Selenoprotein P Concentrations in the Cerebrospinal Fluid and Serum of Individuals Affected by Amyotrophic Lateral Sclerosis, Mild Cognitive Impairment and Alzheimer’s Dementia
Abstract
1. Introduction
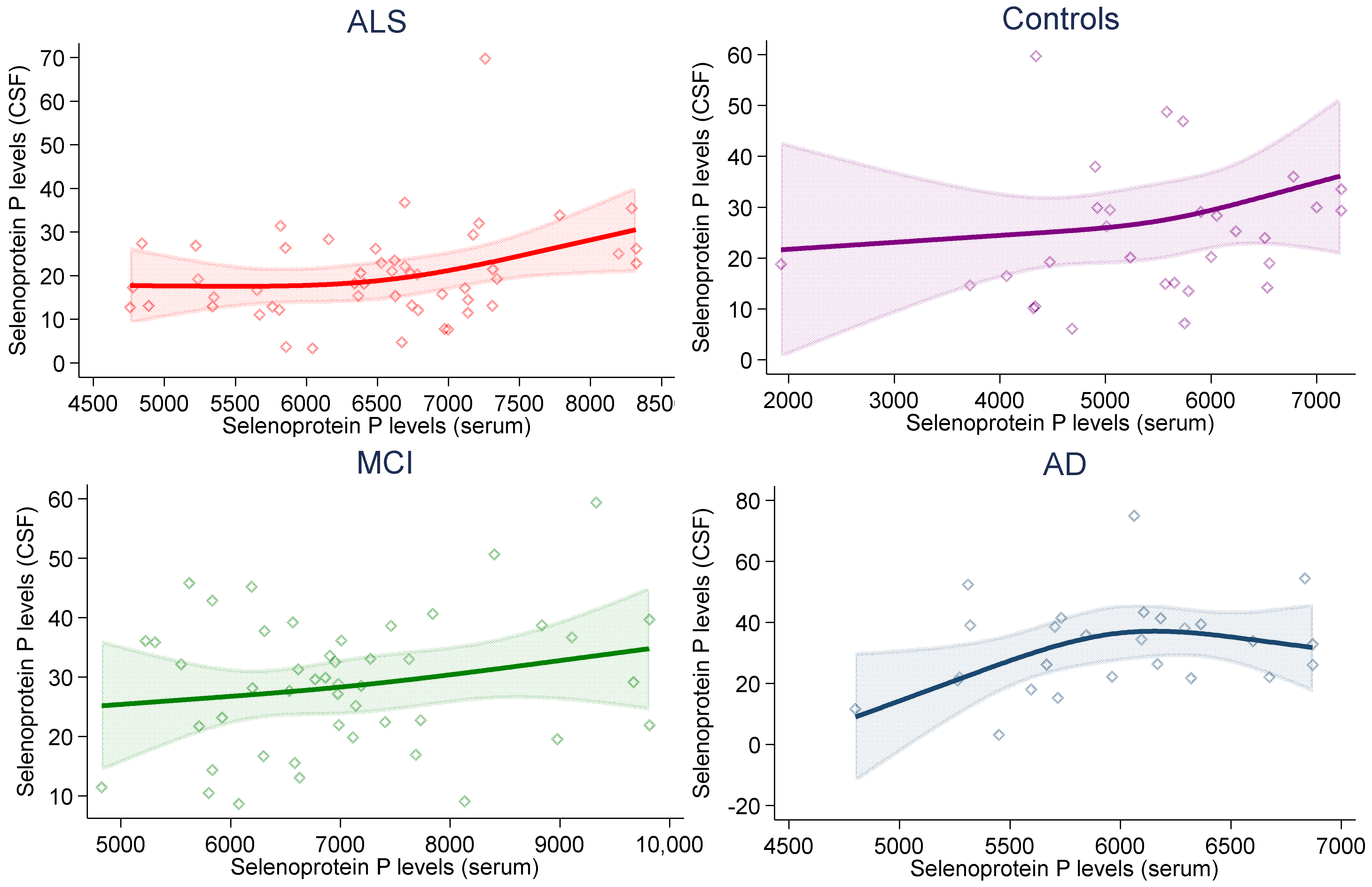
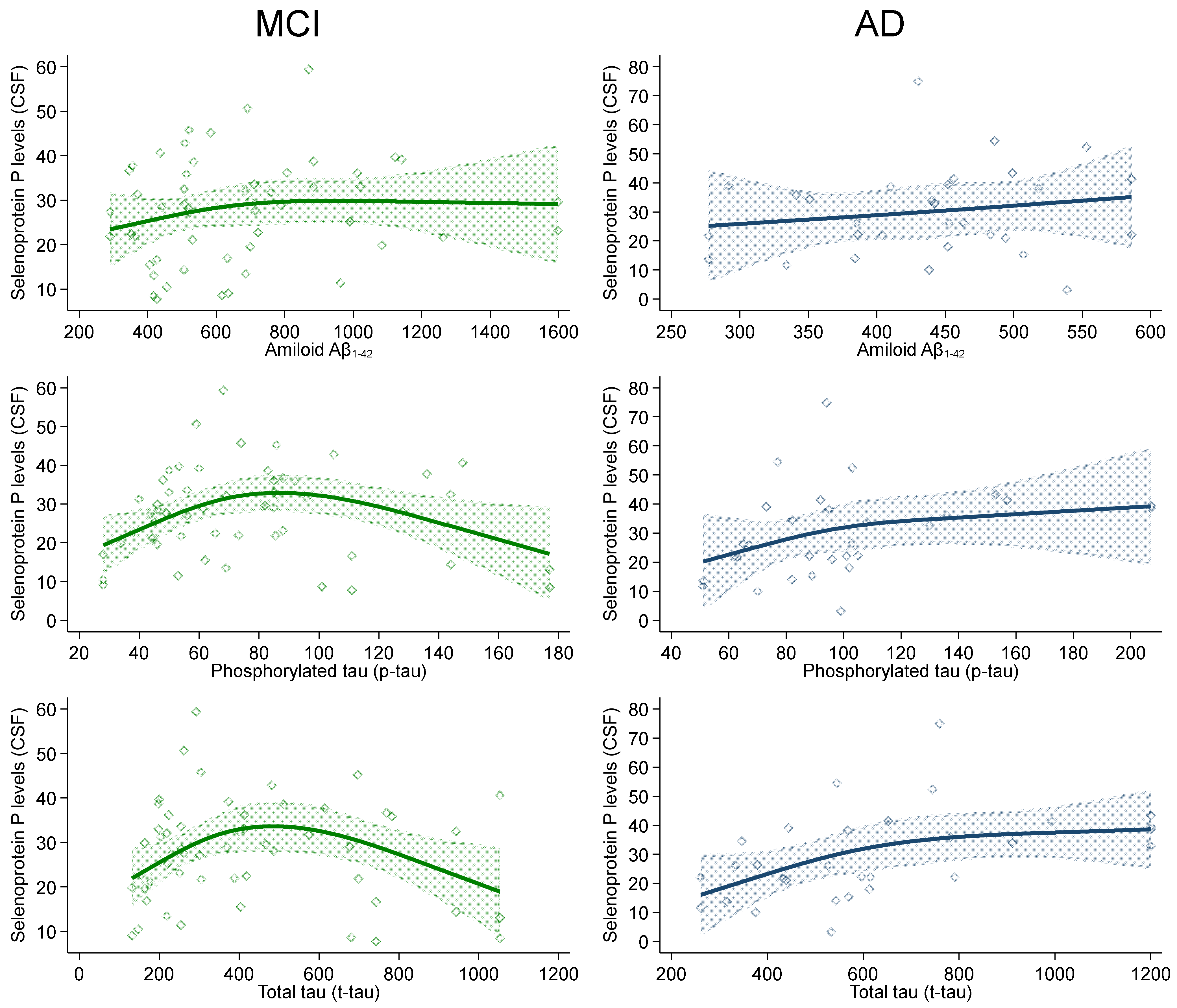
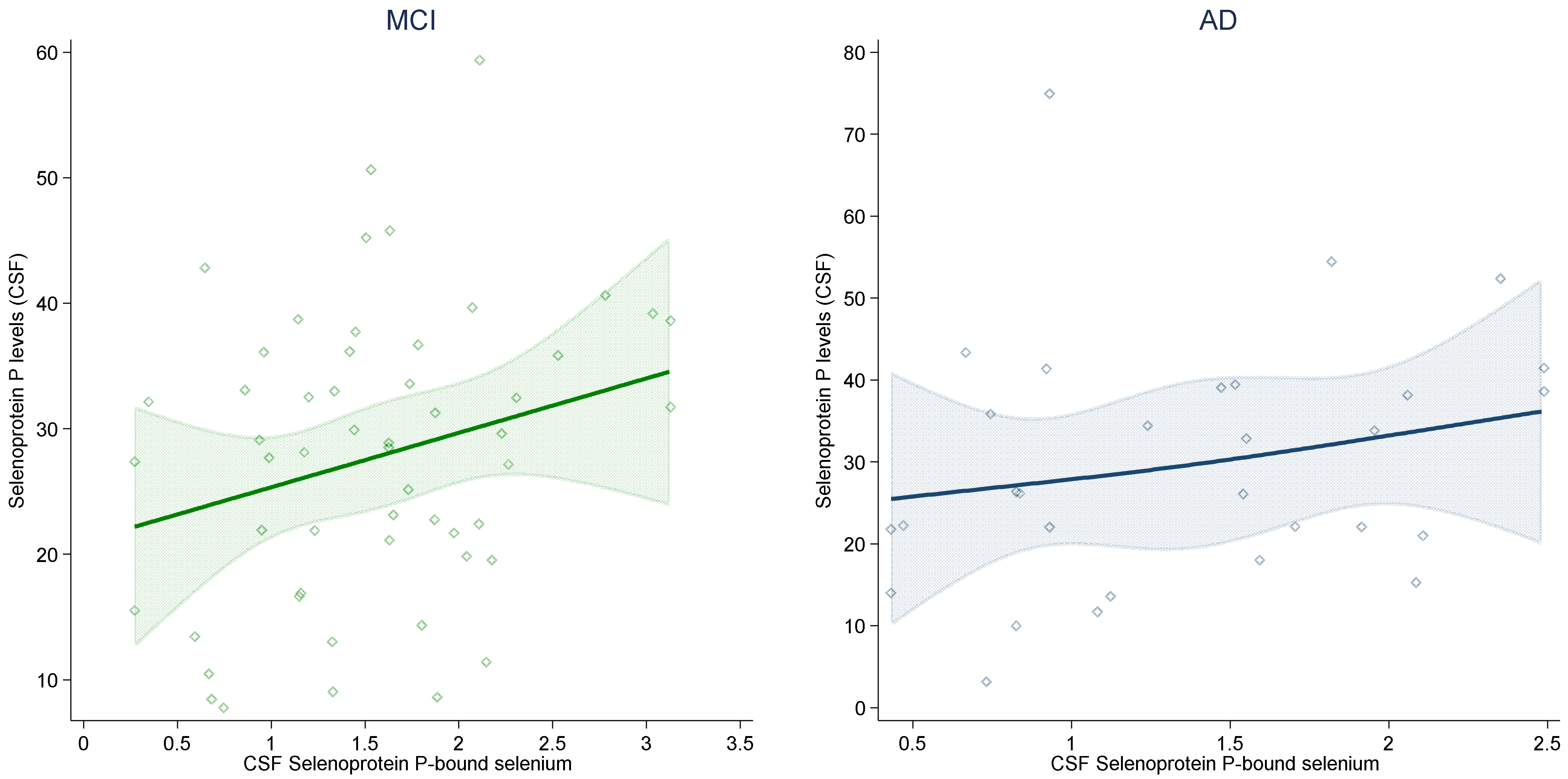
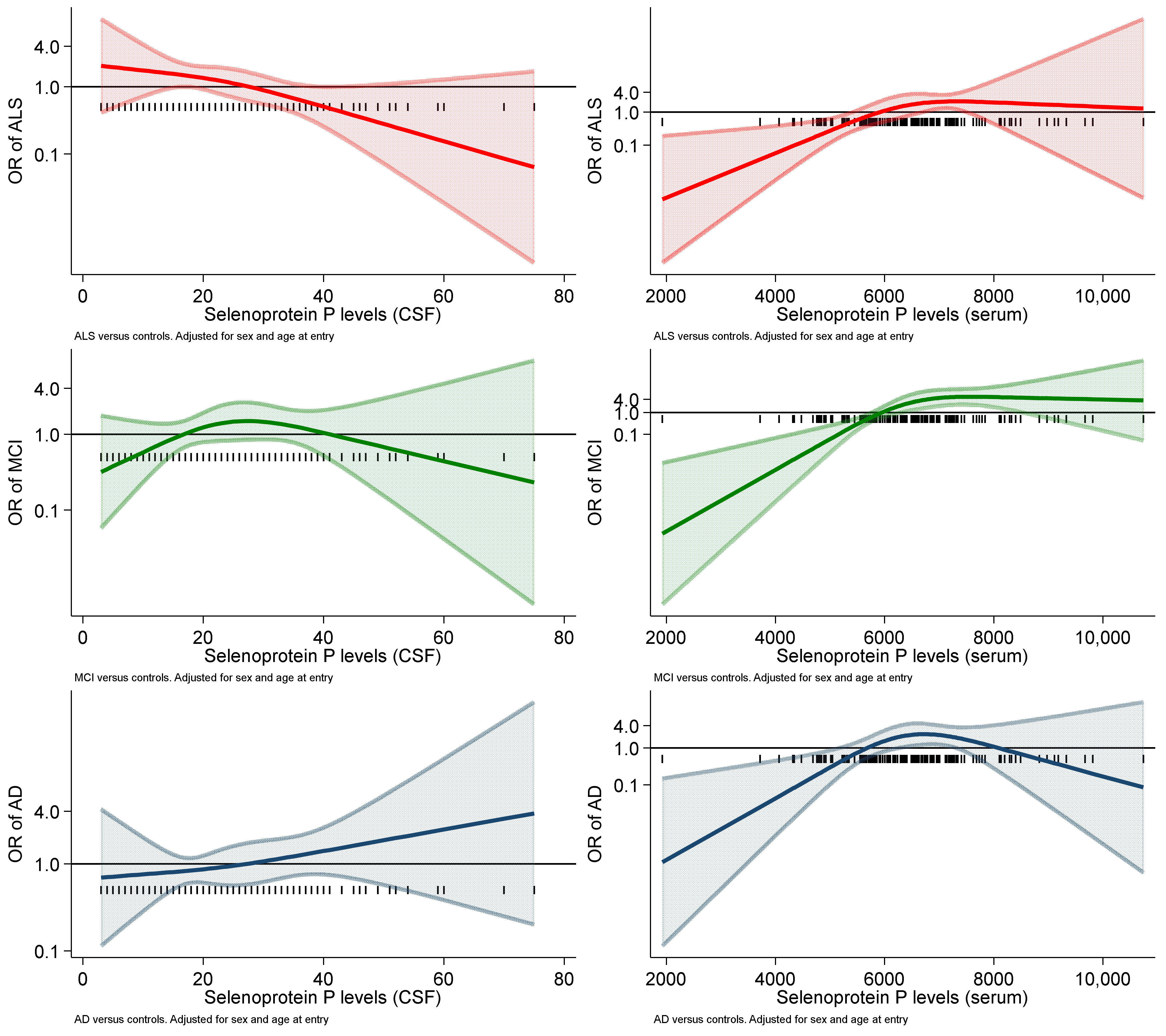
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection
4.3. Selenoprotein P Level Determination
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goutman, S.A.; Hardiman, O.; Al-Chalabi, A.; Chió, A.; Savelieff, M.G.; Kiernan, M.C.; Feldman, E.L. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 2022, 21, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Erkkinen, M.G.; Kim, M.-O.; Geschwind, M.D. Clinical Neurology and Epidemiology of the Major Neurodegenerative Diseases. Cold Spring Harb. Perspect. Biol. 2017, 10, a033118. [Google Scholar] [CrossRef]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Baird, K.; Baillon, S.; Lau, L.S.L.; Storey, M.; Lindesay, J.; Velayudhan, L. Predictive Factors for Conversion to Dementia in Individuals with Early-Onset Mild Cognitive Impairment. Dement. Geriatr. Cogn. Disord. 2021, 50, 548–553. [Google Scholar] [CrossRef]
- Overton, M.; Pihlsgård, M.; Elmståhl, S. Diagnostic Stability of Mild Cognitive Impairment, and Predictors of Reversion to Normal Cognitive Functioning. Dement. Geriatr. Cogn. Disord. 2019, 48, 317–329. [Google Scholar] [CrossRef]
- Vinceti, M.; Bottecchi, I.; Fan, A.; Finkelstein, Y.; Mandrioli, J. Are environmental exposures to selenium, heavy metals, and pesticides risk factors for amyotrophic lateral sclerosis? Rev. Environ. Health 2012, 27, 19–41. [Google Scholar] [CrossRef]
- Filippini, T.; Adani, G.; Malavolti, M.; Garuti, C.; Cilloni, S.; Vinceti, G.; Zamboni, G.; Tondelli, M.; Galli, C.; Costa, M.; et al. Dietary Habits and Risk of Early-Onset Dementia in an Italian Case-Control Study. Nutrients 2020, 12, 3682. [Google Scholar] [CrossRef]
- Adani, G.; Filippini, T.; Garuti, C.; Malavolti, M.; Vinceti, G.; Zamboni, G.; Tondelli, M.; Galli, C.; Costa, M.; Vinceti, M.; et al. Environmental Risk Factors for Early-Onset Alzheimer’s Dementia and Frontotemporal Dementia: A Case-Control Study in Northern Italy. Int. J. Environ. Res. Public Health 2020, 17, 7941. [Google Scholar] [CrossRef]
