The Role of Protein Arginine Methyltransferases in DNA Damage Response
Abstract
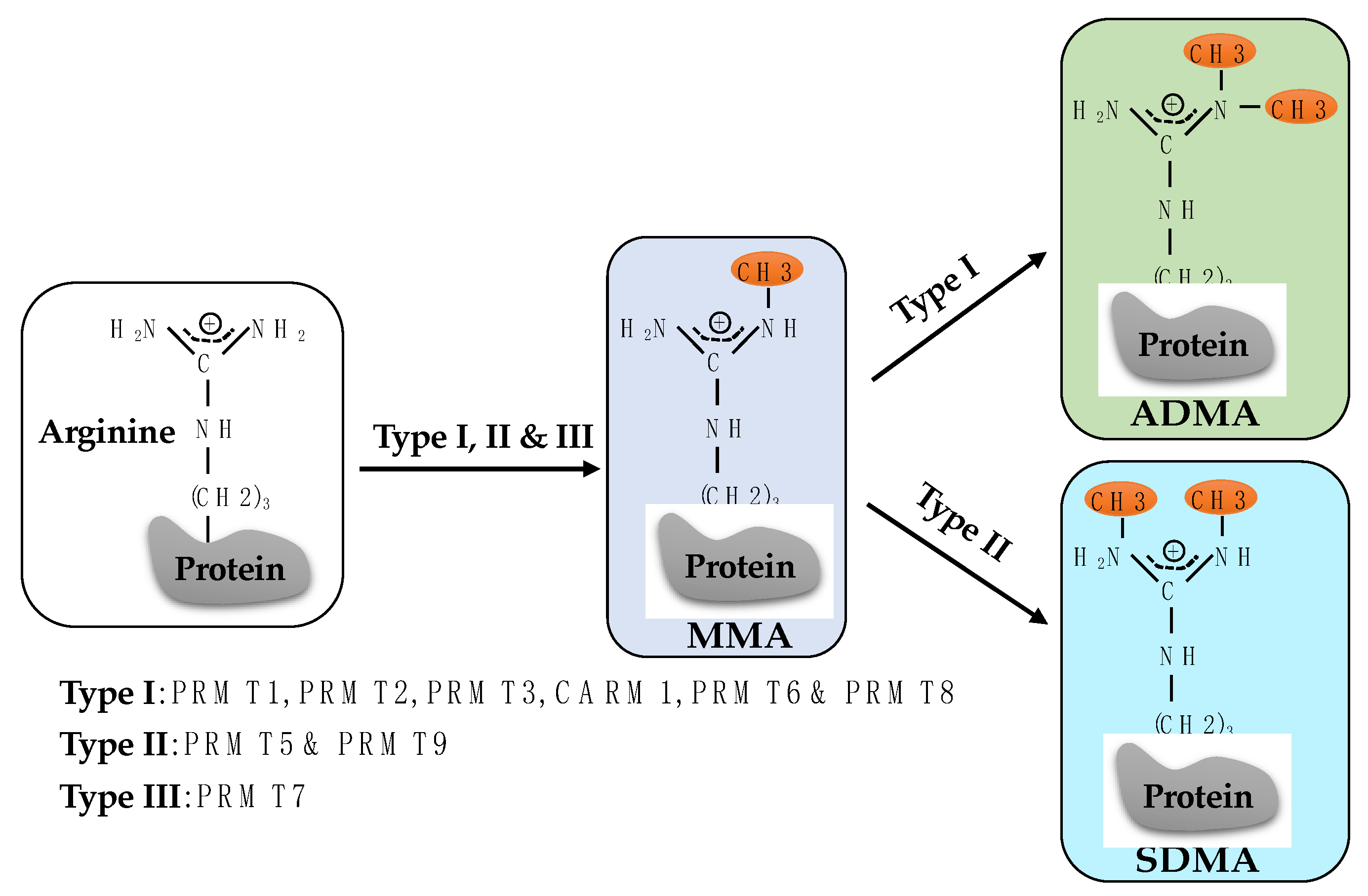
1. Introduction
2. Roles of PRMTs in DDR
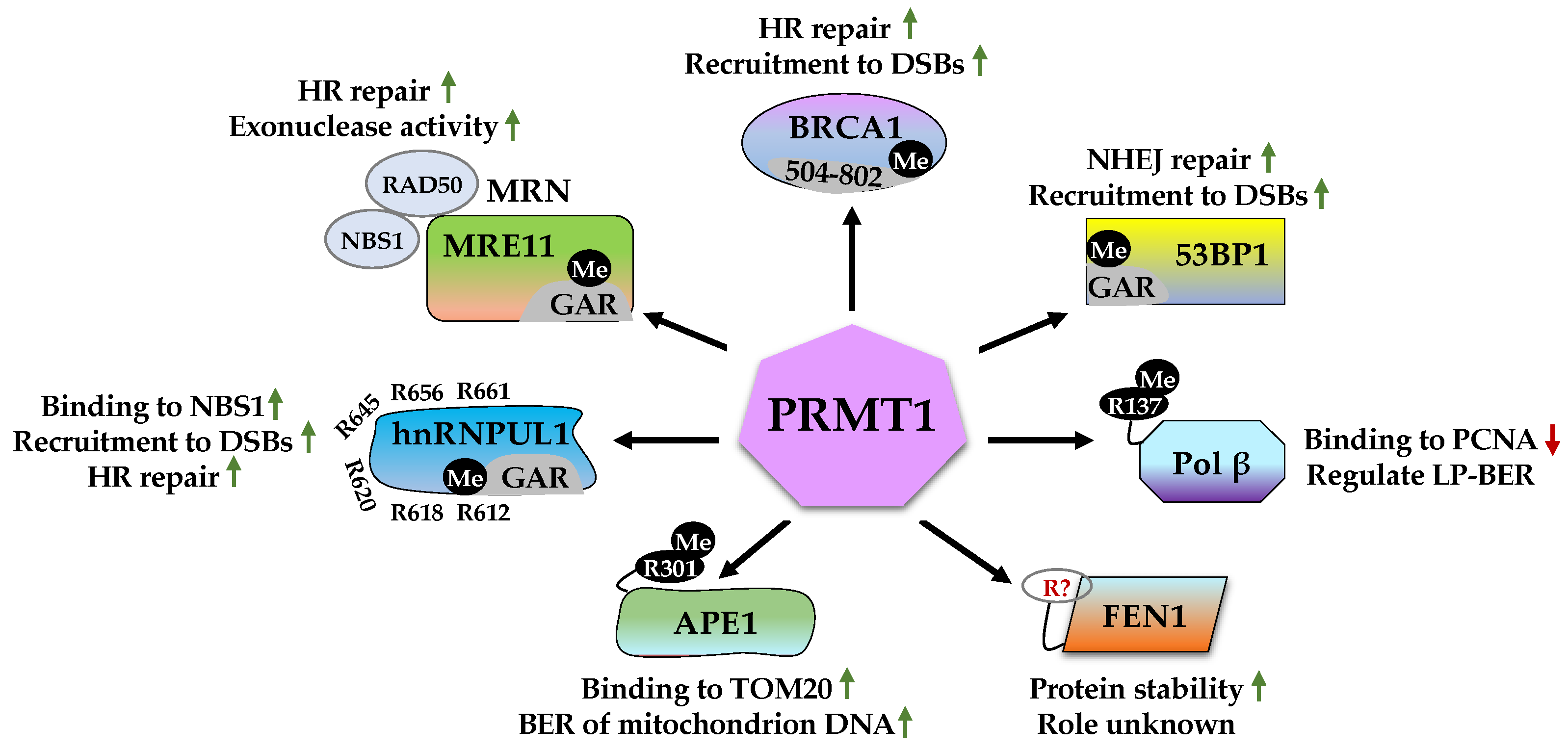
2.1. Roles of PRMT1 in DDR
2.1.1. MRE11 (Meiotic Recombination 11)
2.1.2. BRCA1 (Breast Cancer Type 1 Susceptibility Protein)
2.1.3. 53BP1 (p53-Binding Protein 1)
2.1.4. Pol β (DNA Polymerase β)
2.1.5. FEN1 (Flap Endonuclease 1)
2.1.6. APE1 (Apurinic/Apyrimidinic Endonuclease 1)
2.1.7. hnRNPUL1 (Heterogeneous Nuclear Ribonucleoprotein U-like Protein 1)
2.2. Roles of PRMT2 in DDR
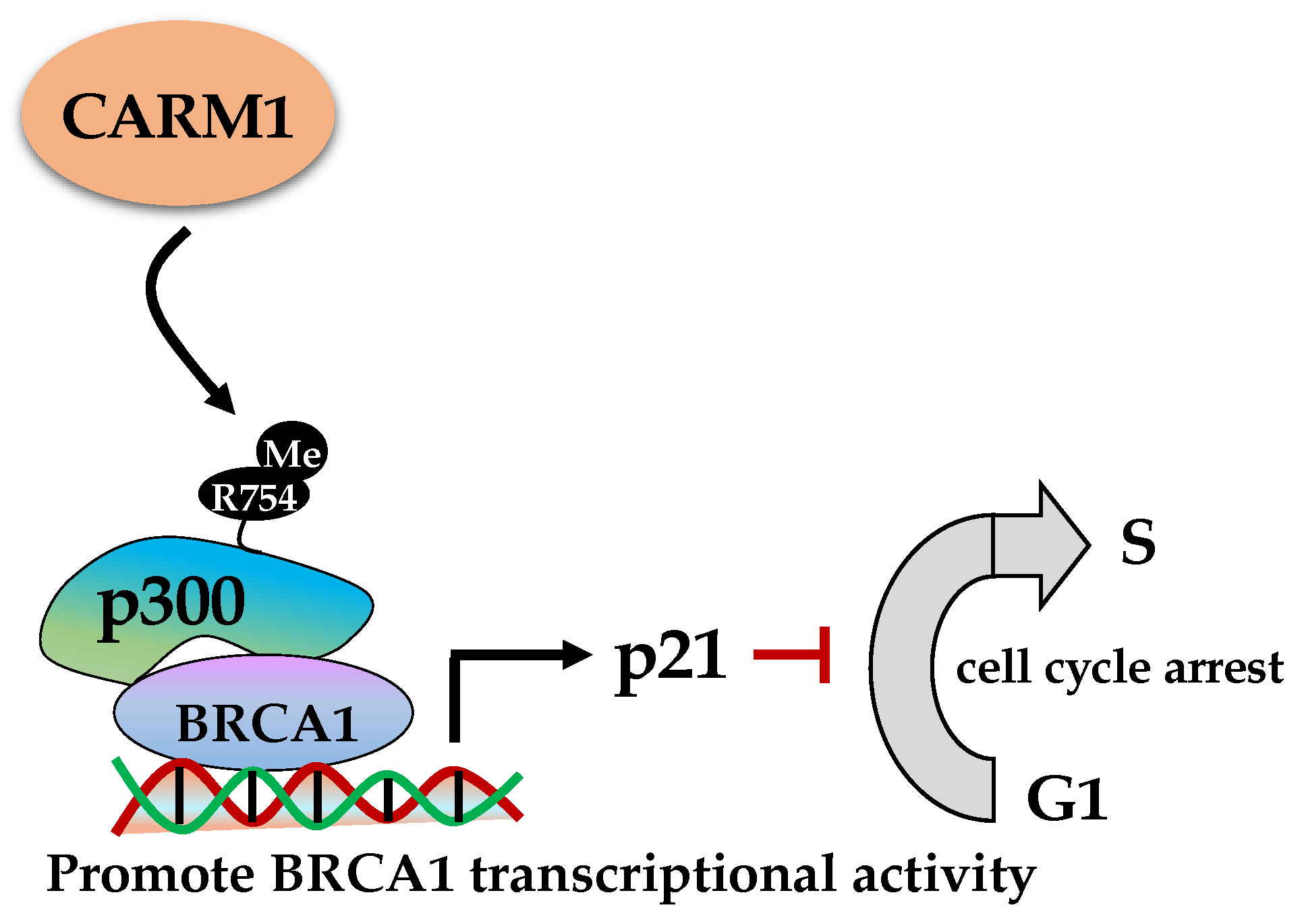
2.3. Roles of CARM1 (PRMT4) in DDR
2.4. Role of PRMT5 in DDR
2.4.1. 53BP1 (p53-Binding Protein 1)
2.4.2. FEN1 (Flap Endonuclease 1)
2.4.3. RAD9
2.4.4. RUVBL1 (RuvB-like 1)
2.4.5. TDP1 (Tyrosyl-DNA Phosphodiesterase 1)
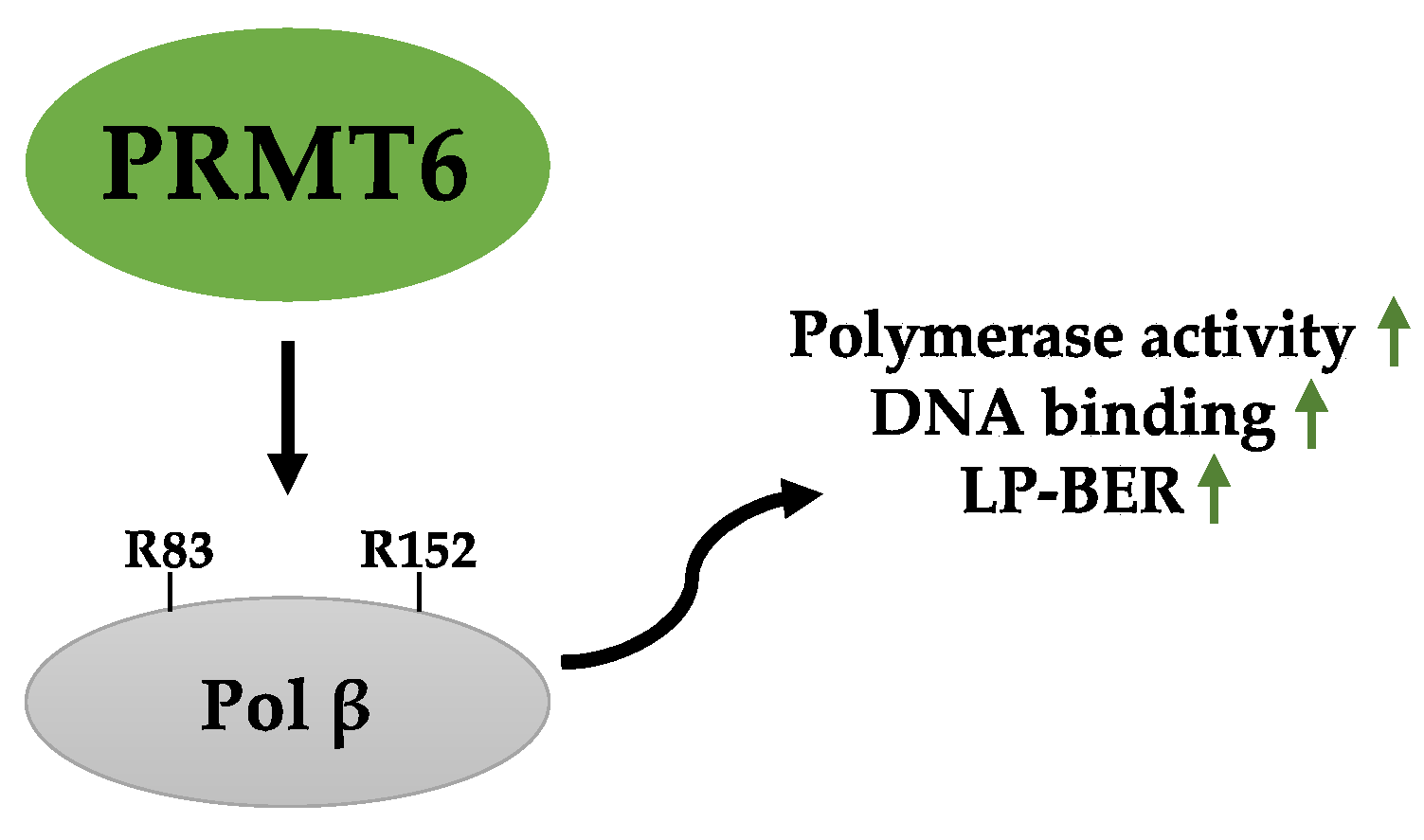
2.5. Role of PRMT6 in DDR
2.6. Role of PRMT7 in DDR
2.7. Role of PRMT8 in DDR
2.8. Role of PRMT3 and PRMT9 in DDR
3. Synergistic Effects of PRMT and DDR
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| 53BP1 | p53-binding protein 1 | Key mediator of NHEJ |
| ALKBH5 | AlkB homolog 5 | RNA demethylase of m6A |
| APE1 | Apurinic/apyrimidinic endonuclease 1 | Key player of BER |
| APEX2 | Apurinic/apyrimidinic endodeoxyribonuclease 2 | Key player of BER |
| ATM | Ataxia-telangiectasia mutated | Master regulator of DDR |
| ATR | ATM- and Rad3-Related | Essential kinase involved in DNA replication stress |
| ATRIP | ATR-interacting partner | Recruitment of ATR to DNA damaging site |
| BARD1 | BRCA1-associated RING domain protein | Interaction with BRCA1 to potentiate BRCA1 E3 ligase function |
| BRCA1 | Breast cancer type 1 susceptibility protein | Key mediator of HR |
| BRCT | BRCA1 C terminus | Phospho-protein binding domain |
| CARM1 | Coactivator-associated arginine methyltransferase 1 | Type I protein arginine methyltransferase |
| CDK2 | Cyclin-dependent kinase 2 | G1/S and S/G2 transition control |
| Chk1 | Checkpoint kinase 1 | Cell cycle checkpoint control during DDR |
| Chk2 | Checkpoint kinase 2 | Cell cycle checkpoint control during DDR |
| CtIP | C-terminal-binding protein (CtBP)-interacting protein | Interaction with MRN complex to facilitate DNA end resection |
| DNA-PK | DNA-dependent protein kinase | Regulator of NHEJ and HR |
| Dvl3 | Dishevelled 3 | Key player of the Wnt signaling pathway |
| eIF2α | Eukaryotic translation initiation factor 2α | Regulation of translation initiation |
| FEN1 | Flap endonuclease 1 | Key enzyme for DNA replication and repair |
| G3BP2 | Ras GTPase-activating protein-binding protein 2 | RNA-binding protein involved in mRNA metabolism and stress granules formation |
| GADD45 | Growth arrest and DNA damage-inducible gene 45 | Regulator of DNA repair, cell cycle control, senescence, and genotoxic stress |
| GFI1 | Growth factor independent 1 | Transcriptional repressor |
| H2AX | H2A histone family member X | Accumulation and recruitment of DNA repair proteins to sites of DSBs |
| hnRNPUL1 | Heterogeneous nuclear ribonucleoprotein U-like protein 1 | RNA-binding protein involved in RNA metabolism and DNA repair |
| Hsp70 | Heat shock protein 70 | Molecular chaperone in protein folding |
| MDC1 | Mediator of DNA damage checkpoint 1 | Recruitment of repair proteins to the site of DNA damage |
| MEP50 | Methylosome protein 50 | Interaction with PRMT5 to promote activity |
| MRE11 | Meiotic recombination 11 | Component of MRN complex for DSB sensing |
| Nav1.2 | Voltage-gated sodium channel type 2 | Sodium channel involved establishing action potential |
| NBS1 | Nijmegen breakage syndrome 1 | Component of MRN complex for DSB sensing |
| NIFK | Nucleolar protein interacting with the forkhead-associated domain of Ki-67 | Regulator of RNA maturation during cell cycle progression |
| NUDT16 | Nudix hydrolase 16 | RNA-binding and decapping enzyme |
| PALB2 | Partner and localizer of BRCA2 | Bridging molecule connecting BRCA1- BRCA2-RAD51 to promote HR |
| PARP1 | Poly (ADP-ribose) polymerase 1 | Poly(ADP-ribosyl)transferase critical for initiation of DNA repair |
| PCNA | Proliferating cell nuclear antigen | Key factor in DNA replication and repair |
| PI3K | Phosphoinositide 3-kinase | Synthesizing PtdIns(3,4,5)P3 (PIP3) to activate AKT/mTOR pathway |
| PNKP | Polynucleotide kinase 3’-phosphatase | Polynucleotide phosphatase/kinase involved in NHEJ and BER |
| Pol β | DNA polymerase β | Catalyzing DNA synthesis |
| POLD1 | DNA polymerase delta 1 | Catalytic subunit of DNA polymerase δ |
| POLD2 | DNA polymerase delta subunit 2 | Regulatory subunit of DNA polymerase δ |
| PTIP | Pax transactivation domain-interacting protein | Interaction with repair proteins to regulator DNA repair |
| RFC2-5 | Replication factor C subunit 2-5 | Accessory proteins involved in the elongation of primed DNA template. |
| RIF1 | Replication timing regulatory factor 1 | Regulation of DSB repair pathway choice and DNA replication timings |
| RING | Really Interesting New Gene | Catalytic domain of RING E3 ubiquitin ligases |
| RNF146 | RING finger protein 146 | Poly(ADP-ribose)-directed E3 ligase |
| RNF168 | RING finger protein 168 | Key E3 ubiquitin ligase required for accumulation of repair proteins to sites of DNA damage |
| RNF8 | RING finger protein 8 | Key E3 ubiquitin ligase required for accumulation of repair proteins to sites of DNA damage |
| RPA | Replication protein A | Single-strand DNA binding protein |
| RUVBL1 | RuvB-like 1 | DNA helicase involved in chromatin remodeling, transcription, and DNA repair |
| RUVBL2 | RuvB-like 2 | DNA helicase involved in chromatin remodeling, transcription, and DNA repair |
| TDP1 | Tyrosyl-DNA phosphodiesterase 1 | Repair of trapped Top I cleavage complexes |
