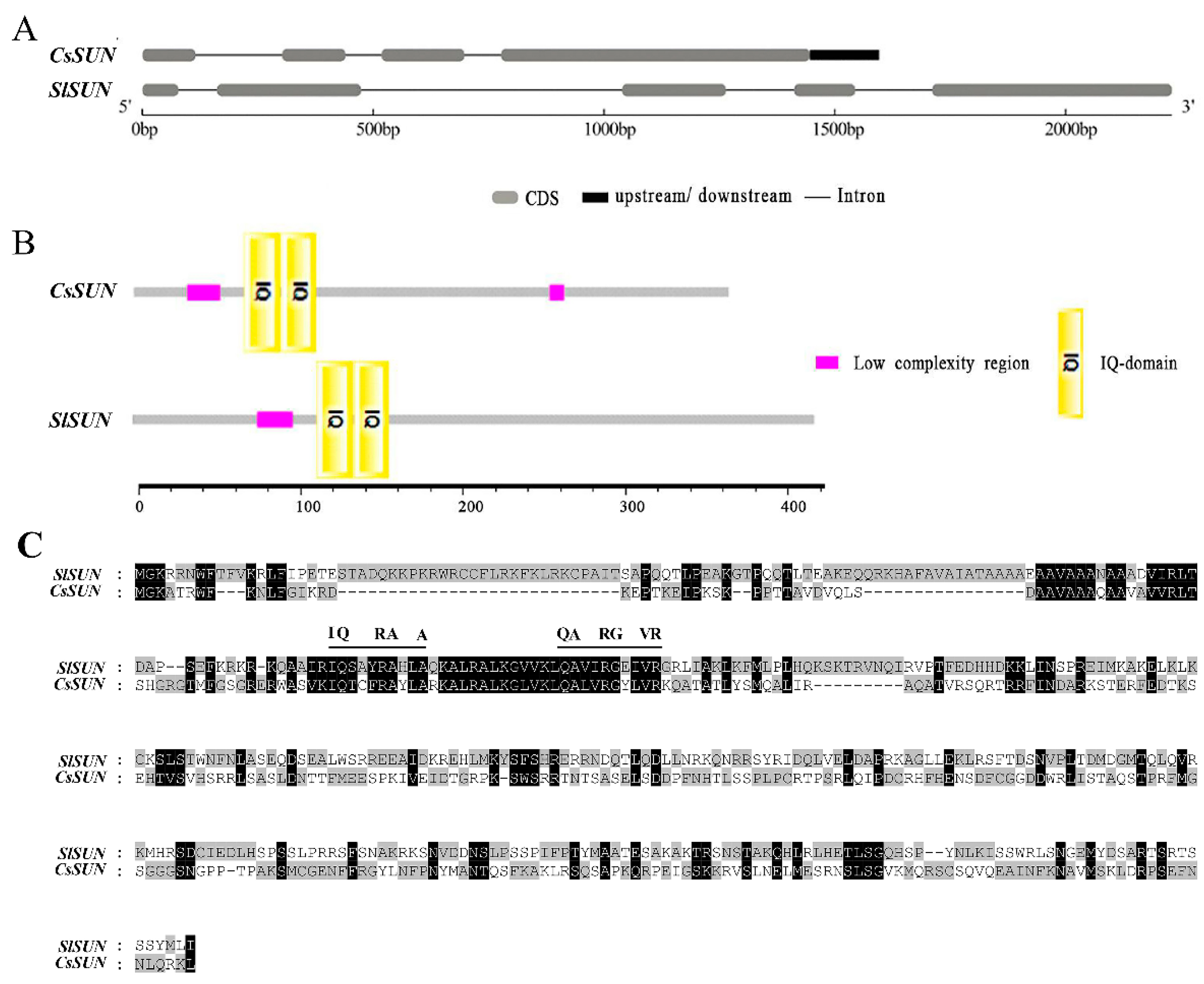
2.1. Sequence Feature Comparison between CsSUN and SlSUN
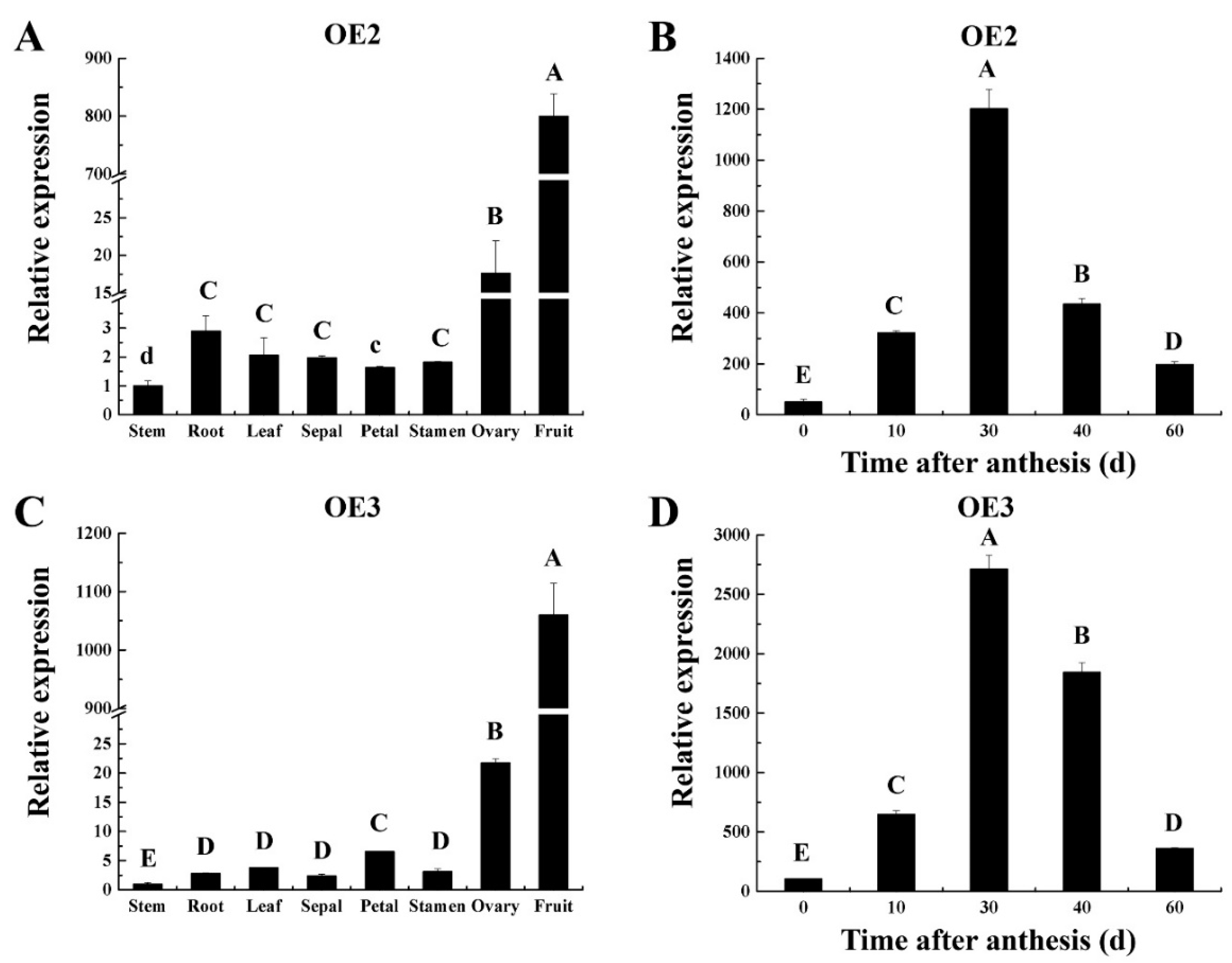
2.2. Identification of CsSUN Transgenic Lines
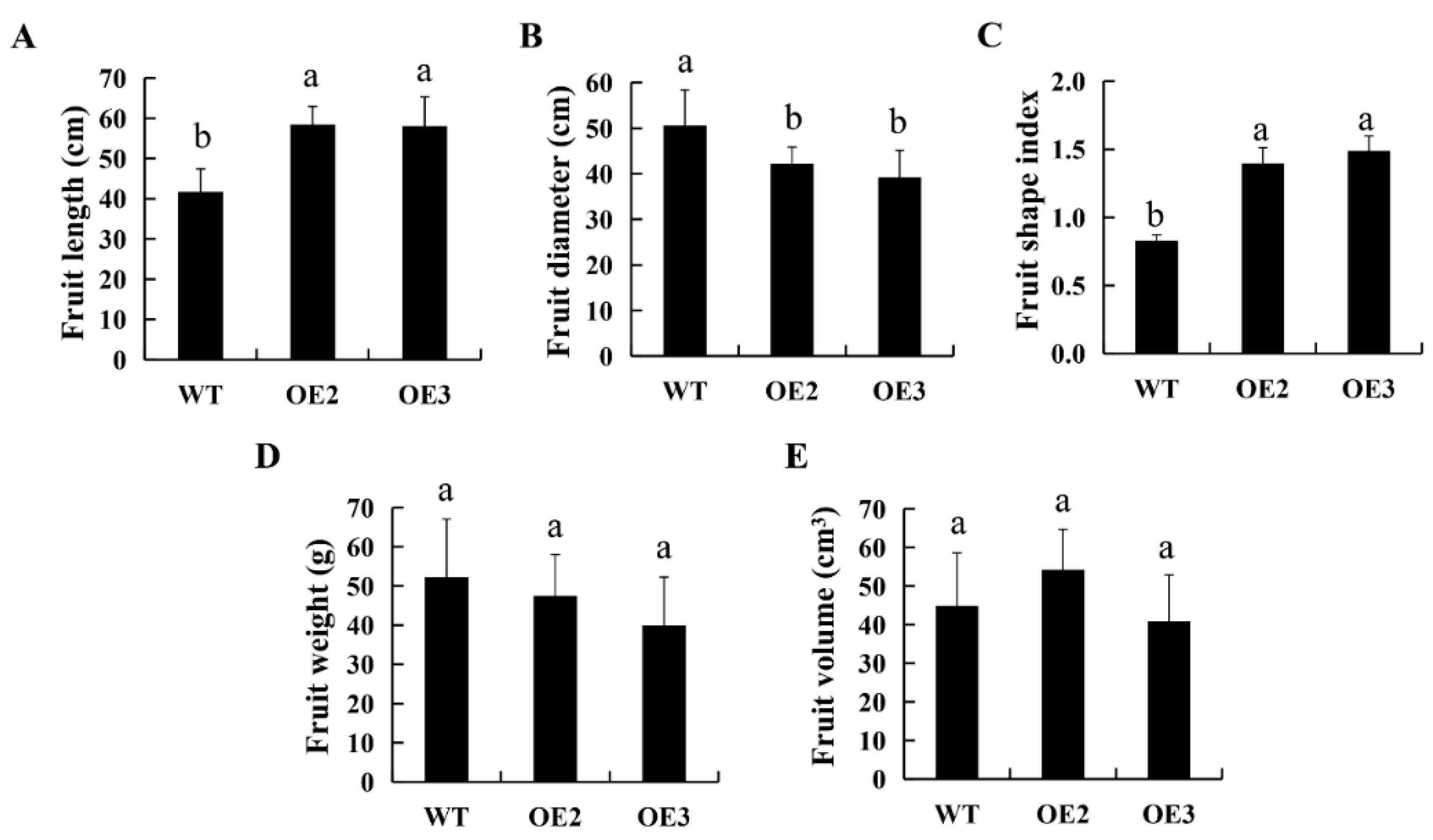
2.3. Ectopic Expression of CsSUN Results in Elongated Tomato Fruits
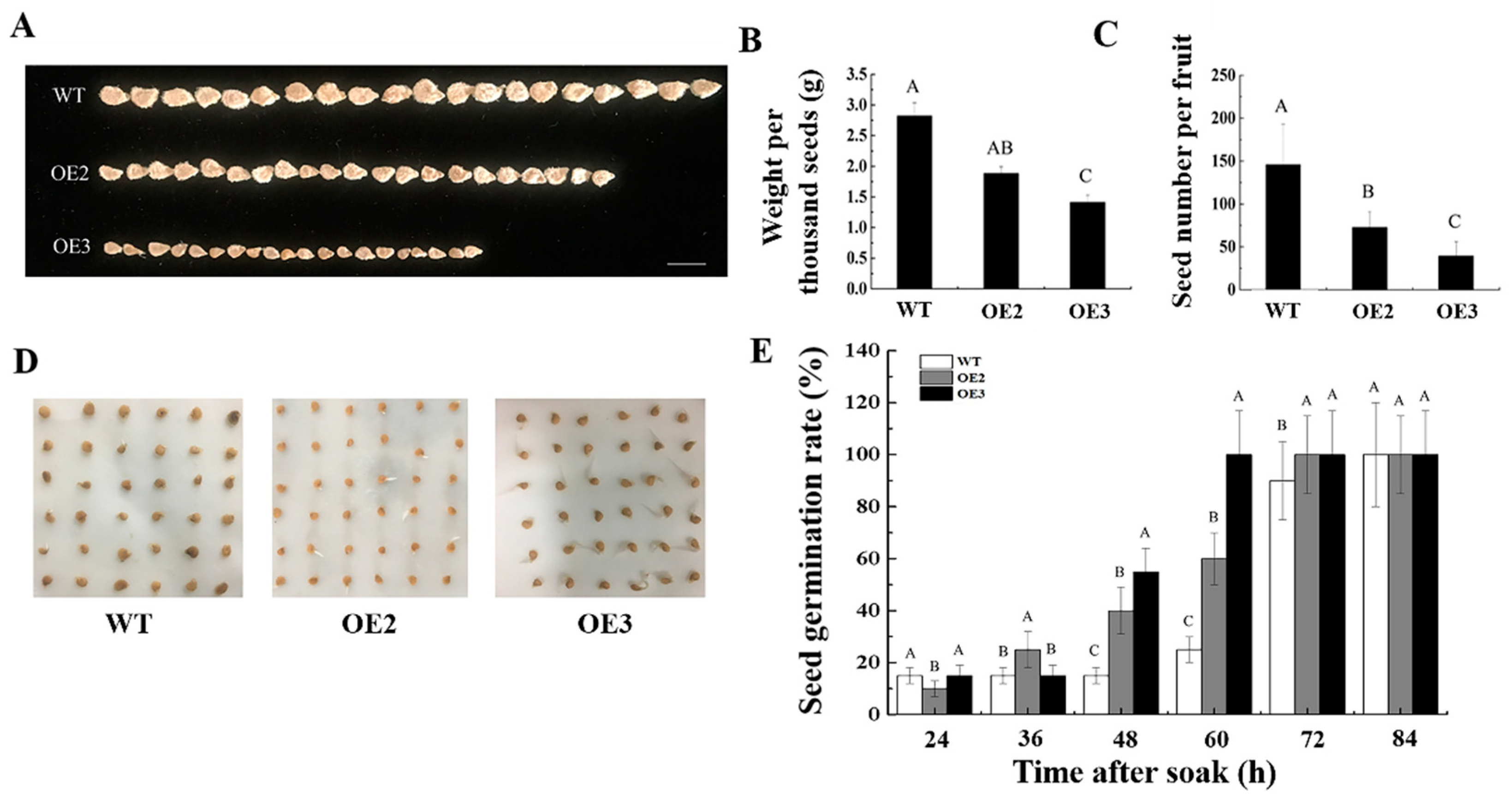
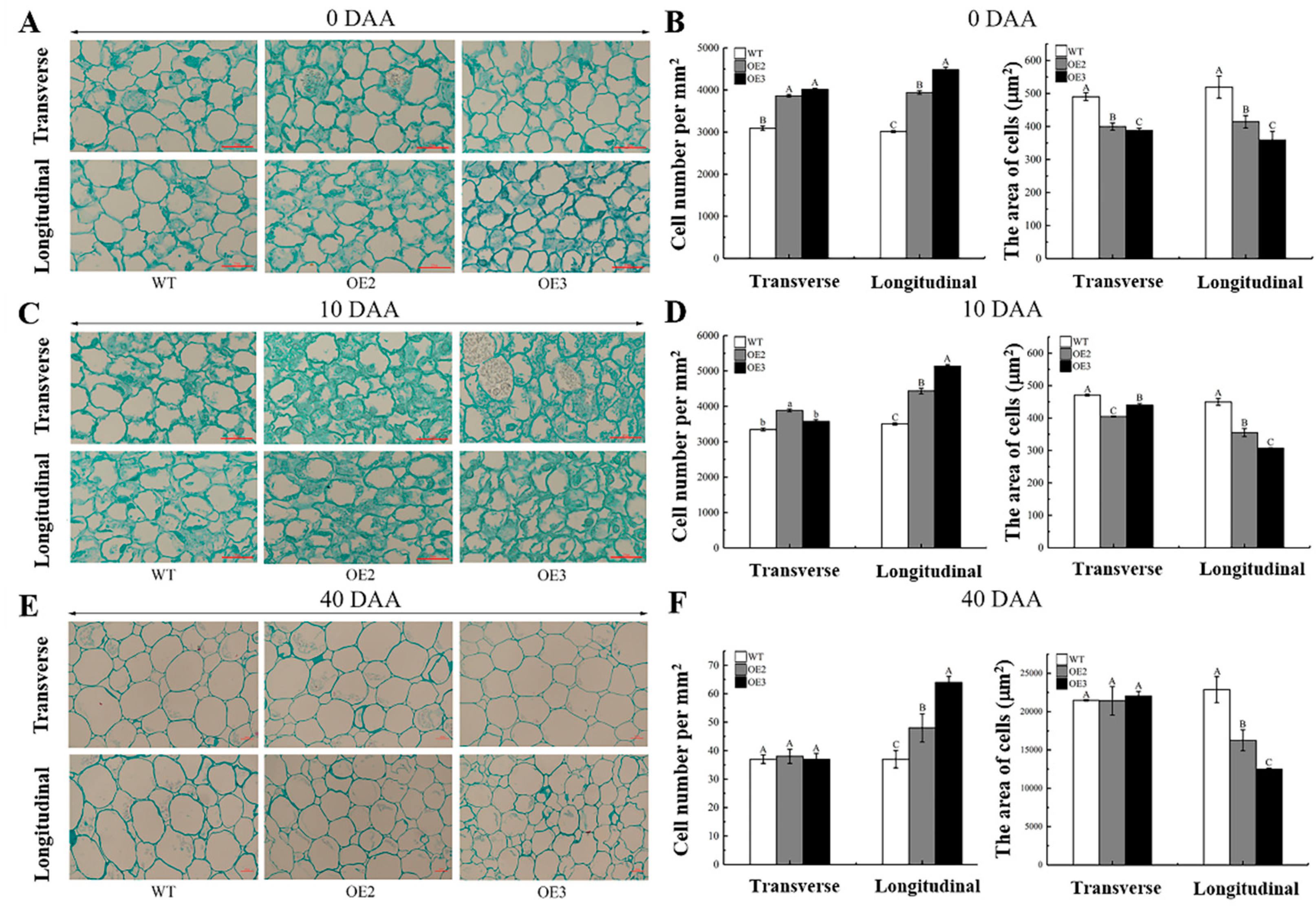
2.4. Ectopic Expression of CsSUN Alters Both Cell Division and Size in Transgenic Tomato Fruits
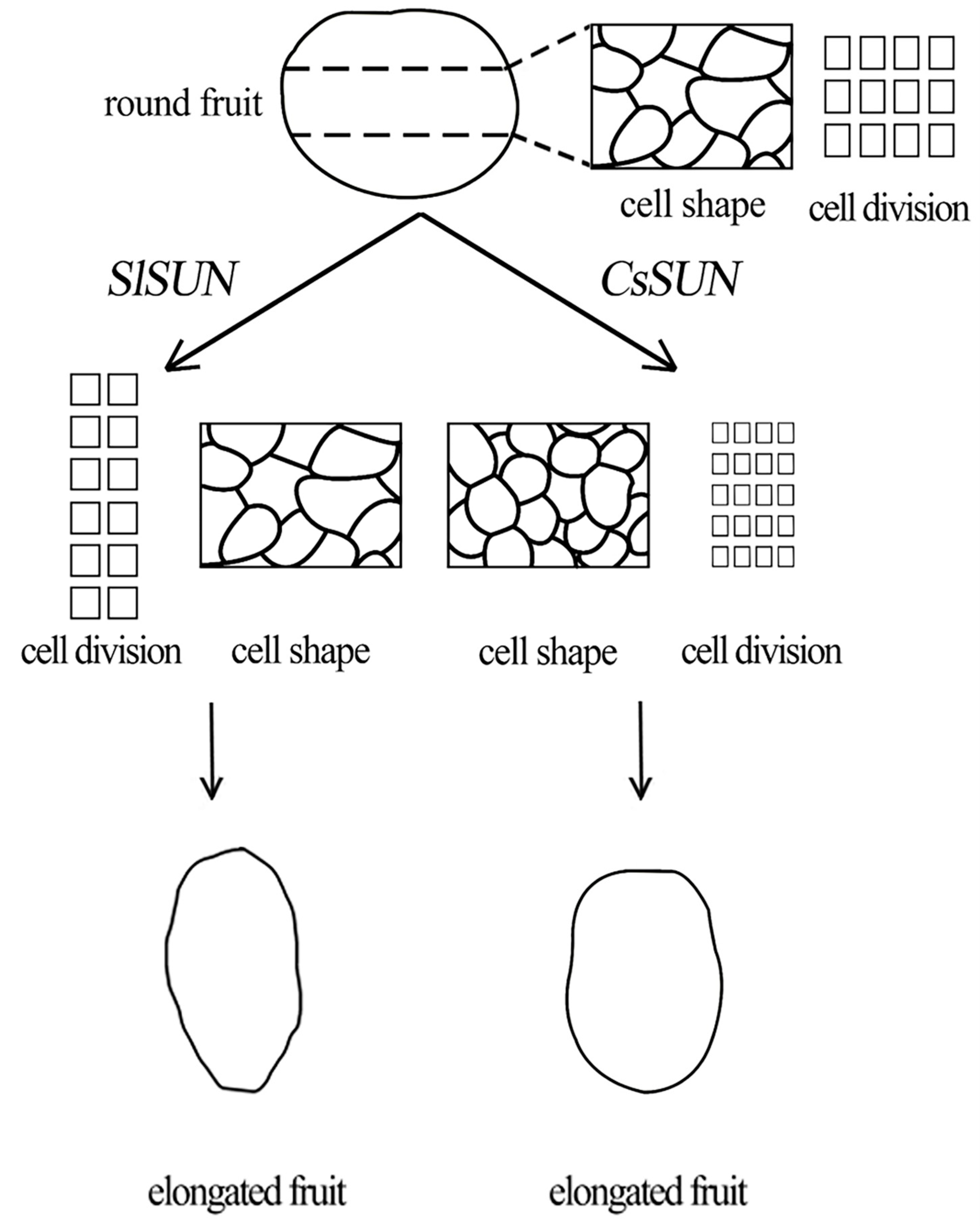
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Sequence Analysis and Cloning of CsSUN Gene
4.3. Binary Vector Preparation and Tomato Plant Transformation
4.4. Expression Analysis of CsSUN by qRT-PCR
4.5. Phenotypic Characterization of Transgenic Tomato Fruits
4.6. Microscopic Investigation of Transgenic Tomato Fruits
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Wang, L.; Zheng, S.; Liu, Z.; Wu, X.; Gao, Z.; Cao, C.; Li, Q.; Ren, Z. A fragment substitution in the promoter of CsHDZIV11/CsGL3 is responsible for fruit spine density in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- Che, G.; Zhang, X. Molecular basis of cucumber