- Filippini, T.; Tesauro, M.; Fiore, M.; Malagoli, C.; Consonni, M.; Violi, F.; Iacuzio, L.; Arcolin, E.; Conti, G.O.; Cristaldi, A.; et al. Environmental and Occupational Risk Factors of Amyotrophic Lateral Sclerosis: A Population-Based Case-Control Study. Int. J. Environ. Res. Public Health 2020, 17, 2882. [Google Scholar] [CrossRef]
- Filippini, T.; Fiore, M.; Tesauro, M.; Malagoli, C.; Consonni, M.; Violi, F.; Arcolin, E.; Iacuzio, L.; Oliveri Conti, G.; Cristaldi, A.; et al. Clinical and Lifestyle Factors and Risk of Amyotrophic Lateral Sclerosis: A Population-Based Case-Control Study. Int. J. Environ. Res. Public Health 2020, 17, 857. [Google Scholar] [CrossRef]
- Aloizou, A.-M.; Siokas, V.; Vogiatzi, C.; Peristeri, E.; Docea, A.O.; Petrakis, D.; Provatas, A.; Folia, V.; Chalkia, C.; Vinceti, M.; et al. Pesticides, cognitive functions and dementia: A review. Toxicol. Lett. 2020, 326, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Vinceti, M.; Bonvicini, F.; Rothman, K.J.; Vescovi, L.; Wang, F. The relation between amyotrophic lateral sclerosis and inorganic selenium in drinking water: A population-based case-control study. Environ. Health 2010, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Michalke, B.; Berthele, A. Contribution to selenium speciation in cerebrospinal fluid samples. J. Anal. At. Spectrom. 2010, 26, 165–170. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Wise, L.A. Environmental Selenium and Human Health: An Update. Curr. Environ. Health Rep. 2018, 5, 464–485. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Cilloni, S.; Bargellini, A.; Vergoni, A.V.; Tsatsakis, A.; Ferrante, M. Health risk assessment of environmental selenium: Emerging evidence and challenges. Mol. Med. Rep. 2017, 15, 3323–3335. [Google Scholar] [CrossRef]
- Vinceti, M.; Mandrioli, J.; Borella, P.; Michalke, B.; Tsatsakis, A.; Finkelstein, Y. Selenium neurotoxicity in humans: Bridging laboratory and epidemiologic studies. Toxicol. Lett. 2014, 230, 295–303. [Google Scholar] [CrossRef]
- Naderi, M.; Puar, P.; Zonouzi-Marand, M.; Chivers, D.P.; Niyogi, S.; Kwong, R.W. A comprehensive review on the neuropathophysiology of selenium. Sci. Total Environ. 2020, 767, 144329. [Google Scholar] [CrossRef]
- Vinceti, M.; Wei, E.; Malagoli, C.; Bergomi, M.; Vivoli, G. Adverse Health Effects of Selenium in Humans. Rev. Environ. Health 2001, 16, 233–251. [Google Scholar] [CrossRef]