| TIP60 | Tat interacting protein 60 kD | Lysine acetyltransferase |
| TOM20 | Translocase of outer mitochondrial membrane 20 | Central component of the TOM receptor complex responsible for translocation of proteins to mitochondria |
| Top I | Topoisomerase I | Enzyme that relaxes supercoiled DNA |
| TopBP1 | Topoisomerase II Binding Protein 1 | Scaffold protein involved in DNA replication, DNA repair, and transcription |
| XRCC1 | X-ray repair cross-complementing protein 1 | Scaffold protein involved in repair of SSBs |
| YY1 | Yin Yang 1 | Transcription factor |
References
- Lindahl, T.; Barnes, D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar]
- De Bont, R.; van Larebeke, N. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 2004, 19, 169–185. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [PubMed]
- Lans, H.; Hoeijmakers, J.H.J.; Vermeulen, W.; Marteijn, J.A. The DNA damage response to transcription stress. Nat. Rev. Mol. Cell Biol. 2019, 20, 766–784. [Google Scholar] [CrossRef] [PubMed]
- Surova, O.; Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 2012, 32, 3789–3797. [Google Scholar] [CrossRef]
- Zhou, B.-B.S.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P.-K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 1–35. [Google Scholar] [CrossRef]
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007, 18, 85–98. [Google Scholar] [CrossRef]
- Caldecott, K.W. Mammalian DNA base excision repair: Dancing in the moonlight. DNA Repair 2020, 93, 102921. [Google Scholar] [CrossRef]
- Spivak, G. Nucleotide excision repair in humans. DNA Repair 2015, 36, 13–18. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.; Klein, H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Lanz, M.C.; DiBitetto, D.; Smolka, M.B. DNA damage kinase signaling: Checkpoint and repair at 30 years. EMBO J. 2019, 38, e101801. [Google Scholar] [CrossRef]
- Rouse, J.; Jackson, S.P. Interfaces between the detection, signaling, and repair of DNA damage. Science 2002, 297, 547–551. [Google Scholar]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Lavin, M.F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 2007, 26, 7749–7758. [Google Scholar] [CrossRef]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar]
- Walker, J.R.; Corpina, R.A.; Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 2001, 412, 607–614. [Google Scholar] [CrossRef]
- Sibanda, B.L.; Chirgadze, D.Y.; Ascher, D.B.; Blundell, T.L. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 2017, 355, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef]
- Huen, M.S.-Y.; Chen, J. The DNA damage response pathways: At the crossroad of protein modifications. Cell Res. 2007, 18, 8–16. [Google Scholar] [CrossRef]
- Dantuma, N.P.; van Attikum, H. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 2016, 35, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM Phosphorylates Histone H2AX in Response to DNA Double-strand Breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Downs, J.A.; Lowndes, N.F.; Jackson, S.P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 2000, 408, 1001–1004. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA Double-stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef]
- Ward, I.M.; Chen, J. Histone H2AX Is Phosphorylated in an ATR-dependent Manner in Response to Replicational Stress. J. Biol. Chem. 2001, 276, 47759–47762. [Google Scholar] [CrossRef] [PubMed]
- Podhorecka, M.; Skladanowski, A.; Bozko, P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J. Nucleic Acids 2010, 2010, 920161. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks in Vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016, 9, rs9. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef]
- Bedford, M.T.; Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef]
- Zhang, F.; Kerbl-Knapp, J.; Colman, M.J.R.; Meinitzer, A.; Macher, T.; Vujić, N.; Fasching, S.; Jany-Luig, E.; Korbelius, M.; Kuentzel, K.B.; et al. Global analysis of protein arginine methylation. Cell Rep. Methods 2021, 1, 100016. [Google Scholar] [CrossRef]
- Bedford, M.T.; Clarke, S.G. Protein Arginine Methylation in Mammals: Who, What, and Why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef]
- Bedford, M.T. Arginine methylation at a glance. J. Cell Sci. 2007, 120, 4243–4246. [Google Scholar] [CrossRef]
- Lee, D.Y.; Teyssier, C.; Strahl, B.D.; Stallcup, M.R. Role of Protein Methylation in Regulation of Transcription. Endocr. Rev. 2005, 26, 147–170. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Stallcup, M.R. Minireview: Protein Arginine Methylation of Nonhistone Proteins in Transcriptional Regulation. Mol. Endocrinol. 2009, 23, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Auclair, Y.; Richard, S. The role of arginine methylation in the DNA damage response. DNA Repair 2013, 12, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ronai, Z.A. PRMT5 function and targeting in cancer. Cell Stress 2020, 4, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Raposo, A.E.; Piller, S.C. Protein arginine methylation: An emerging regulator of the cell cycle. Cell Div. 2018, 13, 3. [Google Scholar] [CrossRef]
- Dhar, S.; Vemulapalli, V.; Patananan, A.N.; Huang, G.L.; Di Lorenzo, A.; Richard, S.; Comb, M.J.; Guo, A.; Clarke, S.G.; Bedford, M.T. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 2013, 3, 1311. [Google Scholar] [CrossRef]
- Tang, J.; Frankel, A.; Cook, R.J.; Kim, S.; Paik, W.K.; Williams, K.R.; Clarke, S.; Herschman, H.R. PRMT1 Is the Predominant Type I Protein Arginine Methyltransferase in Mammalian Cells. J. Biol. Chem. 2000, 275, 7723–7730. [Google Scholar] [CrossRef]
