Transcriptome Analysis Reveals the Multiple Functions of pBD2 in IPEC-J2 Cells against E. coli
Abstract
:1. Introduction
2. Results
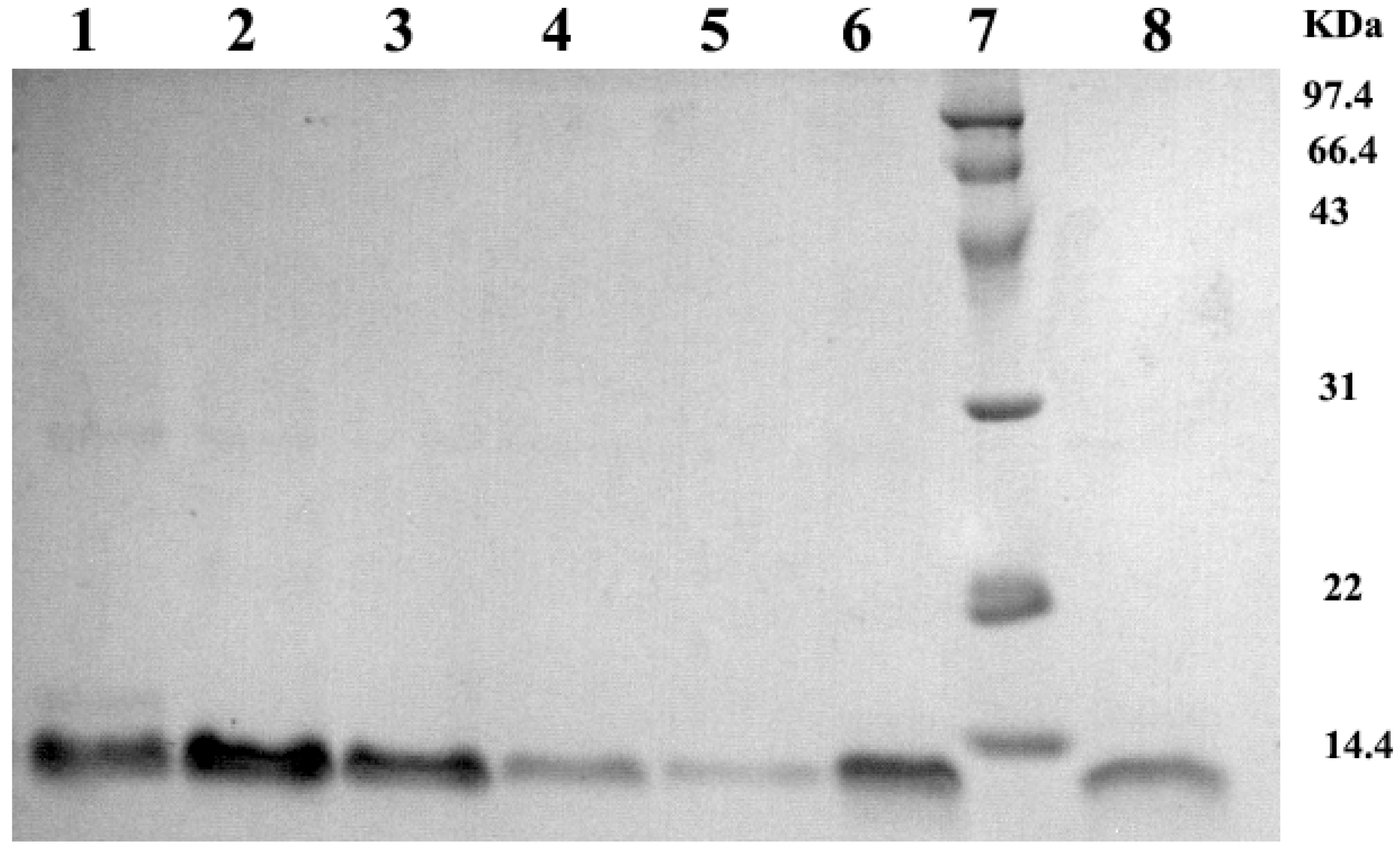
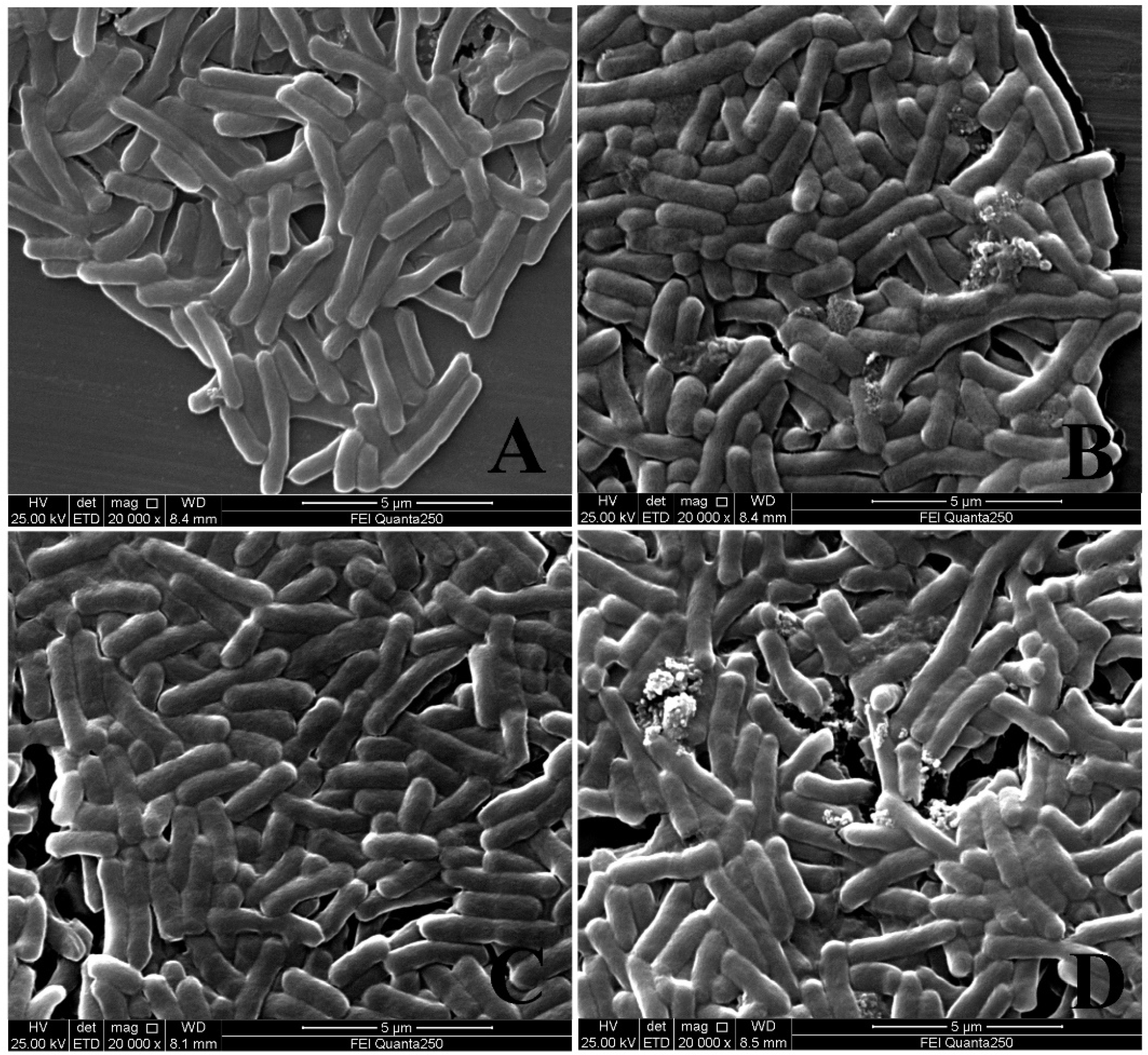