- Morris, J.S.; Crane, S.B. Selenium Toxicity from a Misformulated Dietary Supplement, Adverse Health Effects, and the Temporal Response in the Nail Biologic Monitor. Nutrients 2013, 5, 1024–1057. [Google Scholar] [CrossRef]
- Rae, W.; Kitley, J.; Pinto, A. Selenium Toxicity Associated With Reversible Leukoencephalopathy and Cortical Blindness. JAMA Neurol. 2018, 75, 1282–1283. [Google Scholar] [CrossRef]
- Solovyev, N.; Drobyshev, E.; Blume, B.; Michalke, B. Selenium at the neural barriers: A review. Front. Neurosci. 2021, 15, 630016. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, B.R.; Roberts, B.R.; Bush, A.I.; Hare, D.J. Selenium, selenoproteins and neurodegenerative diseases. Metallomics 2015, 7, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, U.; Fabiano, M. Selenoproteins in brain development and function. Free Radic. Biol. Med. 2022, 190, 105–115. [Google Scholar] [CrossRef]
- Varlamova, E.G.; Gudkov, S.V.; Plotnikov, E.Y.; Turovsky, E.A. Size-Dependent Cytoprotective Effects of Selenium Nanoparticles during Oxygen-Glucose Deprivation in Brain Cortical Cells. Int. J. Mol. Sci. 2022, 23, 7464. [Google Scholar] [CrossRef]
- Khalil, H.M.; Azouz, R.A.; Hozyen, H.F.; Aljuaydi, S.H.; AbuBakr, H.O.; Emam, S.R.; Al-Mokaddem, A.K. Selenium nanoparticles impart robust neuroprotection against deltamethrin-induced neurotoxicity in male rats by reversing behavioral alterations, oxidative damage, apoptosis, and neuronal loss. NeuroToxicology 2022, 91, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Vilela, D.D.; Justino, A.B.; Caixeta, D.C.; de Souza, A.V.; Teixeira, R.R.; Franco, R.R.; Saraiva, A.L.; Fonseca, B.B.; Dantas, N.O.; Silva, A.C.A.; et al. Increased selenium concentration in the synthesis of CdSe m agic-sized quantum dots affects how the brain responds to oxidative stress. J. Biomed. Mater. Res. Part B Appl. Biomater. 2021, 110, 1140–1150. [Google Scholar] [CrossRef]
- Bashir, D.W.; Rashad, M.M.; Ahmed, Y.H.; Drweesh, E.A.; Elzahany, E.A.; Abou-El-Sherbini, K.S.; El-Leithy, E.M. The ameliorative effect of nanoselenium on histopathological and biochemical alterations induced by melamine toxicity on the brain of adult male albino rats. NeuroToxicology 2021, 86, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Fuster, E.; Candela, H.; Estévez, J.; Vilanova, E.; Sogorb, M.A. A Transcriptomic Analysis of T98G Human Glioblastoma Cells after Exposure to Cadmium-Selenium Quantum Dots Mainly Reveals Alterations in Neuroinflammation Processes and Hypothalamus Regulation. Int. J. Mol. Sci. 2022, 23, 2267. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Qu, G.; Wen, R.; Liu, X.; Wang, Y.; Gao, J.; Yin, Y.; Shi, J.; Zhou, Q.; He, B.; et al. Occurrence of silver-containing particles in rat brains upon intranasal exposure of silver nanoparticles. Metallomics 2022, 14, mfab077. [Google Scholar] [CrossRef]
