Morphological, Transcriptome, and Hormone Analysis of Dwarfism in Tetraploids of Populus alba × P. glandulosa
Abstract
:1. Introduction
2. Results
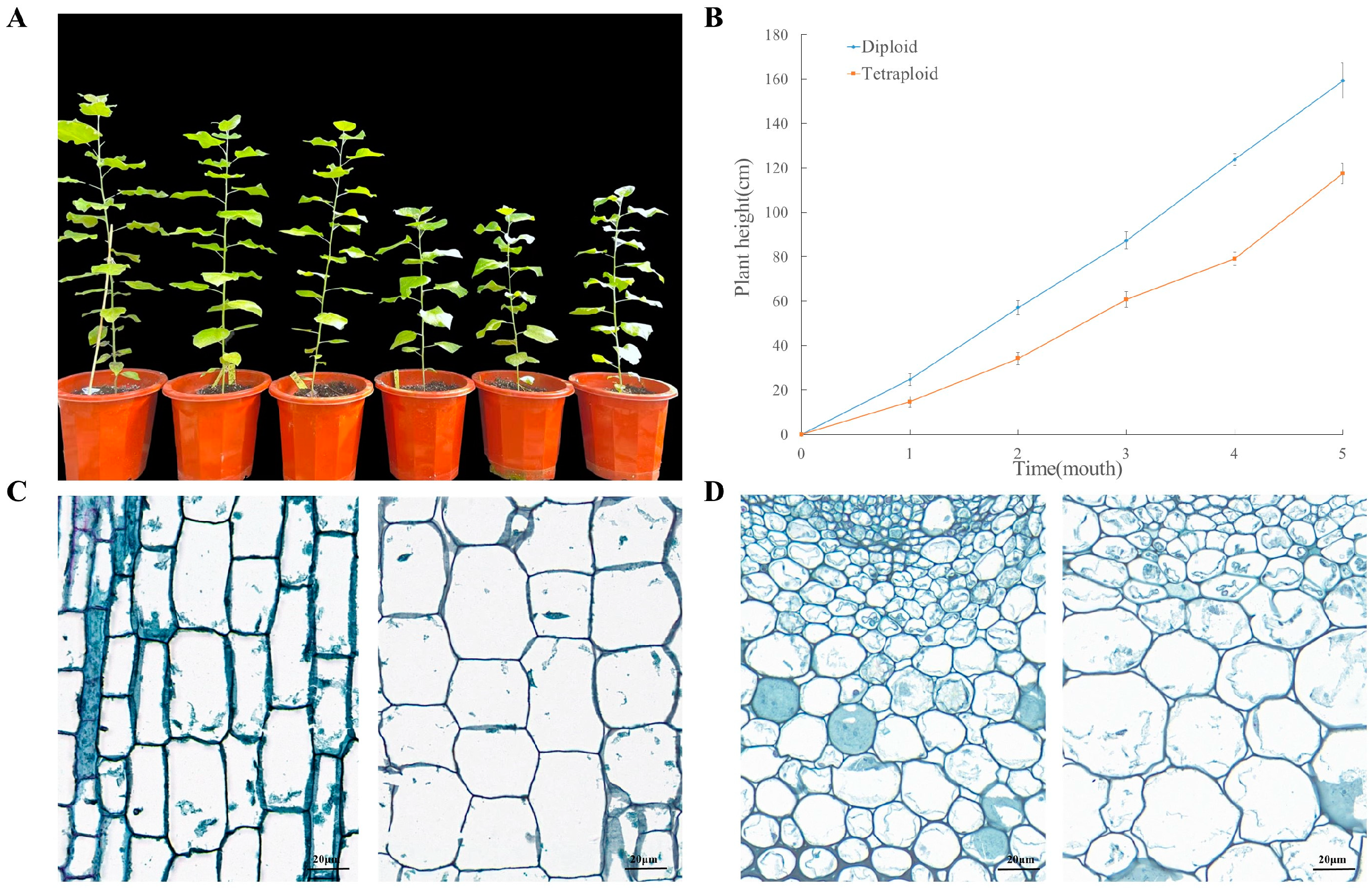
2.1. Comparative Analysis of Phenotypic and Anatomical Characteristics between Tetraploids and Diploids
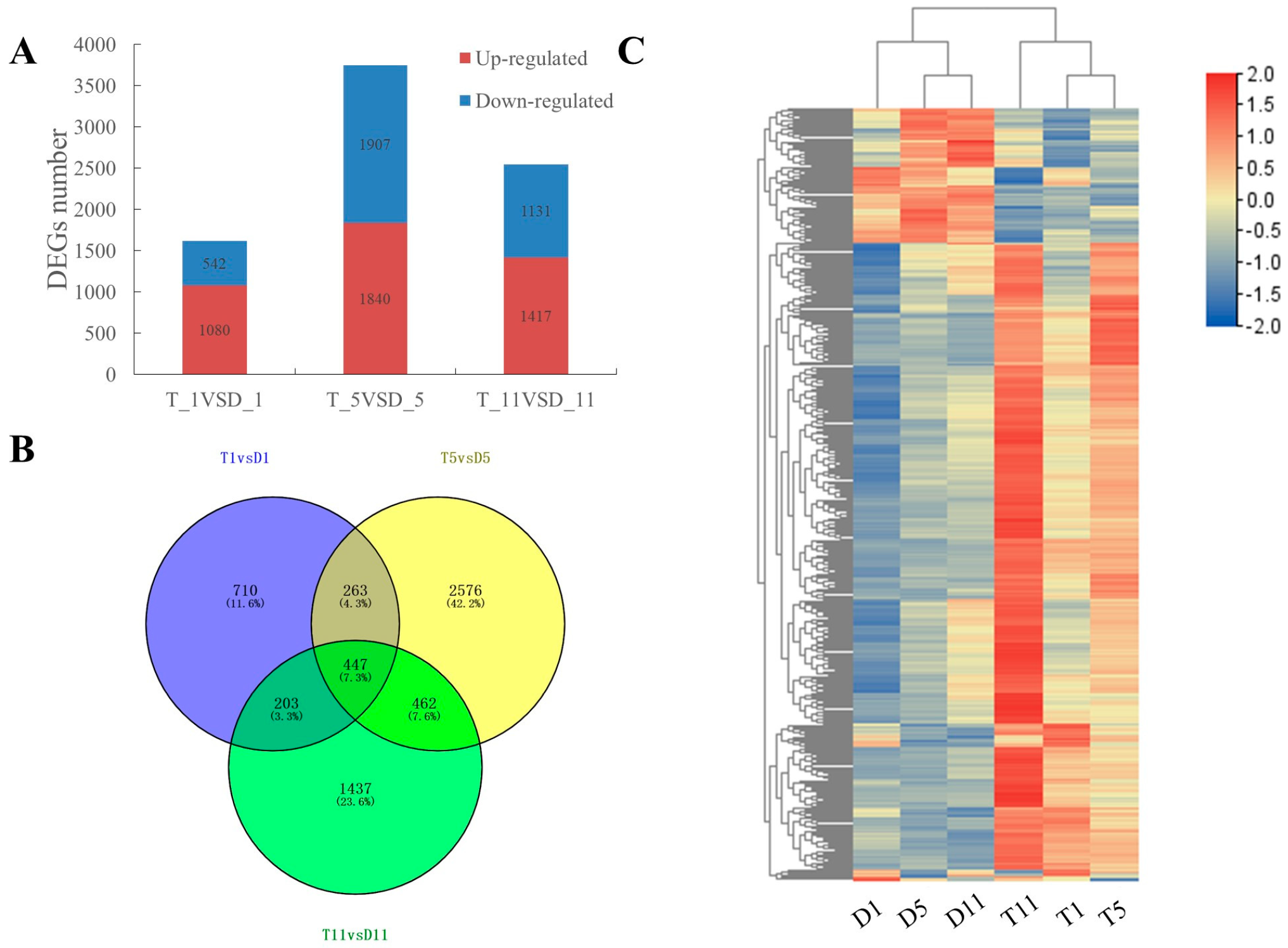
2.2. Transcriptome Sequencing Data Analysis
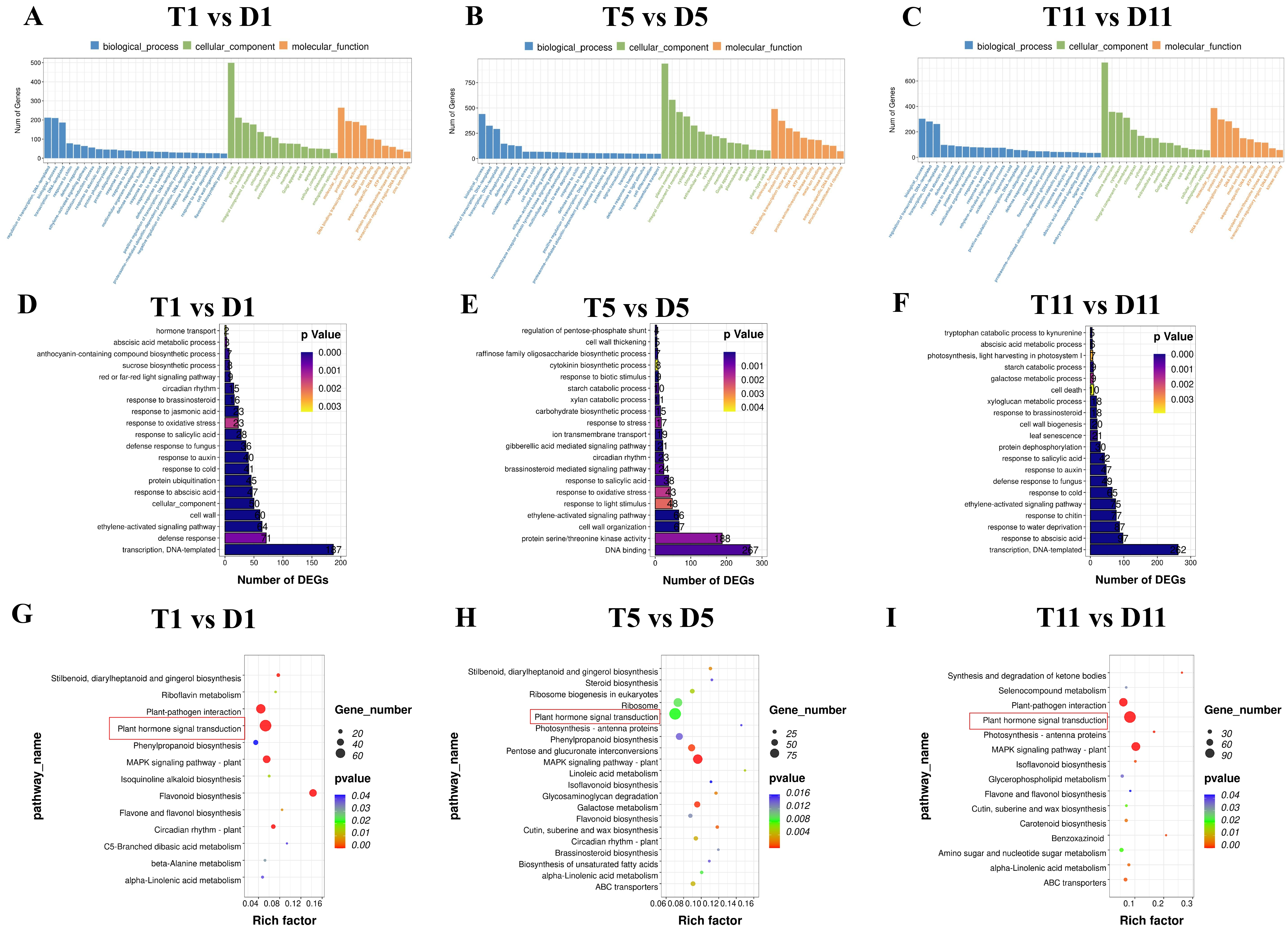
2.3. Functional Enrichment Analysis of DEGs in the Transcriptomes
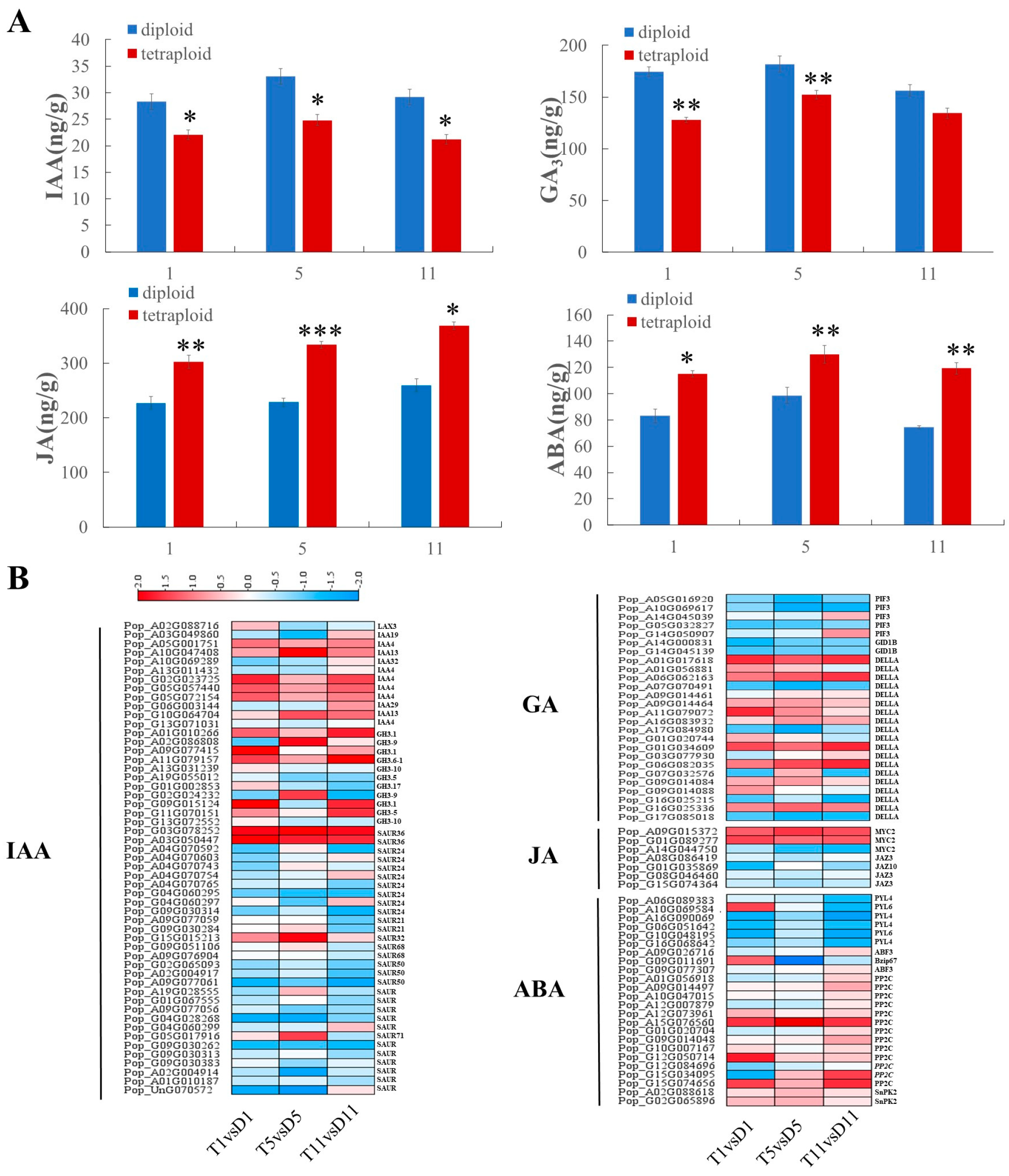
2.4. Determination of Endogenous Hormones and Expression of Hormone-Signaling Genes in Diploids and Tetraploids
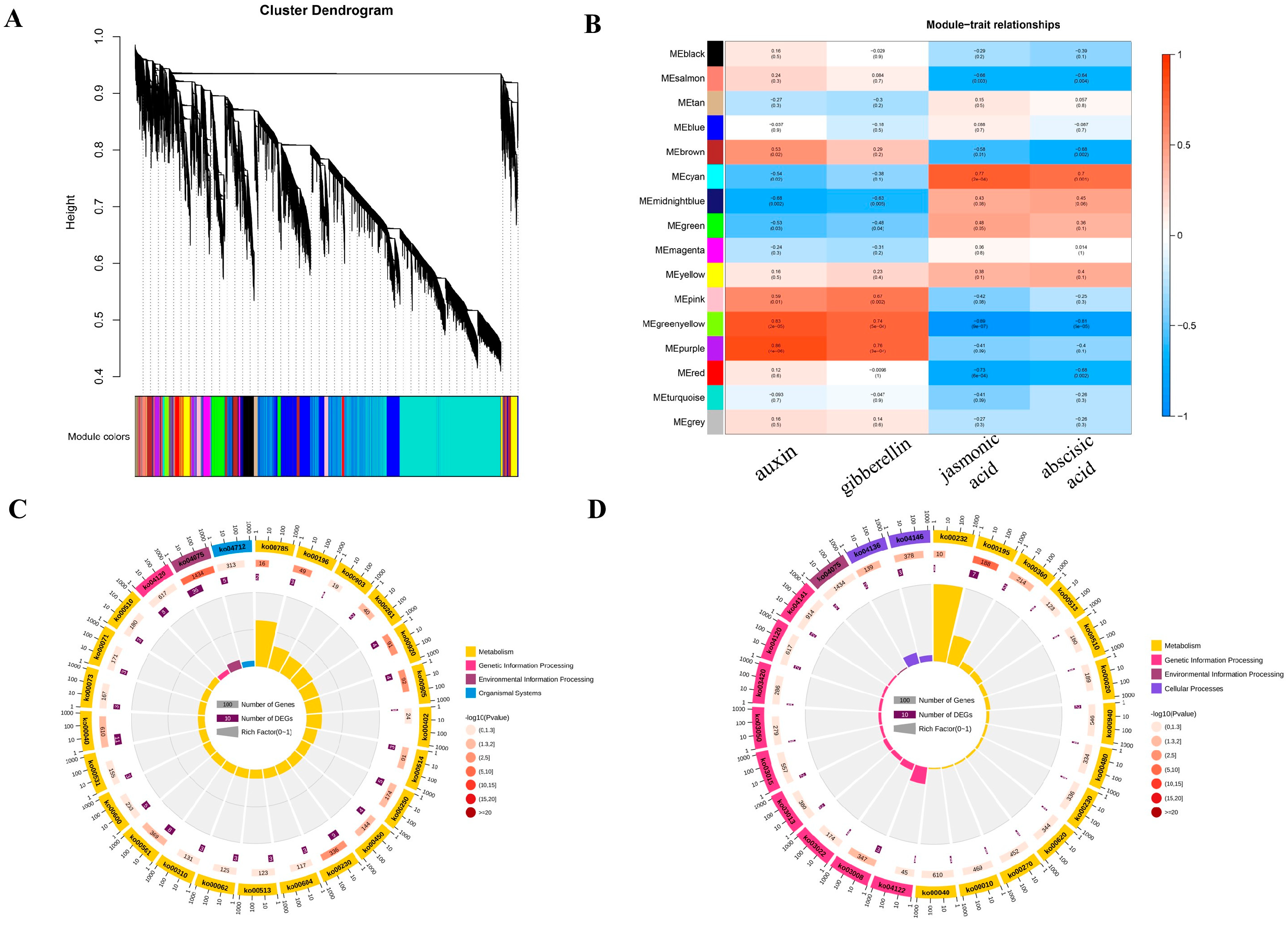
2.5. Co-Expression Modules Associated with Hormone Content
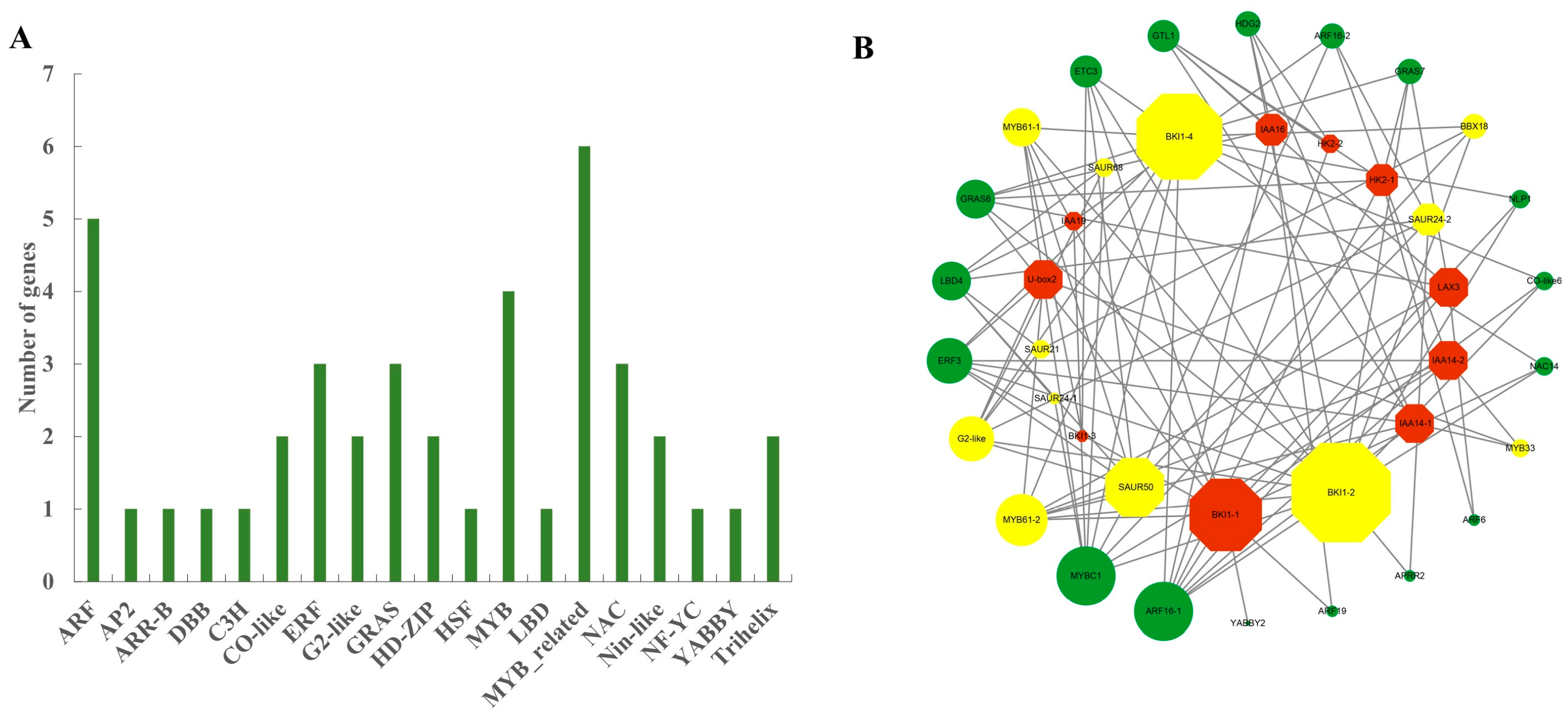
2.6. Association Analysis of Differentially Expressed Transcription Factors and Core Genes
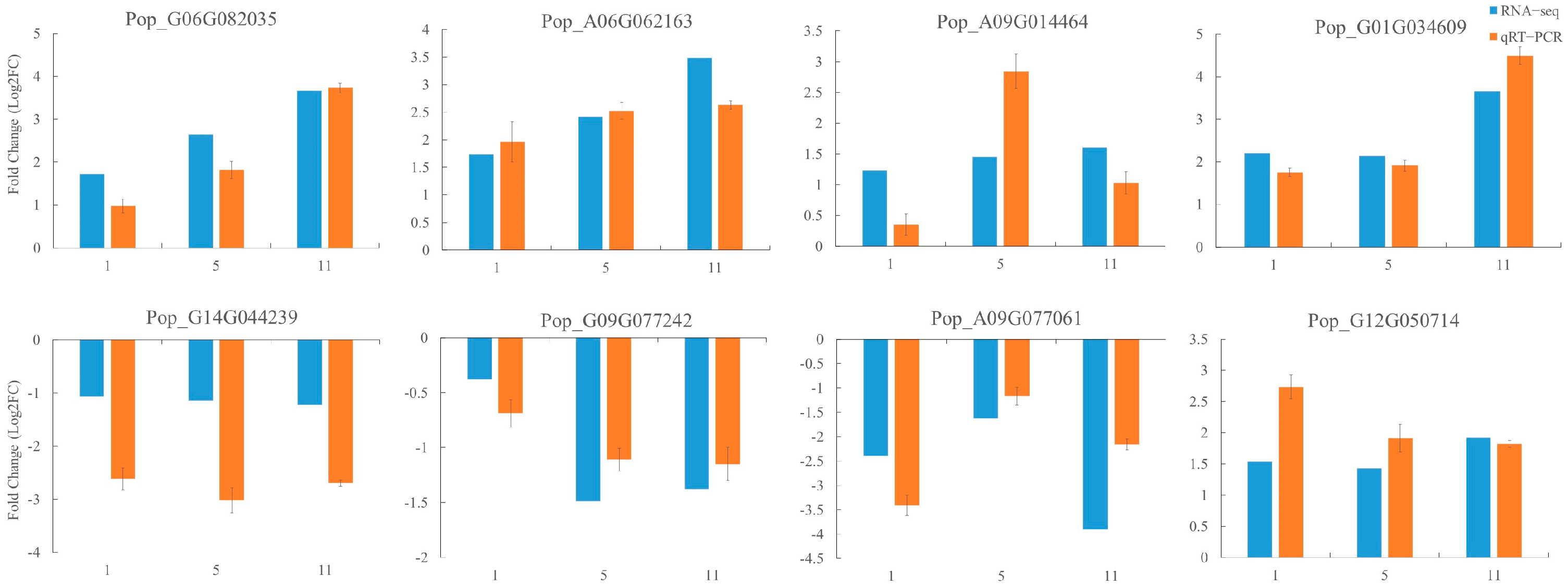
2.7. qRT−PCR Validation
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Measurement of Phenotypic Traits and Anatomical Observations
4.3. Total RNA Extraction and Transcriptome Sequencing
4.4. Enrichment Analysis of DEGs
4.5. Determination of Endogenous Hormone Contens
4.6. Division of Co-Expression Modules and Visualization of Gene Expression
4.7. qRT−PCR Verification of Differentially Expressed Genes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Masterson, J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science 1994, 264, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Svačina, R.; Sourdille, P.; Kopecký, D.; Bartoš, J. Chromosome Pairing in Polyploid Grasses. Front. Plant Sci. 2020, 11, 1056. [Google Scholar] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836. [Google Scholar]