2.1. Analysis of Bactericidal Activity
2.2. Transcriptome Profiling
2.3. Characterization of Differentially Expressed Genes by RNA Sequencing
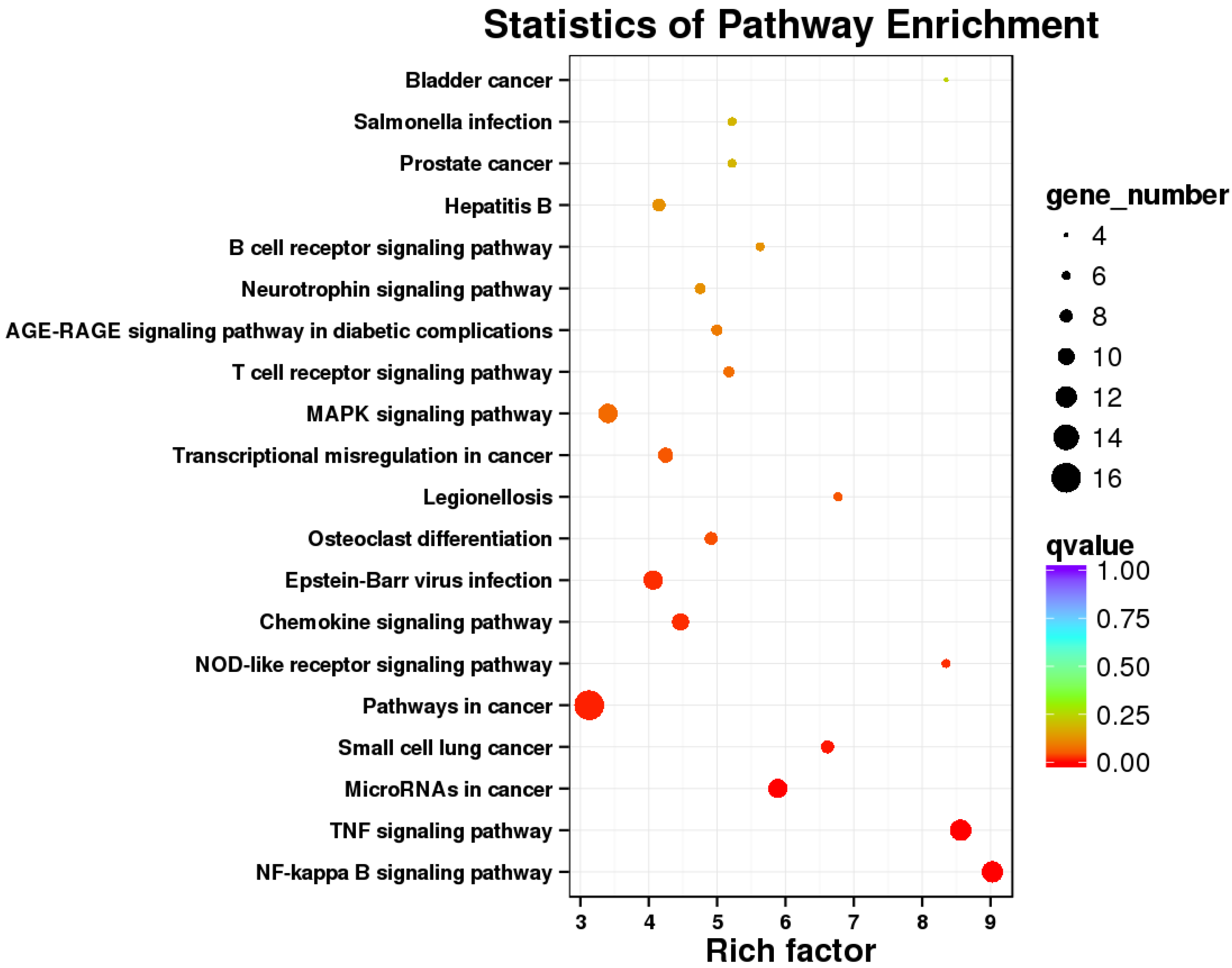