- Saito, Y. Selenium transport mechanism via selenoprotein P-Its physiological role and related diseases. Front. Nutr. 2021, 8, 685517. [Google Scholar] [CrossRef]
- Tsutsumi, R.; Saito, Y. Selenoprotein P; P for Plasma, Prognosis, Prophylaxis, and More. Biol. Pharm. Bull. 2020, 43, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y. Selenoprotein P as an in vivo redox regulator: Disorders related to its deficiency and excess. J. Clin. Biochem. Nutr. 2020, 66, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vinceti, M.; Filippini, T.; Jablonska, E.; Saito, Y.; Wise, L.A. Safety of selenium exposure and limitations of selenoprotein maximization: Molecular and epidemiologic perspectives. Environ. Res. 2022, 211, 113092. [Google Scholar] [CrossRef]
- Saito, Y.; Takahashi, K. Characterization of selenoprotein P as a selenium supply protein. JBIC J. Biol. Inorg. Chem. 2002, 269, 5746–5751. [Google Scholar] [CrossRef]
- Mita, Y.; Uchida, R.; Yasuhara, S.; Kishi, K.; Hoshi, T.; Matsuo, Y.; Yokooji, T.; Shirakawa, Y.; Toyama, T.; Urano, Y.; et al. Identification of a novel endogenous long non-coding RNA that inhibits selenoprotein P translation. Nucleic Acids Res. 2021, 49, 6893–6907. [Google Scholar] [CrossRef]
- Misu, H.; Takamura, T.; Takayama, H.; Hayashi, H.; Matsuzawa-Nagata, N.; Kurita, S.; Ishikura, K.; Ando, H.; Takeshita, Y.; Ota, T.; et al. A Liver-Derived Secretory Protein, Selenoprotein P, Causes Insulin Resistance. Cell Metab. 2010, 12, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, N.; Satoh, K.; Satoh, T.; Yaoita, N.; Siddique, M.A.H.; Omura, J.; Kurosawa, R.; Nogi, M.; Sunamura, S.; Miyata, S.; et al. Diagnostic and Prognostic Significance of Serum Levels of SeP (Selenoprotein P) in Patients With Pulmonary Hypertension. Arter. Thromb. Vasc. Biol. 2019, 39, 2553–2562. [Google Scholar] [CrossRef]
- Kikuchi, N.; Satoh, K.; Kurosawa, R.; Yaoita, N.; Al Mamun, E.; Siddique, M.A.H.; Omura, J.; Satoh, T.; Nogi, M.; Sunamura, S.; et al. Selenoprotein P Promotes the Development of Pulmonary Arterial Hypertension. Circulation 2018, 138, 600–623. [Google Scholar] [CrossRef]