- Liu, L.M.; Sun, W.Z.; Fan, X.Z.; Xu, Y.L.; Cheng, M.B.; Zhang, Y. Methylation of C/EBPalpha by PRMT1 Inhibits Its Tumor-Suppressive Function in Breast Cancer. Cancer Res. 2019, 79, 2865–2877. [Google Scholar] [CrossRef]
- Tang, S.; Sethunath, V.; Metaferia, N.Y.; Nogueira, M.F.; Gallant, D.S.; Garner, E.R.; Lairson, L.A.; Penney, C.M.; Li, J.; Gelbard, M.K.; et al. A genome-scale CRISPR screen reveals PRMT1 as a critical regulator of androgen receptor signaling in prostate cancer. Cell Rep. 2022, 38, 110417. [Google Scholar] [CrossRef]
- Yao, B.; Gui, T.; Zeng, X.; Deng, Y.; Wang, Z.; Wang, Y.; Yang, D.; Li, Q.; Xu, P.; Hu, R.; et al. PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 2021, 13, 58. [Google Scholar] [CrossRef]
- Nicholson, T.B.; Chen, T.; Richard, S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol. Res. 2009, 60, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Yoshimatsu, M.; Toyokawa, G.; Hayami, S.; Unoki, M.; Tsunoda, T.; Field, H.I.; Kelly, J.D.; Neal, D.E.; Maehara, Y.; Ponder, B.A.; et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 2010, 128, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, T.; Hébert, J.; Li, E.; Richard, S. A Mouse PRMT1 Null Allele Defines an Essential Role for Arginine Methylation in Genome Maintenance and Cell Proliferation. Mol. Cell. Biol. 2009, 29, 2982–2996. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Gellert, M. The 3′ to 5′ Exonuclease Activity of Mre11 Facilitates Repair of DNA Double-Strand Breaks. Mol. Cell 1998, 1, 969–979. [Google Scholar] [CrossRef]
- Garcia, V.; Phelps, S.E.L.; Gray, S.; Neale, M.J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T. 20 Years of Mre11 Biology: No End in Sight. Mol. Cell 2018, 71, 419–427. [Google Scholar] [CrossRef]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: Implications for cancer treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef]
- Jacobsen, E.; Beach, T.; Shen, Y.; Li, R.; Chang, Y. Deficiency of the Mre11 DNA repair complex in Alzheimer’s disease brains. Brain Res. Mol. Brain Res. 2004, 128, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhang, H.; Jiang, Y.-N.; Wang, Z.-Q.; Sun, L.; Zhou, Z.-W. Post-Translational Modification of MRE11: Its Implication in DDR and Diseases. Genes 2021, 12, 1158. [Google Scholar] [CrossRef]
- Hopfner, K.-P.; Karcher, A.; Craig, L.; Woo, T.T.; Carney, J.P.; Tainer, J.A. Structural Biochemistry and Interaction Architecture of the DNA Double-Strand Break Repair Mre11 Nuclease and Rad50-ATPase. Cell 2001, 105, 473–485. [Google Scholar] [CrossRef]
- Park, Y.B.; Chae, J.; Kim, Y.C.; Cho, Y. Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations. Structure 2011, 19, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.-M.; Déry, U.; Masson, J.-Y.; Richard, S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 2005, 19, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Vogel, G.; Coulombe, Y.; Dubeau, D.; Spehalski, E.; Hébert, J.; Ferguson, D.O.; Masson, J.Y.; Richard, S. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 2011, 22, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Vadnais, C.; Chen, R.; Fraszczak, J.; Yu, Z.; Boulais, J.; Pinder, J.; Frank, D.; Khandanpour, C.; Hébert, J.; Dellaire, G.; et al. GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat. Commun. 2018, 9, 1418. [Google Scholar] [CrossRef]
- Prakash, R.; Zhang, Y.; Feng, W.; Jasin, M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb. Perspect. Biol. 2015, 7, a016600. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef]
- Liu, Y.; Strumpfer, J.; Freddolino, P.L.; Gruebele, M.; Schulten, K. Structural Characterization of lambda-Repressor Folding from All-Atom Molecular Dynamics Simulations. J. Phys. Chem. Lett. 2012, 3, 1117–1123. [Google Scholar] [CrossRef]
- Wu, L.C.; Wang, Z.W.; Tsan, J.T.; Spillman, M.A.; Phung, A.; Xu, X.L.; Yang, M.-C.W.; Hwang, L.-Y.; Bowcock, A.; Baer, R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996, 14, 430–440. [Google Scholar] [CrossRef]
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-derived Mutation. J. Biol. Chem. 2001, 276, 14537–14540. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, J.; Wu, J.; Ye, L.; Cai, H.; Xia, B.; Yu, X. PALB2 Links BRCA1 and BRCA2 in the DNA-Damage Response. Curr. Biol. 2009, 19, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Sy, S.M.H.; Huen, M.S.Y.; Chen, J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA 2009, 106, 7155–7160. [Google Scholar] [CrossRef] [PubMed]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.-T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 Inhibits Homologous Recombination in Brca1-Deficient Cells by Blocking Resection of DNA Breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Garcia, A.; Lopez-Saavedra, A.; Huertas, P. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014, 9, 451–459. [Google Scholar] [CrossRef]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef]
- Guendel, I.; Carpio, L.; Pedati, C.; Schwartz, A.; Teal, C.; Kashanchi, F.; Kehn-Hall, K. Methylation of the Tumor Suppressor Protein, BRCA1, Influences Its Transcriptional Cofactor Function. PLoS ONE 2010, 5, e11379. [Google Scholar] [CrossRef]
- Montenegro, M.F.; González-Guerrero, R.; Sánchez-Del-Campo, L.; Piñero-Madrona, A.; Cabezas-Herrera, J.; Rodríguez-López, J.N. PRMT1-dependent methylation of BRCA1 contributes to the epigenetic defense of breast cancer cells against ionizing radiation. Sci. Rep. 2020, 10, 13275. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Escribano-Díaz, C.; Orthwein, A.; Fradet-Turcotte, A.; Xing, M.; Young, J.T.; Tkáč, J.; Cook, M.A.; Rosebrock, A.P.; Munro, M.; Canny, M.D.; et al. A Cell Cycle-Dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Mol. Cell 2013, 49, 872–883. [Google Scholar] [CrossRef]