- Xu, T.T.; Zhang, S.W.; Du, K.; Yang, J.; Kang, X.Y. Insights into the Molecular Regulation of Lignin Content in Triploid Poplar Leaves. Int. J. Mol. Sci. 2022, 23, 4603. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Liao, T.; Ren, Y.Y.; Geng, X.N.; Kang, X.Y. Molecular mechanism of vegetative growth advantage in allotriploid Populus. Int. J. Mol. Sci. 2020, 21, 441. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, J.; Song, L.J.; Qi, Q.; Du, K.; Han, Q.; Kang, X.Y. Study of variation in the growth, photosynthesis, and content of secondary metabolites in Eucommia triploids. Trees 2019, 33, 817–826. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.Z.; Liu, Z.; Xiong, T.; Lan, J.; Han, Q.; Li, Y.; Kang, X.Y. Megaspore chromosome doubling in Eucalyptus urophylla ST Blake induced by colchicine treatment to produce triploids. Forests 2018, 9, 728. [Google Scholar] [CrossRef]
- Chen, T.T.; Sheng, Y.; Hao, Z.D.; Long, X.F.; Fu, F.F.; Liu, Y.; Tang, Z.H.; Ali, A.; Peng, Y.; Liu, Y.; et al. Transcriptome and proteome analysis suggest enhanced photosynthesis in tetraploid Liriodendron sino-americanum. Tree Physiol. 2021, 41, 1953–1971. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Qiu, B.; Lu, T.; Kang, X.Y.; Yang, J. Induction and Characterization of Tetraploid Through Zygotic Chromosome Doubling in Eucalyptus urophylla. Front. Plant Sci. 2022, 13, 870698. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, L.; Zhang, F.; Ou, C.Q.; Wang, F.; Zhang, Z.H. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus x domestica). Sci. Rep. 2016, 6, 26719. [Google Scholar] [CrossRef]
- Wu, J.; Sang, Y.R.; Zhou, Q.; Zhang, P.D. Colchicine in vitro tetraploid induction of Populus hopeiensis from leaf blades. Plant Cell Tissue Organ Cult. 2020, 141, 339–349. [Google Scholar]
- Zeng, Q.Q.; Wang, P.Q.; Ren, Y.Y.; Kang, X.Y. Transcriptomic analyses reveal potential mechanisms of premature senescence in hexaploid Populus. Plant Cell Tissue Organ Cult. 2021, 147, 255–270. [Google Scholar]
- Wu, J.; Cheng, X.T.; Kong, B.; Zhou, Q.; Sang, Y.R.; Zhang, P.D. In vitro octaploid induction of Populus hopeiensis with colchicine. BMC Plant Biol. 2022, 22, 176. [Google Scholar]