- Takeishi, R.; Misaka, T.; Ichijo, Y.; Ishibashi, S.; Matsuda, M.; Yamadera, Y.; Ohara, H.; Sugawara, Y.; Hotsuki, Y.; Watanabe, K.; et al. Increases in hepatokine selenoprotein P levels are associated with hepatic hypoperfusion and predict adverse prognosis in patients with heart failure. J. Am. Heart Assoc. 2022, 11, e024901. [Google Scholar] [CrossRef]
- Kamoshita, K.; Tsugane, H.; Ishii, K.-A.; Takayama, H.; Yao, X.; Abuduwaili, H.; Tanida, R.; Taniguchi, Y.; Oo, H.K.; Gafiyatullina, G.; et al. Lauric acid impairs insulin-induced Akt phosphorylation by upregulating SELENOP expression via HNF4α induction. Am. J. Physiol. Metab. 2022, 322, E556–E568. [Google Scholar] [CrossRef]
- Bellinger, F.P.; He, Q.-P.; Bellinger, M.T.; Lin, Y.; Raman, A.V.; White, L.R.; Berry, M.J. Association of Selenoprotein P with Alzheimer’s Pathology in Human Cortex. J. Alzheimer’s Dis. 2008, 15, 465–472. [Google Scholar] [CrossRef]
- Solovyev, N.; Drobyshev, E.; Bjørklund, G.; Dubrovskii, Y.; Lysiuk, R.; Rayman, M.P. Selenium, selenoprotein P, and Alzheimer’s disease: Is there a link? Free Radic. Biol. Med. 2018, 127, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.; Rheinstein, P.H. The association between selenium, selenoprotein P (SEPP1), fluid intelligence, and exercise in the UK Biobank cohort. Cureus 2022, 14, e25353. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, U.; Bohleber, S.; Zhao, W.; Fradejas-Villar, N. The Neurobiology of Selenium: Looking Back and to the Future. Front. Neurosci. 2021, 15, 652099. [Google Scholar] [CrossRef] [PubMed]
- Leiter, O.; Zhuo, Z.; Rust, R.; Wasielewska, J.M.; Grönnert, L.; Kowal, S.; Overall, R.W.; Adusumilli, V.S.; Blackmore, D.G.; Southon, A.; et al. Selenium mediates exercise-induced adult neurogenesis and reverses learning deficits induced by hippocampal injury and aging. Cell Metab. 2022, 34, 408–423.e8. [Google Scholar] [CrossRef]
- Ball, H.A.; McWhirter, L.; Ballard, C.; Bhome, R.; Blackburn, D.J.; Edwards, M.J.; Fleming, S.M.; Fox, N.C.; Howard, R.; Huntley, J.; et al. Functional cognitive disorder: Dementia’s blind spot. Brain 2020, 143, 2895–2903. [Google Scholar] [CrossRef]
- Mandrioli, J.; Michalke, B.; Solovyev, N.; Grill, P.; Violi, F.; Lunetta, C.; Conte, A.; Sansone, V.A.; Sabatelli, M.; Vinceti, M. Elevated Levels of Selenium Species in Cerebrospinal Fluid of Amyotrophic Lateral Sclerosis Patients with Disease-Associated Gene Mutations. Neurodegener. Dis. 2017, 17, 171–180. [Google Scholar] [CrossRef]