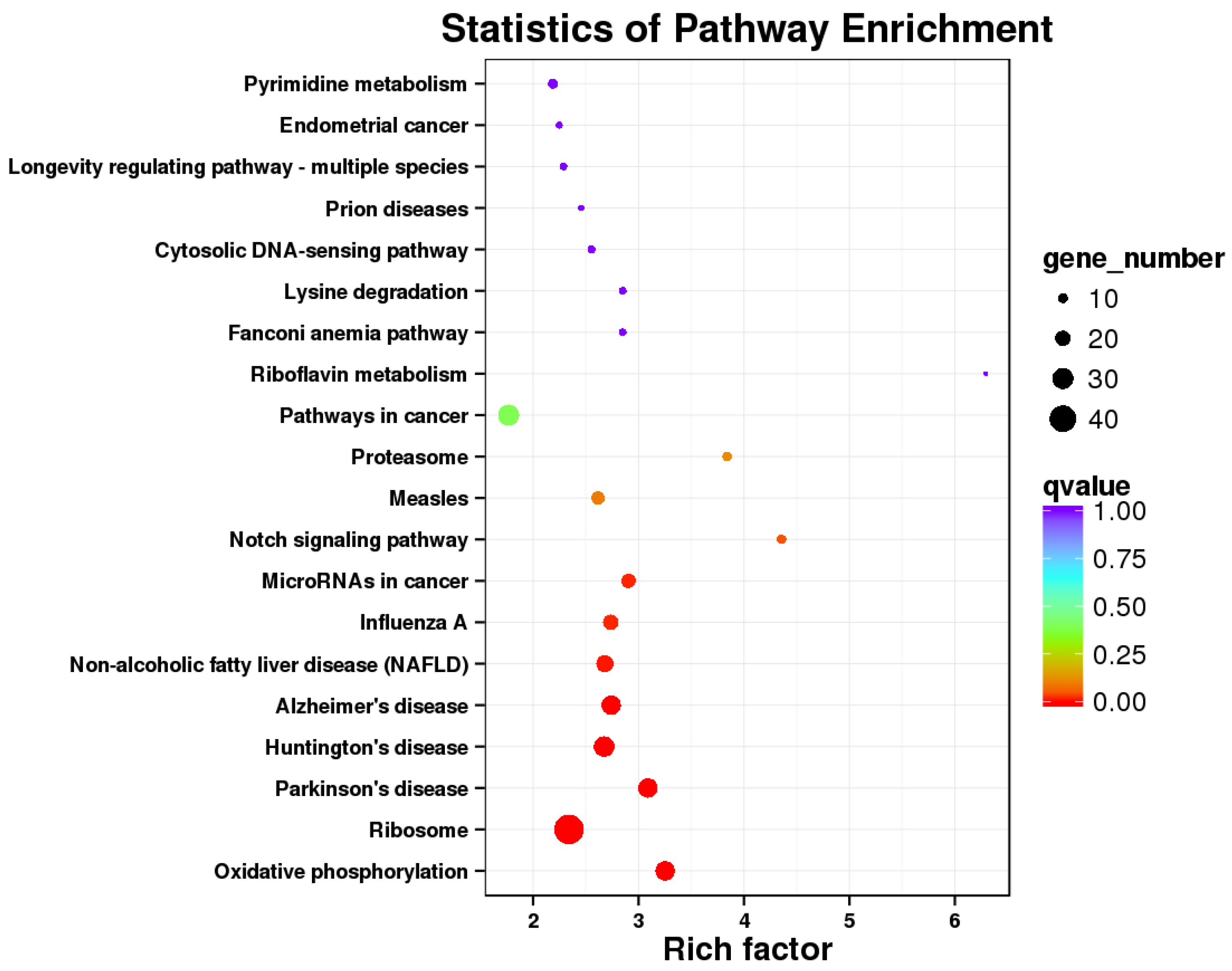
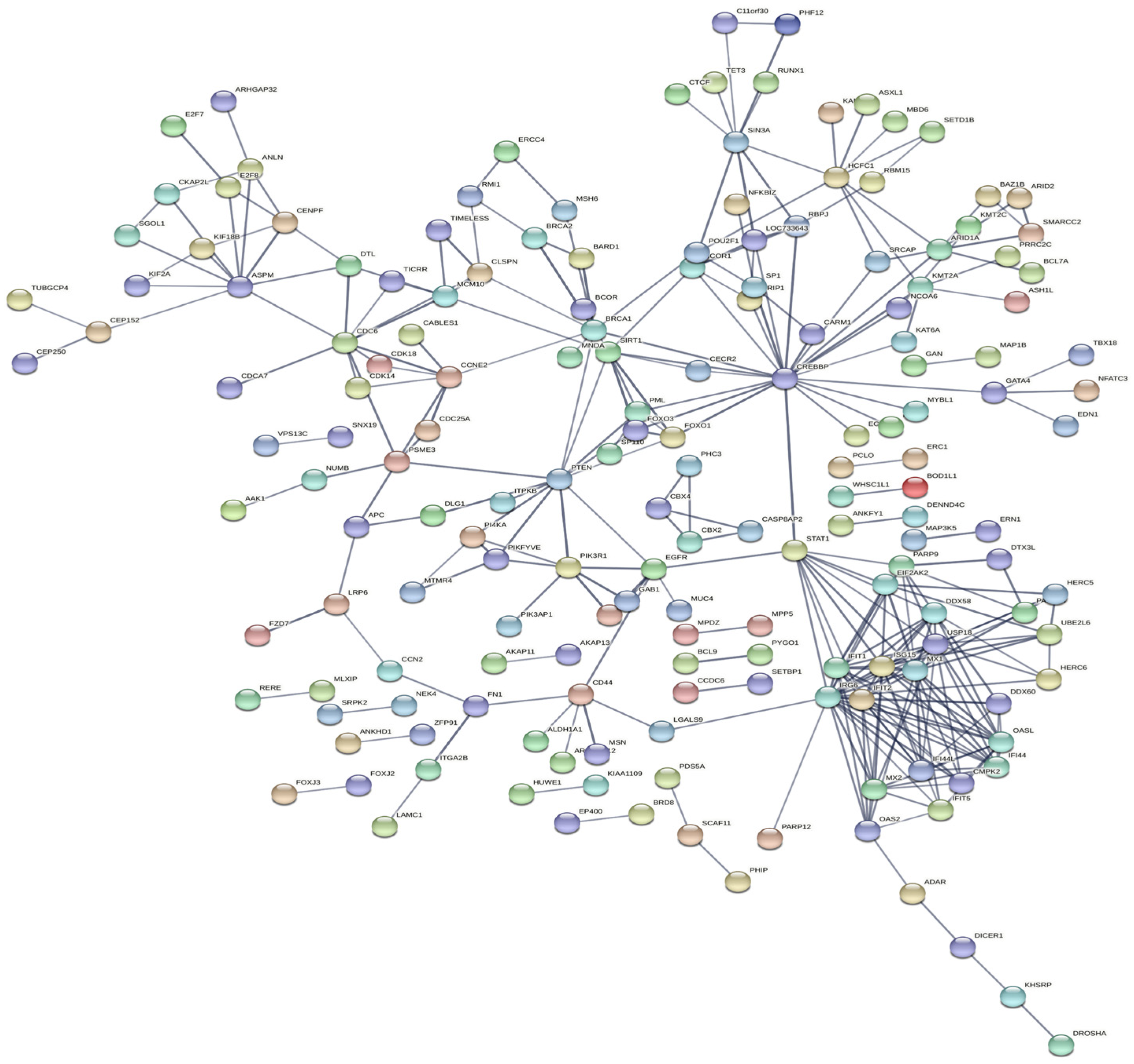
2.4. KEGG Analysis