fruit domestication. Curr Opin Plant Biol 2019, 47, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Ma, Z.; Chen, L.; Wang, Z.; Wang, C.; Wang, L.; Chen, C.; Ren, Z.; Cao, C. Morphological Characterization and Integrated Transcriptome and Proteome Analysis of Organ Development Defective 1 (odd1) Mutant in Cucumis sativus L. Int. J. Mol. Sci. 2022, 23, 5843. [Google Scholar] [CrossRef] [PubMed]
- Gou, C.; Zhu, P.; Meng, Y.; Yang, F.; Xu, Y.; Xia, P.; Chen, J.; Li, J. Evaluation and Genetic Analysis of Parthenocarpic Germplasms in Cucumber. Genes 2022, 13, 225. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, C.; Zheng, S.; Zhang, H.; Liu, P.; Ge, Q.; Li, J.; Ren, Z. Transcriptomic analysis of short-fruit 1 (sf1) reveals new insights into the variation of fruit-related traits in Cucumis sativus. Sci. Rep. 2017, 7, 2950. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef]
- Weng, Y.; Colle, M.; Wang, Y.; Yang, L.; Rubinstein, M.; Sherman, A.; Ophir, R.; Grumet, R. QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 2015, 128, 1747–1763. [Google Scholar] [CrossRef]