- Xiong, L.; McCoy, M.; Komuro, H.; West, X.Z.; Yakubenko, V.; Gao, D.; Dudiki, T.; Milo, A.; Chen, J.; Podrez, E.A.; et al. Inflammation-dependent oxidative stress metabolites as a hallmark of amyotrophic lateral sclerosis. Free Radic. Biol. Med. 2021, 178, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Scharpf, M.; Schweizer, U.; Arzberger, T.; Roggendorf, W.; Schomburg, L.; Köhrle, J. Neuronal and ependymal expression of selenoprotein P in the human brain. J. Neural Transm. 2007, 114, 877–884. [Google Scholar] [CrossRef]
- Solovyev, N.; Berthele, A.; Michalke, B. Selenium speciation in paired serum and cerebrospinal fluid samples. Anal. Bioanal. Chem. 2012, 405, 1875–1884. [Google Scholar] [CrossRef]
- Saito, Y.; Hayashi, T.; Tanaka, A.; Watanabe, Y.; Suzuki, M.; Saito, E.; Takahashi, K. Selenoprotein P in Human Plasma as an Extracellular Phospholipid Hydroperoxide Glutathione Peroxidase. J. Biol. Chem. 1999, 274, 2866–2871. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Caprioli, R.M.; Hill, K.E.; Burk, R.F. Loss of selenium from selenoproteins: Conversion of selenocysteine to dehydroalanine in vitro. J. Am. Soc. Mass Spectrom. 2003, 14, 593–600. [Google Scholar] [CrossRef]
- Kurokawa, S.; Bellinger, F.P.; Hill, K.E.; Burk, R.F.; Berry, M.J. Isoform-specific Binding of Selenoprotein P to the β-Propeller Domain of Apolipoprotein E Receptor 2 Mediates Selenium Supply. J. Biol. Chem. 2014, 289, 9195–9207. [Google Scholar] [CrossRef] [PubMed]
- Turanov, A.A.; Everley, R.A.; Hybsier, S.; Renko, K.; Schomburg, L.; Gygi, S.P.; Hatfield, D.L.; Gladyshev, V.N. Regulation of Selenocysteine Content of Human Selenoprotein P by Dietary Selenium and Insertion of Cysteine in Place of Selenocysteine. PLoS ONE 2015, 10, e0140353. [Google Scholar] [CrossRef]
- Hybsier, S.; Schulz, T.; Wu, Z.; Demuth, I.; Minich, W.B.; Renko, K.; Rijntjes, E.; Köhrle, J.; Strasburger, C.J.; Steinhagen-Thiessen, E.; et al. Sex-specific and inter-individual differences in biomarkers of selenium status identified by a calibrated ELISA for selenoprotein P. Redox Biol. 2016, 11, 403–414. [Google Scholar] [CrossRef]
- Lamarche, J.; Ronga, L.; Szpunar, J.; Lobinski, R. Characterization and Quantification of Selenoprotein P: Challenges to Mass Spectrometry. Int. J. Mol. Sci. 2021, 22, 6283. [Google Scholar] [CrossRef]