- Rao, S.P.; Kang, X.Y.; Li, J.; Chen, J.H. Induction, identification and characterization of tetraploidy in Lycium ruthenicum. Breed Sci. 2019, 69, 160–168. [Google Scholar] [PubMed] [Green Version]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [PubMed]
- Braynen, J.; Yang, Y.; Yuan, J.C.; Xie, Z.Q.; Cao, G.Q.; Wei, X.C.; Shi, G.Y.; Zhang, X.W.; Wei, F.; Tian, B.M. Comparative transcriptome analysis revealed differential gene expression in multiple signaling pathways at flowering in polyploid Brassica rapa. Cell Biosci. 2021, 11, 17. [Google Scholar]
- Wu, W.Q.; Li, J.; Wang, Q.; Lv, K.W.; Du, K.; Zhang, W.L.; Li, Q.Z.; Kang, X.Y.; Wei, H.R. Growth-regulating factor 5 (GRF5) -mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 2021, 230, 612–628. [Google Scholar]
- Dudits, D.; Török, K.; Cseri, A.; Paul, K.; Nagy, A.V.; Nagy, B.; Sass, L.; Ferenc, G.; Vankova, R.; Dobrev, P.; et al. Response of Organ Structure and Physiology to Autotetraploidization in Early Development of Energy Willow Salix viminalis. Plant Physiol. 2016, 170, 1504–1523. [Google Scholar]
- Mu, H.Z.; Liu, Z.J.; Lin, L.; Li, H.Y.; Jiang, J.; Liu, G.F. Transcriptomic analysis of phenotypic changes in birch (Betula platyphylla) autotetraploids. Int. J. Mol. Sci. 2020, 13, 13012–13029. [Google Scholar]
- Dai, F.W.; Wang, Z.J.; Luo, G.Q.; Tang, C.M. Phenotypic and Transcriptomic Analyses of Autotetraploid and Diploid Mulberry (Morus alba L.). Int. J. Mol. Sci. 2015, 16, 22938–22956. [Google Scholar]
- Xu, C.P.; Zhang, Y.; Han, Q.; Kang, X.Y. Molecular Mechanism of Slow Vegetative Growth in Populus Tetraploid. Genes 2020, 11, 1417. [Google Scholar]
- Wang, Y.H.; Huang, S.G.; Liu, Z.Y.; Tang, X.Y.; Feng, H. Changes in endogenous phytohormones regulated by microRNA-target mRNAs contribute to the development of Dwarf Autotetraploid Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Mol. Genet. Genom. 2018, 293, 1535–1546. [Google Scholar]
- Shu, W.B.; Zhou, H.J.; Jiang, C.; Zhao, S.T.; Wang, L.Q.; Li, Q.Z.; Yang, Z.Q.; Groover, A.; Lu, M.Z. The auxin receptor TIR 1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/ IAA 28 in Populus. Plant Biotechnol. J. 2019, 17, 338–349. [Google Scholar]