- Bouwman, P.; Aly, A.; Escandell, J.M.; Pieterse, M.; Bartkova, J.; Van Der Gulden, H.; Hiddingh, S.; Thanasoula, M.; Kulkarni, A.; Yang, Q.; et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010, 17, 688–695. [Google Scholar] [CrossRef]
- Jaspers, J.E.; Kersbergen, A.; Boon, U.; Sol, W.; van Deemter, L.; Zander, S.A.; Drost, R.; Wientjens, E.; Ji, J.; Aly, A.; et al. Loss of 53BP1 Causes PARP Inhibitor Resistance in Brca1-Mutated Mouse Mammary Tumors. Cancer Discov. 2013, 3, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Mirza-Aghazadeh-Attari, M.; Mohammadzadeh, A.; Yousefi, B.; Mihanfar, A.; Karimian, A.; Majidinia, M. 53BP1: A key player of DNA damage response with critical functions in cancer. DNA Repair 2018, 73, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2013, 15, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.M.; Wang, B.; Xia, Z.; Morales, J.C.; Lu, X.; Donehower, L.A.; Bochar, D.A.; Elledge, S.J.; Carpenter, P.B. 53BP1 Oligomerization is Independent of its Methylation by PRMT1. Cell Cycle 2005, 4, 1854–1861. [Google Scholar] [CrossRef]
- Boisvert, F.-M.; Rhie, A.; Richard, S.; Doherty, A.J. The GAR Motif of 53BP1 is Arginine Methylated by PRMT1 and is Necessary for 53BP1 DNA Binding Activity. Cell Cycle 2005, 4, 1834–1841. [Google Scholar] [CrossRef]
- Mentegari, E.; Kissova, M.; Bavagnoli, L.; Maga, G.; Crespan, E. DNA Polymerases lambda and beta: The Double-Edged Swords of DNA Repair. Genes 2016, 7, 57. [Google Scholar] [CrossRef]
- Beard, W.A.; Wilson, S.H. Structure and mechanism of DNA polymerase beta. Biochemistry 2014, 53, 2768–2780. [Google Scholar] [CrossRef]
- Ray, S.; Breuer, G.; DeVeaux, M.; Zelterman, D.; Bindra, R.; Sweasy, J.B. DNA polymerase beta participates in DNA End-joining. Nucleic Acids Res. 2017, 46, 242–255. [Google Scholar] [CrossRef]
- El-Andaloussi, N.; Valovka, T.; Toueille, M.; Hassa, P.O.; Gehrig, P.; Covic, M.; Hubscher, U.; Hottiger, M.O. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 2007, 21, 26–34. [Google Scholar] [CrossRef]
- Klungland, A.; Lindahl, T. Second pathway for completion of human DNA base excision-repair: Reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 1997, 16, 3341–3348. [Google Scholar] [CrossRef]
- Shen, B.; Singh, P.; Liu, R.; Qiu, J.; Zheng, L.; Finger, L.D.; Alas, S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. BioEssays 2005, 27, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, L.; Bambara, R.A. Flap endonuclease 1. Annu. Rev. Biochem. 2013, 82, 119–138. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.M.; Russell, R.; Albarakati, N.; Maloney, D.J.; Dorjsuren, D.; Rueda, O.M.; Moseley, P.; Mohan, V.; Sun, H.; Abbotts, R.; et al. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol. Oncol. 2014, 8, 1326–1338. [Google Scholar] [CrossRef]
- Zheng, L.; Jia, J.; Finger, L.D.; Guo, Z.; Zer, C.; Shen, B. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 2010, 39, 781–794. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hu, Z.; Sun, Y.; Zhang, M.; Zhu, H.; Jiang, L.; Zhang, Q.; Mu, D.; Zhang, J.; Gu, L.; et al. PRMT1 is critical to FEN1 expression and drug resistance in lung cancer cells. DNA Repair 2020, 95, 102953. [Google Scholar] [CrossRef] [PubMed]
- Abbotts, R.; Madhusudan, S. Human AP endonuclease 1 (APE1): From mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010, 36, 425–435. [Google Scholar] [CrossRef]
- Tell, G.; Quadrifoglio, F.; Tiribelli, C.; Kelley, M.R. The many functions of APE1/Ref-1: Not only a DNA repair enzyme. Antioxid. Redox Signal 2009, 11, 601–620. [Google Scholar] [CrossRef]
- Whitaker, A.M.; Freudenthal, B.D. APE1: A skilled nucleic acid surgeon. DNA Repair 2018, 71, 93–100. [Google Scholar] [CrossRef]
- Hegde, M.L.; Hazra, T.K.; Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008, 18, 27–47. [Google Scholar] [CrossRef]
- Thakur, S.; Sarkar, B.; Cholia, R.P.; Gautam, N.; Dhiman, M.; Mantha, A.K. APE1/Ref-1 as an emerging therapeutic target for various human diseases: Phytochemical modulation of its functions. Exp. Mol. Med. 2014, 46, e106. [Google Scholar] [CrossRef]
- Busso, C.S.; Lake, M.W.; Izumi, T. Posttranslational modification of mammalian AP endonuclease (APE1). Experientia 2010, 67, 3609–3620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.; Li, L.; Mu, D.; Hua, K.; Ci, S.; Shen, L.; Zheng, L.; Shen, B.; Guo, Z. Arginine methylation of APE1 promotes its mitochondrial translocation to protect cells from oxidative damage. Free Radic. Biol. Med. 2020, 158, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Park, S.B. Label-free target identification reveals oxidative DNA damage as the mechanism of a selective cytotoxic agent. Chem. Sci. 2019, 10, 3449–3458. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Yuan, J.; Yuan, B.-F.; Wang, Y. DNA–Protein Cross-Linking Sequencing for Genome-Wide Mapping of Thymidine Glycol. J. Am. Chem. Soc. 2022, 144, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Qual. Life Res. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Blackford, A.N.; Chapman, J.R.; Baskcomb, L.; Gravel, S.; Rusch, A.; Thomas, A.; Blundred, R.; Smith, P.; Kzhyshkowska, J.; et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell 2012, 45, 505–516. [Google Scholar] [CrossRef]
- Gurunathan, G.; Yu, Z.; Coulombe, Y.; Masson, J.-Y.; Richard, S. Arginine methylation of hnRNPUL1 regulates interaction with NBS1 and recruitment to sites of DNA damage. Sci. Rep. 2015, 5, 10475. [Google Scholar] [CrossRef]
- Katsanis, N.; Yaspo, M.-L.; Fisher, E. Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mamm. Genome 1997, 8, 526–529. [Google Scholar] [CrossRef]
- Scott, H.; Antonarakis, S.E.; Lalioti, M.D.; Rossiera, C.; Silver, P.A.; Henry, M.F. Identification and Characterization of Two Putative Human Arginine Methyltransferases (HRMT1L1 and HRMT1L2). Genomics 1998, 48, 330–340. [Google Scholar] [CrossRef]