- Rueli, R.H.; Parubrub, A.C.; Dewing, A.S.; Hashimoto, A.C.; Bellinger, M.T.; Weeber, E.J.; Uyehara-Lock, J.H.; White, L.R.; Berry, M.J.; Bellinger, F.P. Increased Selenoprotein P in Choroid Plexus and Cerebrospinal Fluid in Alzheimer’s Disease Brain. J. Alzheimer’s Dis. 2015, 44, 379–383. [Google Scholar] [CrossRef]
- Vinceti, M.; Chiari, A.; Eichmuller, M.; Rothman, K.J.; Filippini, T.; Malagoli, C.; Weuve, J.; Tondelli, M.; Zamboni, G.; Nichelli, P.F.; et al. A selenium species in cerebrospinal fluid predicts conversion to Alzheimer’s dementia in persons with mild cognitive impairment. Alzheimers Res. Ther. 2017, 9, 100. [Google Scholar] [CrossRef]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Winblad, B.; Palmer, K.; Kivipelto, M.; Jelic, V.; Fratiglioni, L.; Wahlund, L.-O.; Nordberg, A.; Backman, L.J.; Albert, M.S.; Almkvist, O.; et al. Mild cognitive impairment—beyond controversies, towards a consensus: Report of the International Working Group on Mild Cognitive Impairment. J. Intern. Med. 2004, 256, 240–246. [Google Scholar] [CrossRef]
- Limongi, F.; for the MCI Working Group; Noale, M.; Bianchetti, A.; Ferrara, N.; Padovani, A.; Scarpini, E.; Trabucchi, M.; Maggi, S. The instruments used by the Italian centres for cognitive disorders and dementia to diagnose mild cognitive impairment (MCI). Aging 2018, 31, 101–107. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Tondelli, M.; Bedin, R.; Chiari, A.; Molinari, M.A.; Bonifacio, G.; Lelli, N.; Trenti, T.; Nichelli, P. Role of cerebrospinal fluid biomarkers to predict conversion to dementia in patients with mild cognitive impairment: A clinical cohort study. Clin. Chem. Lab. Med. (CCLM) 2015, 53, 453–460. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saito, Y.; Misu, H.; Takayama, H.; Takashima, S.-I.; Usui, S.; Takamura, M.; Kaneko, S.; Takamura, T.; Noguchi, N. Comparison of Human Selenoprotein P Determinants in Serum between Our Original Methods and Commercially Available Kits. Biol. Pharm. Bull. 2018, 41, 828–832. [Google Scholar] [CrossRef]
| Group Ɨ | ||||
|---|---|---|---|---|
| AD (n = 30) | MCI (n = 54) | ALS (n = 50) | Controls (n = 30) | |
| Selenoprotein P (ng/mL) | ||||
| CSF | 26.3 (21.0–39.1) | 28.7 (19.8–36.1) | 18.7 (13.1–26.1) | 22.1 (14.9–29.9) |
| Serum | 6012 (5595–6319) | 6952 (6191–7686) | 6623 (5817–7136) | 5615 (4683–6235) |