- Ren, Y.Y.; Jing, Y.C.; Kang, X.Y. In vitro induction of tetraploid and resulting trait variation in Populus alba× Populus glandulosa clone 84 K. Plant Cell Tissue Organ Cult. 2020, 146, 285–296. [Google Scholar]
- Yang, T.; Davies, P.J.; Reid, J.B. Genetic Dissection of the Relative Roles of Auxin and Gibberellin in the Regulation of Stem Elongation in Intact Light-Grown Peas. Plant Physiol. 1996, 110, 1029–1034. [Google Scholar] [PubMed]
- Wojciechowska, N.; Sobieszczuk-Nowicka, E.; Bagniewska-Zadworna, A. Plant organ senescence—Regulation by manifold pathways. Plant Biol. 2018, 20, 167–181. [Google Scholar] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar]
- Kang, X.Y.; Wei, H.R. Breeding polyploid Populus: Progress and perspective. For. Res. 2022, 2, 4. [Google Scholar]
- Corneillie, S.; De Storme, N.; Van Acker, R.; Fangel, J.U.; De Bruyne, M.; De Rycke, R.; Geelen, D.; Willats, W.; Vanholme, B.; Boerjan, W. Polyploidy Affects Plant Growth and Alters Cell Wall Composition. Plant Physiol. 2019, 179, 74–87. [Google Scholar]
- Rosellini, D.; Ferradini, N.; Allegrucci, S.; Capomaccio, S.; Zago, E.D.; Leonetti, P.; Balech, B.; Aversano, R.; Carputo, D.; Reale, L.; et al. Sexual Polyploidization in Medicago sativa L.: Impact on the Phenotype, Gene Transcription, and Genome Methylation. G3 2016, 6, 925–938. [Google Scholar]
- Yu, X.M.; Wang, X.X.; Hyldgaard, B.; Zhu, Z.B.; Zhou, R.; Kjaer, K.H.; Ouzounis, T.; Lou, Q.F.; Li, J.; Cai, Q.S.; et al. Allopolyploidization in Cucumis contributes to delayed leaf maturation with repression of redundant homoeologous genes. Plant J. 2018, 94, 393–404. [Google Scholar] [PubMed]
- Wang, T.; Liu, L.; Wang, X.J.; Liang, L.X.; Yue, J.J.; Li, L.B. Comparative Analyses of Anatomical Structure, Phytohormone Levels, and Gene Expression Profiles Reveal Potential Dwarfing Mechanisms in Shengyin Bamboo (Phyllostachys edulis f. tubaeformis). Int. J. Mol. Sci. 2018, 19, 1697. [Google Scholar]