- Cura, V.; Cavarelli, J. Structure, Activity and Function of the PRMT2 Protein Arginine Methyltransferase. Life 2021, 11, 1263. [Google Scholar] [CrossRef]
- Cura, V.; Marechal, N.; Troffer-Charlier, N.; Strub, J.-M.; Van Haren, M.J.; Martin, N.I.; Cianferani, S.; Bonnefond, L.; Cavarelli, J. Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors. FEBS J. 2016, 284, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, T.; Boehm, M.; Olive, M.; Crook, M.F.; San, H.; Langenickel, T.; Nabel, E.G. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp. Cell Res. 2006, 312, 2040–2053. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Yan, C.; Xie, P.; Cao, Y.; Shao, J.; Ge, J. PRMT2 accelerates tumorigenesis of hepatocellular carcinoma by activating Bcl2 via histone H3R8 methylation. Exp. Cell Res. 2020, 394, 112152. [Google Scholar] [CrossRef]
- Zhong, J.; Cao, R.-X.; Liu, J.-H.; Liu, Y.-B.; Wang, J.; Liu, L.-P.; Chen, Y.-J.; Yang, J.; Zhang, Q.-H.; Wu, Y.; et al. Nuclear loss of protein arginine N-methyltransferase 2 in breast carcinoma is associated with tumor grade and overexpression of cyclin D1 protein. Oncogene 2013, 33, 5546–5558. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oh, T.G.; Bailey, P.; Dray, E.; Smith, A.G.; Goode, J.; Eriksson, N.; Funder, J.W.; Fuller, P.J.; Simpson, E.R.; Tilley, W.D.; et al. PRMT2 and RORgamma expression are associated with breast cancer survival outcomes. Mol. Endocrinol. 2014, 28, 1166–1185. [Google Scholar] [CrossRef] [PubMed]
- Schurter, B.T.; Koh, S.S.; Chen, D.; Bunick, G.J.; Harp, J.M.; Hanson, B.L.; Henschen-Edman, A.; Mackay, D.R.; Stallcup, M.R.; Aswad, D.W. Methylation of Histone H3 by Coactivator-Associated Arginine Methyltransferase 1. Biochemistry 2001, 40, 5747–5756. [Google Scholar] [CrossRef]
- Bauer, U.; Daujat, S.; Nielsen, S.J.; Nightingale, K.; Kouzarides, T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002, 3, 39–44. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Z.; Meyer, M.B.; Saha, S.; Yu, M.; Guo, A.; Wisinski, K.B.; Huang, W.; Cai, W.; Pike, J.W.; et al. CARM1 Methylates Chromatin Remodeling Factor BAF155 to Enhance Tumor Progression and Metastasis. Cancer Cell 2014, 25, 21–36. [Google Scholar] [CrossRef]
- Suresh, S.; Huard, S.; Dubois, T. CARM1/PRMT4: Making Its Mark beyond Its Function as a Transcriptional Coactivator. Trends Cell Biol. 2021, 31, 402–417. [Google Scholar] [CrossRef]
- Yadav, N.; Lee, J.; Kim, J.; Shen, J.; Hu, M.C.-T.; Aldaz, C.M.; Bedford, M.T. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 2003, 100, 6464–6468. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, H.; Liao, P.; Xu, B.; Hu, R.; Yang, Y.; Li, Y. Systematic pan-cancer landscape identifies CARM1 as a potential prognostic and immunological biomarker. BMC Genom. Data 2022, 23, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Bedford, M.T.; Stallcup, M.R. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev. 2011, 25, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell Mol. Life 2015, 72, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Karkhanis, V.; Hu, Y.-J.; Baiocchi, R.A.; Imbalzano, A.N.; Sif, S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 2011, 36, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Antonysamy, S.; Bonday, Z.; Campbell, R.M.; Doyle, B.; Druzina, Z.; Gheyi, T.; Han, B.; Jungheim, L.N.; Qian, Y.; Rauch, C.; et al. Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Chen, X.; Liu, L.; Shu, Y.; Zhang, M.; Zhong, Y. Role of protein arginine methyltransferase 5 in human cancers. Biomed. Pharmacother. 2019, 114, 108790. [Google Scholar] [CrossRef]
- Hwang, J.W.; Kim, S.-N.; Myung, N.; Song, D.; Han, G.; Bae, G.-U.; Bedford, M.T.; Kim, Y.K. PRMT5 promotes DNA repair through methylation of 53BP1 and is regulated by Src-mediated phosphorylation. Commun. Biol. 2020, 3, 428. [Google Scholar] [CrossRef]
- Zhang, F.; Lou, L.; Peng, B.; Song, X.; Reizes, O.; Almasan, A.; Gong, Z. Nudix Hydrolase NUDT16 Regulates 53BP1 Protein by Reversing 53BP1 ADP-Ribosylation. Cancer Res. 2020, 80, 999–1010. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, C.; Huang, K.; Xia, F.; Parvin, J.D.; Mondal, N. Regulation of 53BP1 Protein Stability by RNF8 and RNF168 Is Important for Efficient DNA Double-Strand Break Repair. PLoS ONE 2014, 9, e110522. [Google Scholar] [CrossRef]
- Guo, Z.; Zheng, L.; Xu, H.; Dai, H.; Zhou, M.; Pascua, M.R.; Chen, Q.M.; Shen, B. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 2010, 6, 766–773. [Google Scholar] [CrossRef]
- Guo, Z.; Kanjanapangka, J.; Liu, N.; Liu, S.; Liu, C.; Wu, Z.; Wang, Y.; Loh, T.; Kowolik, C.; Jamsen, J.; et al. Sequential Posttranslational Modifications Program FEN1 Degradation during Cell-Cycle Progression. Mol. Cell 2012, 47, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, H.B. Rad9, an evolutionarily conserved gene with multiple functions for preserving genomic Integrity. J. Cell. Biochem. 2006, 97, 690–697. [Google Scholar] [CrossRef]
- Hang, H.; Lieberman, H.B. Physical Interactions among Human Checkpoint Control Proteins HUS1p, RAD1p, and RAD9p, and Implications for the Regulation of Cell Cycle Progression. Genomics 2000, 65, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, H.B.; Panigrahi, S.K.; Hopkins, K.M.; Wang, L.; Broustas, C.G. p53 and RAD9, the DNA Damage Response, and Regulation of Transcription Networks. Radiat. Res. 2017, 187, 424–432. [Google Scholar] [CrossRef]
- Zou, L.; Cortez, D.; Elledge, S.J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002, 16, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Liu, D.; Elledge, S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 2003, 100, 13827–13832. [Google Scholar] [CrossRef]