- Wang, Y.; Bo, K.; Gu, X.; Pan, J.; Li, Y.; Chen, J.; Wen, C.; Ren, Z.; Ren, H.; Chen, X.; et al. Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic. Res. 2020, 7, 3. [Google Scholar] [CrossRef]
- Pan, Y.; Liang, X.; Gao, M.; Liu, H.; Meng, H.; Weng, Y.; Cheng, Z. Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 2017, 130, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodriguez, G.R.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Diaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S.; et al. A Functional Allele of CsFUL1 Regulates Fruit Length through Repressing CsSUP and Inhibiting Auxin Transport in Cucumber. Plant Cell 2019, 31, 1289–1307. [Google Scholar] [CrossRef] [PubMed]
- Xin, T.; Zhang, Z.; Li, S.; Zhang, S.; Li, Q.; Zhang, Z.H.; Huang, S.; Yang, X. Genetic Regulation of Ethylene Dosage for Cucumber Fruit Elongation. Plant Cell 2019, 31, 1063–1076. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, B.; Wang, S.; Lin, T.; Yang, L.; Zhao, Z.; Zhang, Z.; Huang, S.; Yang, X. Genome-wide Target Mapping Shows Histone Deacetylase Complex1 Regulates Cell Proliferation in Cucumber Fruit. Plant Physiol. 