- Sun, Z.X.; Wang, X.F.; Liu, R.H.; Du, W.; Ma, M.C.; Han, Y.H.; Li, H.Y.; Liu, L.L.; Hou, S.Y. Comparative transcriptomic analysis reveals the regulatory mechanism of the gibberellic acid pathway of Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.) dwarf mutants. BMC Plant Biol. 2021, 21, 206. [Google Scholar]
- Jackson, S.; Chen, Z.J. Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 2010, 13, 153–159. [Google Scholar] [PubMed]
- Qiu, D.Y.; Bai, S.L.; Ma, J.C.; Zhang, L.S.; Shao, F.J.; Zhang, K.K.; Yang, Y.F.; Sun, T.; Huang, J.L.; Zhou, Y.; et al. The genome of Populus alba x Populus tremula var. glandulosa clone 84K. DNA Res. 2019, 26, 423–431. [Google Scholar]
- Xu, F.Y.; Xue, S.Q.; Deng, L.M.; Zhang, S.F.; Li, Y.X.; Zhao, X. The piperazine compound ASP activates an auxin response in Arabidopsis thaliana. BMC Genom. 2020, 21, 788. [Google Scholar]
- Qiu, T.; Qi, M.Y.; Ding, X.H.; Zheng, Y.Y.; Zhou, T.J.; Chen, Y.; Han, N.; Zhu, M.Y.; Bian, H.W.; Wang, J.H. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2020, 125, 805–819. [Google Scholar]
- Abel, S.; Oeller, P.W.; Theologis, A. Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 326–330. [Google Scholar]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar]
- Wojtaczka, P.; Ciarkowska, A.; Starzynska, E.; Ostrowski, M. The GH3 amidosynthetases family and their role in metabolic crosstalk modulation of plant signaling compounds. Phytochemistry 2022, 194, 113039. [Google Scholar] [PubMed]
- Zhang, S.W.; Li, C.H.; Cao, J.; Zhang, Y.C.; Zhang, S.Q.; Xia, Y.F.; Sun, D.Y.; Sun, Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [PubMed]
- Du, H.; Wu, N.; Fu, J.; Wang, S.P.; Li, X.H.; Xiao, J.H.; Xiong, L.Z. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [PubMed]
- Porco, S.; Larrieu, A.; Du, Y.J.; Gaudinier, A.; Goh, T.; Swarup, K.; Swarup, R.; Kuempers, B.; Bishopp, A.; Lavenus, J.; et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 2016, 143, 3340–3349. [Google Scholar] [PubMed]