| Selenoprotein-P-bound selenium in CSF (ng/mL) | 1.4 (0.8–1.9) | 1.6 (1.1–2.0) | - | - |
| Neurodegeneration biomarkers (ng/L) | ||||
| Amyloid-beta1–42 | 447 (385–494) | 600 (441–806) | - | - |
| Phosphorylated tau | 94 (73–105) | 68 (49–88) | - | - |
| Total tau | 568 (433–782) | 337 (219–614) | - | - |
| Univariate Model | Adjusted Ɨ Model | |||||
|---|---|---|---|---|---|---|
| AD | MCI | ALS | AD | MCI | ALS | |
| Selenoprotein P (ng/mL) | ||||||
| CSF | 1.33 (0.90–1.97) | 1.28 (0.86–1.90) | 0.72 (0.48–1.08) | 1.26 (0.83–1.91) | 1.12 (0.69–1.79) | 0.63 (0.40–1.00) |
| Serum | 1.01 (1.00–1.01) | 1.01 (1.01–1.02) | 1.01 (1.00–1.02) | 1.01 (1.00–1.01) | 1.02 (1.01–1.03) | 1.01 (1.00–1.02) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbano, T.; Vinceti, M.; Mandrioli, J.; Chiari, A.; Filippini, T.; Bedin, R.; Tondelli, M.; Simonini, C.; Zamboni, G.; Shimizu, M.; et al. Selenoprotein P Concentrations in the Cerebrospinal Fluid and Serum of Individuals Affected by Amyotrophic Lateral Sclerosis, Mild Cognitive Impairment and Alzheimer’s Dementia. Int. J. Mol. Sci. 2022, 23, 9865. https://doi.org/10.3390/ijms23179865
Urbano T, Vinceti M, Mandrioli J, Chiari A, Filippini T, Bedin R, Tondelli M, Simonini C, Zamboni G, Shimizu M, et al. Selenoprotein P Concentrations in the Cerebrospinal Fluid and Serum of Individuals Affected by Amyotrophic Lateral Sclerosis, Mild Cognitive Impairment and Alzheimer’s Dementia. International Journal of Molecular Sciences. 2022; 23(17):9865. https://doi.org/10.3390/ijms23179865
Chicago/Turabian StyleUrbano, Teresa, Marco Vinceti, Jessica Mandrioli, Annalisa Chiari, Tommaso Filippini, Roberta Bedin, Manuela Tondelli, Cecilia Simonini, Giovanna Zamboni, Misaki Shimizu, and et al. 2022. "Selenoprotein P Concentrations in the Cerebrospinal Fluid and Serum of Individuals Affected by Amyotrophic Lateral Sclerosis, Mild Cognitive Impairment and Alzheimer’s Dementia" International Journal of Molecular Sciences 23, no. 17: 9865. https://doi.org/10.3390/ijms23179865
APA StyleUrbano, T., Vinceti, M., Mandrioli, J., Chiari, A., Filippini, T., Bedin, R., Tondelli, M., Simonini, C., Zamboni, G., Shimizu, M., & Saito, Y. (2022). Selenoprotein P Concentrations in the Cerebrospinal Fluid and Serum of Individuals Affected by Amyotrophic Lateral Sclerosis, Mild Cognitive Impairment and Alzheimer’s Dementia. International Journal of Molecular Sciences, 23(17), 9865. https://doi.org/10.3390/ijms23179865