2020, 182, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, N.; Wang, Y.; Fan, J.; Li, Y.; Ding, J. Down-Regulation of Cytokinin Receptor Gene SlHK2 Improves Plant Tolerance to Drought, Heat, and Combined Stresses in Tomato. Plants 2022, 11, 154. [Google Scholar] [CrossRef]
- Van der Knaap, E.; Ostergaard, L. Shaping a fruit: Developmental pathways that impact growth patterns. Semin. Cell Dev. Biol. 2018, 79, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Van der Knaap, E.; Chakrabarti, M.; Chu, Y.H.; Clevenger, J.P.; Illa-Berenguer, E.; Huang, Z.; Keyhaninejad, N.; Mu, Q.; Sun, L.; Wang, Y.; et al. What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 2014, 5, 227. [Google Scholar] [CrossRef]
- Sun, L.; Rodriguez, G.R.; Clevenger, J.P.; Illa-Berenguer, E.; Lin, J.; Blakeslee, J.J.; Liu, W.; Fei, Z.; Wijeratne, A.; Meulia, T.; et al. Candidate gene selection and detailed morphological evaluations of fs8.1, a quantitative trait locus controlling tomato fruit shape. J. Exp. Bot. 2015, 66, 6471–6482. [Google Scholar] [CrossRef]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef]
- Jiang, N.; Gao, D.; Xiao, H.; van der Knaap, E. Genome organization of the tomato sun locus and characterization of the unusual retrotransposon Rider. Plant J. 2009, 60, 181–193. [Google Scholar] [CrossRef]