- Dong, C.J.; Liu, X.Y.; Xie, L.L.; Wang, L.L.; Shang, Q.M. Salicylic acid regulates adventitious root formation via competitive inhibition of the auxin conjugation enzyme CsGH3.5 in cucumber hypocotyls. Planta 2020, 252, 75. [Google Scholar]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.B.; Miao, Y.C.; Liu, Y.Y.; Botella, J.R.; Li, W.Q.; Li, K.; Song, C.P. Function of Protein Kinases in Leaf Senescence of Plants. Front. Plant Sci. 2022, 13, 864215. [Google Scholar]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar]
- Zhao, G.J.; Luo, C.X.; Luo, J.N.; Li, J.X.; Gong, H.; Zheng, X.M.; Liu, X.X.; Guo, J.J.; Zhou, L.Y.; Wu, H.B. A mutation in LacDWARF1 results in a GA-deficient dwarf phenotype in sponge gourd (Luffa acutangula). Theor. Appl. Genet. 2021, 134, 3443–3457. [Google Scholar]
- Lantzouni, O.; Alkofer, A.; Falter-Braun, P.; Schwechheimer, C. GROWTH-REGULATING FACTORS Interact with DELLAs and Regulate Growth in Cold Stress. Plant Cell 2020, 32, 1018–1034. [Google Scholar]
- Wu, K.; Xu, H.; Gao, X.H.; Fu, X.D. New insights into gibberellin signaling in regulating plant growth-metabolic coordination. Curr. Opin. Plant Biol. 2021, 63, 102074. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, J.; Briones-Moreno, A.; Blázquez, M.A. Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 2021, 109, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Wang, W.; Huang, J.F.; Wang, Y.T.; Hu, L.; Yuan, Y.; Lyu, M.L.; Wu, B.H. Role of gibberellin and its three GID1 receptors in Jasminum sambac stem elongation and flowering. Planta 2021, 255, 17. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Kato, J. Isolation and identification of a senescence-promoting substance from Wormwood (Artemisia absinthium L.). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef]