- Gembka, A.; Toueille, M.; Smirnova, E.; Poltz, R.; Ferrari, E.; Villani, G.; Hubscher, U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 2007, 35, 2596–2608. [Google Scholar] [CrossRef]
- He, W.; Ma, X.; Yang, X.; Zhao, Y.; Qiu, J.; Hang, H. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 2011, 39, 4719–4727. [Google Scholar] [CrossRef]
- Jha, S.; Dutta, A. RVB1/RVB2: Running Rings around Molecular Biology. Mol. Cell 2009, 34, 521–533. [Google Scholar] [CrossRef]
- Gorynia, S.; Bandeiras, T.M.; Pinho, F.G.; McVey, C.E.; Vonrhein, C.; Round, A.; Svergun, D.I.; Donner, P.; Matias, P.M.; Carrondo, M.A. Structural and functional insights into a dodecameric molecular machine—the RuvBL1/RuvBL2 complex. J. Struct. Biol. 2011, 176, 279–291. [Google Scholar] [CrossRef]
- Ikura, T.; Tashiro, S.; Kakino, A.; Shima, H.; Jacob, N.; Amunugama, R.; Yoder, K.; Izumi, S.; Kuraoka, I.; Tanaka, K.; et al. DNA Damage-Dependent Acetylation and Ubiquitination of H2AX Enhances Chromatin Dynamics. Mol. Cell. Biol. 2007, 27, 7028–7040. [Google Scholar] [CrossRef]
- Jha, S.; Shibata, E.; Dutta, A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell Biol. 2008, 28, 2690–2700. [Google Scholar] [CrossRef]
- Tang, J.; Cho, N.W.; Cui, G.; Manion, E.M.; Shanbhag, N.M.; Botuyan, M.V.; Mer, G.; Greenberg, R.A. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013, 20, 317–325. [Google Scholar] [CrossRef] [PubMed]
- López-Perrote, A.; Alatwi, H.E.; Torreira, E.; Ismail, A.; Ayora, S.; Downs, J.A.; Llorca, O. Structure of Yin Yang 1 Oligomers That Cooperate with RuvBL1-RuvBL2 ATPases. J. Biol. Chem. 2014, 289, 22614–22629. [Google Scholar] [CrossRef] [PubMed]
- Gospodinov, A.; Tsaneva, I.; Anachkova, B. RAD51 foci formation in response to DNA damage is modulated by TIP49. Int. J. Biochem. Cell Biol. 2009, 41, 925–933. [Google Scholar] [CrossRef]
- Clarke, T.L.; Sanchez-Bailon, M.P.; Chiang, K.; Reynolds, J.J.; Herrero-Ruiz, J.; Bandeiras, T.M.; Matias, P.M.; Maslen, S.L.; Skehel, J.M.; Stewart, G.S.; et al. PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination. Mol. Cell 2017, 65, 900–916.e7. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Huang, S.Y.; Gao, R.; DAS, B.B.; Murai, J.; Marchand, C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair 2014, 19, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Ashour, M.E.; Atteya, R.; El-Khamisy, S.F. Topoisomerase-mediated chromosomal break repair: An emerging player in many games. Nat. Cancer 2015, 15, 137–151. [Google Scholar] [CrossRef]
- Kawale, A.S.; Povirk, L.F. Tyrosyl–DNA phosphodiesterases: Rescuing the genome from the risks of relaxation. Nucleic Acids Res. 2018, 46, 520–537. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R.; Interthal, H.; Champoux, J.J.; Hol, W.G. The Crystal Structure of Human Tyrosyl-DNA Phosphodiesterase, Tdp1. Structure 2002, 10, 237–248. [Google Scholar] [CrossRef]
- Rehman, I.; Basu, S.M.; Das, S.K.; Bhattacharjee, S.; Ghosh, A.; Pommier, Y.; Das, B.B. PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes. Nucleic Acids Res. 2018, 46, 5601–5617. [Google Scholar] [CrossRef]
- Casadio, F.; Lu, X.; Pollock, S.B.; LeRoy, G.; Garcia, B.A.; Muir, T.W.; Roeder, R.G.; Allis, C.D. H3R42me2a is a histone modification with positive transcriptional effects. Proc. Natl. Acad. Sci. USA 2013, 110, 14894–14899. [Google Scholar] [CrossRef]
- Waldmann, T.; Izzo, A.; Kamieniarz, K.; Richter, F.; Vogler, C.; Sarg, B.; Lindner, H.; Young, N.L.; Mittler, G.; Garcia, B.A.; et al. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 2011, 4, 11. [Google Scholar] [CrossRef]
- Cheng, D.; Gao, G.; Di Lorenzo, A.; Jayne, S.; Hottiger, M.O.; Richard, S.; Bedford, M.T. Genetic evidence for partial redundancy between the arginine methyltransferases CARM1 and PRMT6. J. Biol. Chem. 2020, 295, 17060–17070. [Google Scholar] [CrossRef]
- Guccione, E.; Bassi, C.; Casadio, F.; Martinato, F.; Cesaroni, M.; Schuchlautz, H.; Lüscher, B.; Amati, B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007, 449, 933–937. [Google Scholar] [CrossRef]
- Hyllus, D.; Stein, C.; Schnabel, K.; Schiltz, E.; Imhof, A.; Dou, Y.; Hsieh, J.; Bauer, U.-M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007, 21, 3369–3380. [Google Scholar] [CrossRef]
- Neault, M.; Mallette, F.A.; Vogel, G.; Michaud-Levesque, J.; Richard, S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012, 40, 9513–9521. [Google Scholar] [CrossRef]
- Feng, J.; Dang, Y.; Zhang, W.; Zhao, X.; Zhang, C.; Hou, Z.; Jin, Y.; McNutt, M.A.; Marks, A.R.; Yin, Y. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2019, 116, 6868–6877. [Google Scholar] [CrossRef]
- Raveendran, V.V.; Al-Haffar, K.; Kunhi, M.; Belhaj, K.; Al-Habeeb, W.; Al-Buraiki, J.; Eyjolsson, A.; Poizat, C. Protein arginine methyltransferase 6 mediates cardiac hypertrophy by differential regulation of histone H3 arginine methylation. Heliyon 2020, 6, e03864. [Google Scholar] [CrossRef]
- Dowhan, D.H.; Harrison, M.J.; Eriksson, N.A.; Bailey, P.; Pearen, M.A.; Fuller, P.; Funder, J.W.; Simpson, E.R.; Leedman, P.J.; Tilley, W.; et al. Protein arginine methyltransferase 6-dependent gene expression and splicing: Association with breast cancer outcomes. Endocr.-Relat. Cancer 2012, 19, 509–526. [Google Scholar] [CrossRef]
- El-Andaloussi, N.; Valovka, T.; Toueille, M.; Steinacher, R.; Focke, F.; Gehrig, P.; Covic, M.; Hassa, P.O.; Schar, P.; Hubscher, U.; et al. Arginine methylation regulates DNA polymerase beta. Mol. Cell 2006, 22, 51–62. [Google Scholar] [CrossRef]
- Miranda, T.B.; Miranda, M.; Frankel, A.; Clarke, S. PRMT7 Is a Member of the Protein Arginine Methyltransferase Family with a Distinct Substrate Specificity. J. Biol. Chem. 2004, 279, 22902–22907. [Google Scholar] [CrossRef]