- Rodriguez, G.R.; Munos, S.; Anderson, C.; Sim, S.C.; Michel, A.; Causse, M.; Gardener, B.B.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Van der Knaap, E.; Tanksley, S.D. Identification and characterization of a novel locus controlling early fruit development in tomato. Theor. Appl. Genet. 2001, 103, 353–358. [Google Scholar] [CrossRef]
- Xiao, H.; Radovich, C.; Welty, N.; Hsu, J.; Li, D.; Meulia, T.; van der Knaap, E. Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 2009, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xiao, H.; Cabrera, A.; Meulia, T.; van der Knaap, E. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 2011, 157, 1175–1186. [Google Scholar] [CrossRef]
- Clevenger, J.P.; van Houten, J.; Blackwood, M.; Rodriguez, G.R.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Saito, K.; Visa, S.; van der Knaap, E. Network Analyses Reveal Shifts in Transcript Profiles and Metabolites That Accompany the Expression of SUN and an Elongated Tomato Fruit. Plant Physiol. 2015, 168, 1164–1178. [Google Scholar] [CrossRef]
- Wang, Y.; Clevenger, J.P.; Illa-Berenguer, E.; Meulia, T.; van der Knaap, E.; Sun, L. A Comparison of sun, ovate, fs8.1 and Auxin Application on Tomato Fruit Shape and Gene Expression. Plant Cell Physiol. 2019, 60, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, M.D.; Wu, S.; Snouffer, A.; Wang, Y.; van der Knaap, E. Plant Organ Shapes Are Regulated by Protein Interactions and Associations With Microtubules. Front Plant Sci. 2018, 9, 1766. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Clevenger, J.P.; Sun, L.; Visa, S.; Kamiya, Y.; Jikumaru, Y.; Blakeslee, J.; van der Knaap, E. The control of tomato fruit elongation orchestrated by sun, ovate and fs8.1 in a wild relative of tomato. Plant Sci. 2015, 238, 95–104. [Google Scholar] [CrossRef]
- Ge, Q.; Wang, X.F.; Li, H.; Ren, Z.H.; Wang, L.N. Genome-wide identification and analysis of IQD/SUN gene family in cucumber. Genom. Appl. Biol. 2019, 38, 4110–4119. [Google Scholar] [CrossRef]
- Gupta, A.; Upadhyay, R.K.; Prabhakar, R.; Tiwari, N.; Garg, R.; Sane, V.A.; Sane, A.P. SlDREB3, a negative regulator of ABA responses, controls seed germination, fruit size and the onset of ripening in tomato. Plant Sci. 2022, 319, 111249. [Google Scholar] [CrossRef]