- Paponov, M.; Antonyan, M.; Slimestad, R.; Paponov, I.A. Decoupling of Plant Growth and Accumulation of Biologically Active Compounds in Leaves, Roots, and Root Exudates of Hypericum perforatum L. by the Combination of Jasmonate and Far-Red Lighting. Biomolecules 2021, 11, 1283. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Zhuo, M.N.; Sakuraba, Y.; Yanagisawa, S. A Jasmonate-Activated MYC2-Dof2.1-MYC2 Transcriptional Loop Promotes Leaf Senescence in Arabidopsis. Plant Cell 2020, 32, 242–262. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.Q.; Zhang, W.; Wang, X.X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017, 15, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.W.; Abeysinghe, J.K.; Kamali, M. Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [PubMed]
- Arce-Rodríguez, M.L.; Ochoa-Alejo, N. An R2R3-MYB Transcription Factor Regulates Capsaicinoid Biosynthesis. Plant Physiol. 2017, 174, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.J.; Ma, Z.T.; Chen, H.; Liu, M.Y. MYB Gene Family in Potato (Solanum tuberosum L.): Genome-Wide Identification of Hormone-Responsive Reveals Their Potential Functions in Growth and Development. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef]
- Gocal, G.F.; Sheldon, C.C.; Gubler, F.; Moritz, T.; Bagnall, D.J.; MacMillan, C.P.; Li, S.F.; Parish, R.W.; Dennis, E.S.; Weigel, D.; et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 2001, 127, 1682–1693. [Google Scholar] [CrossRef]
- Matías-Hernández, L.; Jiang, W.M.; Yang, K.; Tang, K.X.; Brodelius, P.E.; Pelaz, S. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 2017, 90, 520–534. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Wang, R.H.; Wang, J.B. Comprehensive Transcriptomic Analysis for Developing Seeds of a Synthetic Brassica Hexaploid. Plants 2020, 9, 1141. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, L.Y.; Li, Y.H.; Zhang, D.P.; Gao, Y. Plant NIGT1/HRS1/HHO Transcription Factors: Key Regulators with Multiple Roles in Plant Growth, Development, and Stress Responses. Int. J. Mol. Sci. 2021, 22, 8685. [Google Scholar] [CrossRef]
- Powell, A.L.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernández-Muñoz, R.; Vicente, A.; et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef]