- Feng, Y.; Maity, R.; Whitelegge, J.P.; Hadjikyriacou, A.; Li, Z.; Zurita-Lopez, C.; Al-Hadid, Q.; Clark, A.T.; Bedford, M.T.; Masson, J.-Y.; et al. Mammalian Protein Arginine Methyltransferase 7 (PRMT7) Specifically Targets RXR Sites in Lysine- and Arginine-rich Regions. J. Biol. Chem. 2013, 288, 37010–37025. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Avasarala, S.; Vanscoyk, M.; Sechler, M.; Kelley, N.; Malbon, C.C.; Winn, R.A. Dishevelled3 is a novel arginine methyl transferase substrate. Sci. Rep. 2012, 2, 805. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Malbon, C.C. Wnt3a-stimulated LRP6 phosphorylation is dependent upon arginine methylation of G3BP2. J. Cell Sci. 2012, 125, 2446–2456. [Google Scholar] [CrossRef]
- Haghandish, N.; Baldwin, R.M.; Morettin, A.; Dawit, H.T.; Adhikary, H.; Masson, J.-Y.; Mazroui, R.; Trinkle-Mulcahy, L.; Côté, J. PRMT7 methylates eukaryotic translation initiation factor 2α and regulates its role in stress granule formation. Mol. Biol. Cell 2019, 30, 778–793. [Google Scholar] [CrossRef]
- Szewczyk, M.M.; Ishikawa, Y.; Organ, S.; Sakai, N.; Li, F.; Halabelian, L.; Ackloo, S.; Couzens, A.L.; Eram, M.; Dilworth, D.; et al. Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Nat. Commun. 2020, 11, 2396. [Google Scholar] [CrossRef]
- Dhar, S.S.; Lee, S.-H.; Kan, P.-Y.; Voigt, P.; Ma, L.; Shi, X.; Reinberg, D.; Lee, M.G. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012, 26, 2749–2762. [Google Scholar] [CrossRef]
- Yao, R.; Jiang, H.; Ma, Y.; Wang, L.; Wang, L.; Du, J.; Hou, P.; Gao, Y.; Zhao, L.; Wang, G.; et al. PRMT7 Induces Epithelial-to-Mesenchymal Transition and Promotes Metastasis in Breast Cancer. Cancer Res. 2014, 74, 5656–5667. [Google Scholar] [CrossRef]
- Karkhanis, V.; Wang, L.; Tae, S.; Hu, Y.J.; Imbalzano, A.N.; Sif, S. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. J. Biol. Chem. 2012, 287, 29801–29814. [Google Scholar] [CrossRef]
- Verbiest, V.; Montaudon, D.; Tautu, M.T.; Moukarzel, J.; Portail, J.-P.; Markovits, J.; Robert, J.; Ichas, F.; Pourquier, P. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008, 582, 1483–1489. [Google Scholar] [CrossRef]
- Lee, J.; Sayegh, J.; Daniel, J.; Clarke, S.; Bedford, M.T. PRMT8, a New Membrane-bound Tissue-specific Member of the Protein Arginine Methyltransferase Family. J. Biol. Chem. 2005, 280, 32890–32896. [Google Scholar] [CrossRef]
- Sayegh, J.; Webb, K.; Cheng, D.; Bedford, M.T.; Clarke, S.G. Regulation of Protein Arginine Methyltransferase 8 (PRMT8) Activity by Its N-terminal Domain. J. Biol. Chem. 2007, 282, 36444–36453. [Google Scholar] [CrossRef]
- Kim, J.-D.; Park, K.-E.; Ishida, J.; Kako, K.; Hamada, J.; Kani, S.; Takeuchi, M.; Namiki, K.; Fukui, H.; Fukuhara, S.; et al. PRMT8 as a phospholipase regulates Purkinje cell dendritic arborization and motor coordination. Sci. Adv. 2015, 1, e1500615. [Google Scholar] [CrossRef]
- Lee, W.-C.; Lin, W.-L.; Matsui, T.; Chen, E.S.-W.; Wei, T.-Y.W.; Hu, H.; Zheng, Y.G.; Tsai, M.-D.; Ho, M.-C. Protein Arginine Methyltransferase 8: Tetrameric Structure and Protein Substrate Specificity. Biochemistry 2015, 54, 7514–7523. [Google Scholar] [CrossRef]
- Baek, J.-H.; Rubinstein, M.; Scheuer, T.; Trimmer, J.S. Reciprocal Changes in Phosphorylation and Methylation of Mammalian Brain Sodium Channels in Response to Seizures. J. Biol. Chem. 2014, 289, 15363–15373. [Google Scholar] [CrossRef]
- Dong, R.; Li, X.; Lai, K.-O. Activity and Function of the PRMT8 Protein Arginine Methyltransferase in Neurons. Life 2021, 11, 1132. [Google Scholar] [CrossRef]
- Simandi, Z.; Pajer, K.; Karolyi, K.; Sieler, T.; Jiang, L.-L.; Kolostyak, Z.; Sari, Z.; Fekecs, Z.; Pap, A.; Patsalos, A.; et al. Arginine Methyltransferase PRMT8 Provides Cellular Stress Tolerance in Aging Motoneurons. J. Neurosci. 2018, 38, 7683–7700. [Google Scholar] [CrossRef]
- Tang, J.; Gary, J.D.; Clarke, S.; Herschman, H.R. PRMT 3, a Type I Protein Arginine N-Methyltransferase That Differs from PRMT1 in Its Oligomerization, Subcellular Localization, Substrate Specificity, and Regulation. J. Biol. Chem. 1998, 273, 16935–16945. [Google Scholar] [CrossRef]
- Frankel, A.; Clarke, S. PRMT3 Is a Distinct Member of the Protein Arginine N-Methyltransferase Family. J. Biol. Chem. 2000, 275, 32974–32982. [Google Scholar] [CrossRef]
- Bachand, F.; A Silver, P. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004, 23, 2641–2650. [Google Scholar] [CrossRef]
- Swiercz, R.; Person, M.D.; Bedford, M.T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 2005, 386, 85–91. [Google Scholar] [CrossRef]
- Fronz, K.; Otto, S.; Kölbel, K.; Kühn, U.; Friedrich, H.; Schierhorn, A.; Beck-Sickinger, A.G.; Ostareck-Lederer, A.; Wahle, E. Promiscuous Modification of the Nuclear Poly(A)-binding Protein by Multiple Protein-arginine Methyltransferases Does Not Affect the Aggregation Behavior. J. Biol. Chem. 2008, 283, 20408–20420. [Google Scholar] [CrossRef]
- Lei, Y.; Han, P.; Chen, Y.; Wang, H.; Wang, S.; Wang, M.; Liu, J.; Yan, W.; Tian, D.; Liu, M. Protein arginine methyltransferase 3 promotes glycolysis and hepatocellular carcinoma growth by enhancing arginine methylation of lactate dehydrogenase A. Clin. Transl. Med. 2022, 12, e686. [Google Scholar] [CrossRef]
- Hsu, M.-C.; Tsai, Y.-L.; Lin, C.-H.; Pan, M.-R.; Shan, Y.-S.; Cheng, T.-Y.; Cheng, S.H.-C.; Chen, L.-T.; Hung, W.-C. Protein arginine methyltransferase 3-induced metabolic reprogramming is a vulnerable target of pancreatic cancer. J. Hematol. Oncol. 2019, 12, 1–14. [Google Scholar] [CrossRef]
- Yang, Y.; Hadjikyriacou, A.; Xia, Z.; Gayatri, S.; Kim, D.; Zuritalopez, C.I.; Kelly, R.; Guo, A.; Li, W.; Clarke, S.G.; et al. PRMT9 is a Type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 2015, 6, 6428. [Google Scholar] [CrossRef]