- Cheng, W.H.; Chiang, M.H.; Hwang, S.G.; Lin, P.C. Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol. Biol. 2009, 71, 61–80. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Cao, H.; Sun, Y.; Li, X.; Chen, F.; Carles, A.; Li, Y.; Ding, M.; Zhang, C.; Deng, X.; et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 2013, 25, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, Z.; Chen, L.; Ren, Z. Comprehensive analysis of multiprotein bridging factor 1 family genes and SlMBF1c negatively regulate the resistance to Botrytis cinerea in tomato. BMC Plant Biol. 2019, 19, 437. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chu, Z.; Jia, R.; Dan, F.; Shen, X.; Li, Y.; Ding, X. SlMYB12 Regulates Flavonol Synthesis in Three Different Cherry Tomato Varieties. Sci. Rep. 2018, 8, 1582. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Primer Name | Forward Primer | Reverse Primer |
|---|---|---|
| CsSUN-cloning | GGATCCATGGGGAAAGCAACGAGATG | ACTAGTTCATAATTTTCTCTGCAAAT |
| β-Actin | TGTTGCTATTCAGGCTGTGC | CTGCTCCTGGCAGTTTGAAT |
| CsSUN-RT | GCACTTAGAGCTTTGAAAGG | CTACTGTGAACGGAAACTGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Han, J.; Chen, L.; Han, N.; Hu, Y.; Ge, Q.; Ren, Z.; Wang, L. Ectopic Expression of CsSUN in Tomato Results in Elongated Fruit Shape via Regulation of Longitudinal Cell Division. Int. J. Mol. Sci. 2022, 23, 9973. https://doi.org/10.3390/ijms23179973
Li H, Han J, Chen L, Han N, Hu Y, Ge Q, Ren Z, Wang L. Ectopic Expression of CsSUN in Tomato Results in Elongated Fruit Shape via Regulation of Longitudinal Cell Division. International Journal of Molecular Sciences. 2022; 23(17):9973. https://doi.org/10.3390/ijms23179973
Chicago/Turabian StyleLi, Hao, Jing Han, Linjie Chen, Ni Han, Yajing Hu, Qian Ge, Zhonghai Ren, and Lina Wang. 2022. "Ectopic Expression of CsSUN in Tomato Results in Elongated Fruit Shape via Regulation of Longitudinal Cell Division" International Journal of Molecular Sciences 23, no. 17: 9973. https://doi.org/10.3390/ijms23179973
APA StyleLi, H., Han, J., Chen, L., Han, N., Hu, Y., Ge, Q., Ren, Z., & Wang, L. (2022). Ectopic Expression of CsSUN in Tomato Results in Elongated Fruit Shape via Regulation of Longitudinal Cell Division. International Journal of Molecular Sciences, 23(17), 9973. https://doi.org/10.3390/ijms23179973