- Qin, M.Y.; Zhang, B.H.; Gu, G.; Yuan, J.Z.; Yang, X.S.; Yang, J.H.; Xie, X.F. Genome-Wide Analysis of the G2-Like Transcription Factor Genes and Their Expression in Different Senescence Stages of Tobacco (Nicotiana tabacum L.). Front. Genet. 2021, 12, 626352. [Google Scholar] [CrossRef]
- Lyu, G.Z.; Li, D.B.; Li, S.S. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1782647. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, C.X.; Hou, L.; Yang, W.C.; Liu, S.S.; Pang, X.M.; Li, Y.Y. Multiple responses contribute to the enhanced drought tolerance of the autotetraploid Ziziphus jujuba Mill. var. spinosa. Cell Biosci. 2021, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Li, F.J.; Wu, Q.; Liao, B.P.; Yu, K.K.; Huo, Y.N.; Meng, L.; Wang, S.M.; Wang, B.M.; Du, M.W.; Tian, X.L.; et al. Thidiazuron Promotes Leaf Abscission by Regulating the Crosstalk Complexities between Ethylene, Auxin, and Cytokinin in Cotton. Int. J. Mol. Sci. 2022, 23, 2696. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Zhang, S.; Xu, T.; Kang, X. Morphological, Transcriptome, and Hormone Analysis of Dwarfism in Tetraploids of Populus alba × P. glandulosa. Int. J. Mol. Sci. 2022, 23, 9762. https://doi.org/10.3390/ijms23179762
Ren Y, Zhang S, Xu T, Kang X. Morphological, Transcriptome, and Hormone Analysis of Dwarfism in Tetraploids of Populus alba × P. glandulosa. International Journal of Molecular Sciences. 2022; 23(17):9762. https://doi.org/10.3390/ijms23179762
Chicago/Turabian StyleRen, Yongyu, Shuwen Zhang, Tingting Xu, and Xiangyang Kang. 2022. "Morphological, Transcriptome, and Hormone Analysis of Dwarfism in Tetraploids of Populus alba × P. glandulosa" International Journal of Molecular Sciences 23, no. 17: 9762. https://doi.org/10.3390/ijms23179762
APA StyleRen, Y., Zhang, S., Xu, T., & Kang, X. (2022). Morphological, Transcriptome, and Hormone Analysis of Dwarfism in Tetraploids of Populus alba × P. glandulosa. International Journal of Molecular Sciences, 23(17), 9762. https://doi.org/10.3390/ijms23179762