- Hadjikyriacou, A.; Yang, Y.; Espejo, A.; Bedford, M.T.; Clarke, S.G. Unique Features of Human Protein Arginine Methyltransferase 9 (PRMT9) and Its Substrate RNA Splicing Factor SF3B2. J. Biol. Chem. 2015, 290, 16723–16743. [Google Scholar] [CrossRef]
- Boshuizen, J.; Peeper, D.S. Rational Cancer Treatment Combinations: An Urgent Clinical Need. Mol. Cell 2020, 78, 1002–1018. [Google Scholar] [CrossRef]
- Mokhtari, R.B.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H. Combination therapy in combating cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar] [CrossRef]
- Arora, S.; Balasubramaniam, S.; Zhang, H.; Berman, T.; Narayan, P.; Suzman, D.; Bloomquist, E.; Tang, S.; Gong, Y.; Sridhara, R.; et al. FDA Approval Summary: Olaparib Monotherapy or in Combination with Bevacizumab for the Maintenance Treatment of Patients with Advanced Ovarian Cancer. Oncologist 2020, 26, e164–e172. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Jarrold, J.; Davies, C.C. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol. Med. 2019, 25, 993–1009. [Google Scholar] [CrossRef]
- Hu, H.; Qian, K.; Ho, M.-C.; Zheng, Y.G. Small Molecule Inhibitors of Protein Arginine Methyltransferases. Expert Opin. Investig. Drugs 2016, 25, 335–358. [Google Scholar] [CrossRef]
- Vinet, M.; Suresh, S.; Maire, V.; Monchecourt, C.; Némati, F.; Lesage, L.; Pierre, F.; Ye, M.; Lescure, A.; Brisson, A.; et al. Protein arginine methyltransferase 5: A novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019, 8, 2414–2428. [Google Scholar] [CrossRef]
- Hu, R.; Zhou, B.; Chen, Z.; Chen, S.; Chen, N.; Shen, L.; Xiao, H.; Zheng, Y. PRMT5 Inhibition Promotes PD-L1 Expression and Immuno-Resistance in Lung Cancer. Front. Immunol. 2022, 12, 5877. [Google Scholar] [CrossRef]
- Ma, D.; Yang, M.; Wang, Q.; Sun, C.; Shi, H.; Jing, W.; Bi, Y.; Shen, X.; Ma, X.; Qin, Z.; et al. Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Sci. Adv. 2021, 7, 13. [Google Scholar] [CrossRef]
- Sengupta, S.; Kennemer, A.; Patrick, K.; Tsichlis, P.; Guerau-De-Arellano, M. Protein Arginine Methyltransferase 5 in T Lymphocyte Biology. Trends Immunol. 2020, 41, 918–931. [Google Scholar] [CrossRef]
- Dominici, C.; Sgarioto, N.; Yu, Z.; Sesma-Sanz, L.; Masson, J.-Y.; Richard, S.; Raynal, N.J.-M. Synergistic effects of type I PRMT and PARP inhibitors against non-small cell lung cancer cells. Clin. Epigenetics 2021, 13, 54. [Google Scholar] [CrossRef]
- Hamard, P.-J.; Santiago, G.E.; Liu, F.; Karl, D.L.; Martinez, C.; Man, N.; Mookhtiar, A.K.; Duffort, S.; Greenblatt, S.; Verdun, R.E.; et al. PRMT5 Regulates DNA Repair by Controlling the Alternative Splicing of Histone-Modifying Enzymes. Cell Rep. 2018, 24, 2643–2657. [Google Scholar] [CrossRef]
- Musiani, D.; Giambruno, R.; Massignani, E.; Ippolito, M.R.; Maniaci, M.; Jammula, S.; Manganaro, D.; Cuomo, A.; Nicosia, L.; Pasini, D.; et al. PRMT1 Is Recruited via DNA-PK to Chromatin Where It Sustains the Senescence-Associated Secretory Phenotype in Response to Cisplatin. Cell Rep. 2020, 30, 1208–1222.e9. [Google Scholar] [CrossRef]
- Wei, X.; Yang, J.; Adair, S.J.; Ozturk, H.; Kuscu, C.; Lee, K.Y.; Kane, W.J.; O’Hara, P.E.; Liu, D.; Demirlenk, Y.M.; et al. Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28068–28079. [Google Scholar] [CrossRef]


| Drug Name | Targeted PRMTs | Trial Number | Disease or Condition | Status |
|---|---|---|---|---|
| GSK3368715 | Type I PRMTs | NCT03666988 | DLBCL and MTAP-deficient solid tumors | Terminated |
| GSK3326595 | PRMT5 | NCT04676516 | Breast cancer | Not yet recruiting |
| JNJ-64619178 | PRMT5 | NCT03573310 | Solid tumor, Non-Hodgkin Lymphoma, myelodysplastic syndromes | Active, not recruiting |
| PF-06939999 | PRMT5 | NCT03854227 | Metastatic NSCLC, HNSCC, esophageal cancer, endometrial cancer, cervical cancer, and bladder cancer | Terminated |
| PRT811 | PRMT5 | NCT04089449 | Advanced solid tumors, CNS lymphoma, and glioma | Recruiting |
| PRT543 | PRMT5 | NCT03886831 | Advanced solid tumors and hematologic malignancies | Active, not recruiting |
| TNG908 | PRMT5 | NCT05275478 | Patients with MTAP-deleted advanced or metastatic solid tumors | Recruiting |
| MRTX1719 | PRTM5-MTA | NCT05245500 | Patients with MTAP-deleted advanced or metastatic solid tumors | Recruiting |
| Inhibition of PRMT1 or PRMT5 | Synergistic Agents | Cancer Type |
|---|---|---|
| Type I PRMT inhibitor (MS023) | PARP inhibitor (BMN-677) | Lung Cancer [199] |
| Type I PRMT inhibitor (MS023) | Cisplatin | Ovarian cancer [201] |
| Knockdown of PRMT1 | Etoposide | Osteosarcoma [53] |
| Knockdown of PRMT1 | TMZ or 5-FU | Lung Cancer [95] |
| PRMT5 inhibitor (EPZ015666) | Gemcitabine | Pancreatic cancer [53] |
| Knockdown of PRMT5 | Etoposide | Breast cancer [127] |
| PRMT5 inhibitor (GSK3186000A) | PARP inhibitor (Olaparib) | AML [200] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brobbey, C.; Liu, L.; Yin, S.; Gan, W. The Role of Protein Arginine Methyltransferases in DNA Damage Response. Int. J. Mol. Sci. 2022, 23, 9780. https://doi.org/10.3390/ijms23179780
Brobbey C, Liu L, Yin S, Gan W. The Role of Protein Arginine Methyltransferases in DNA Damage Response. International Journal of Molecular Sciences. 2022; 23(17):9780. https://doi.org/10.3390/ijms23179780
Chicago/Turabian StyleBrobbey, Charles, Liu Liu, Shasha Yin, and Wenjian Gan. 2022. "The Role of Protein Arginine Methyltransferases in DNA Damage Response" International Journal of Molecular Sciences 23, no. 17: 9780. https://doi.org/10.3390/ijms23179780
APA StyleBrobbey, C., Liu, L., Yin, S., & Gan, W. (2022). The Role of Protein Arginine Methyltransferases in DNA Damage Response. International Journal of Molecular Sciences, 23(17), 9780. https://doi.org/10.3390/ijms23179780