Revealed That Immune Responses of Cells Were Trigger by E. coli
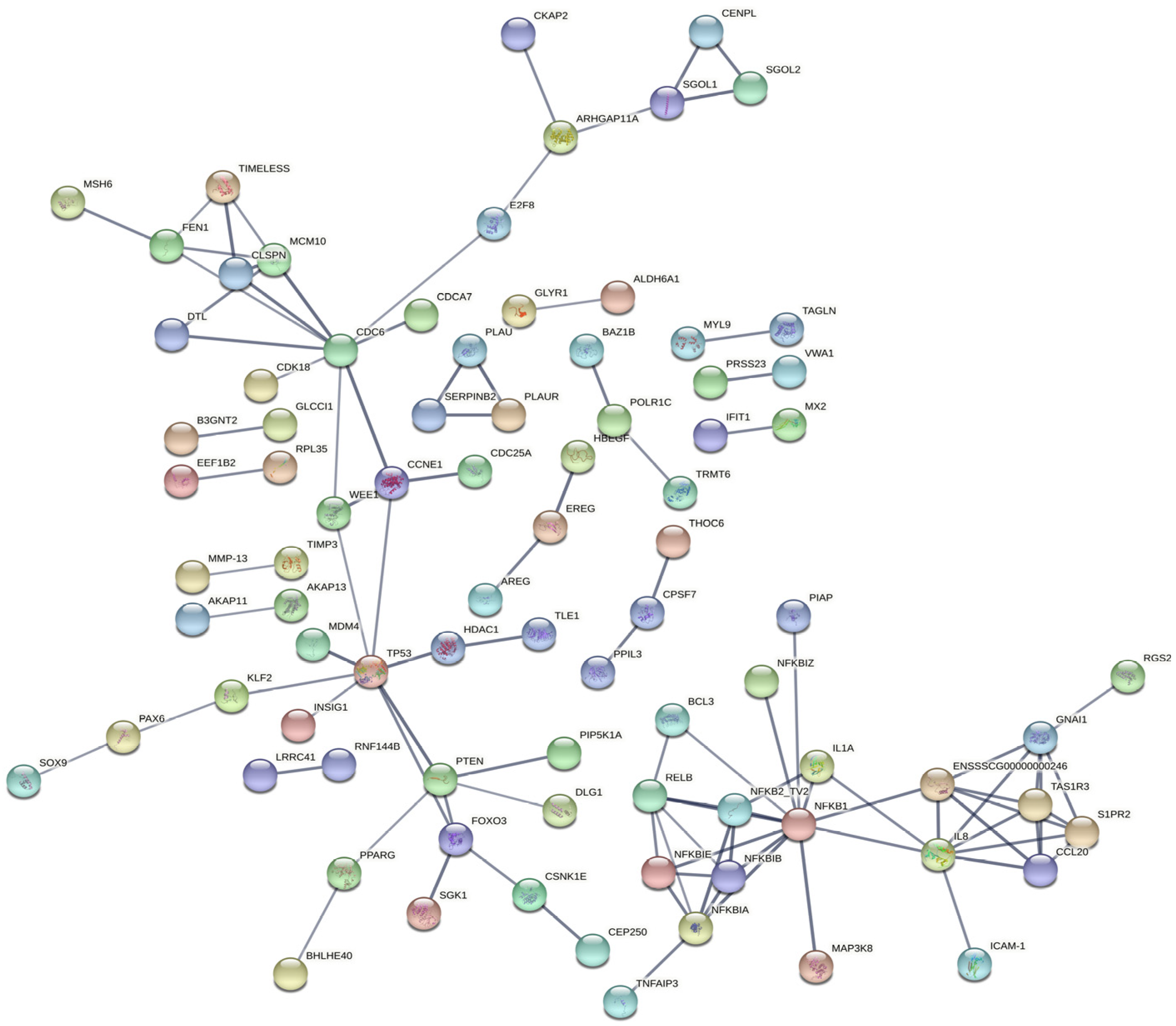
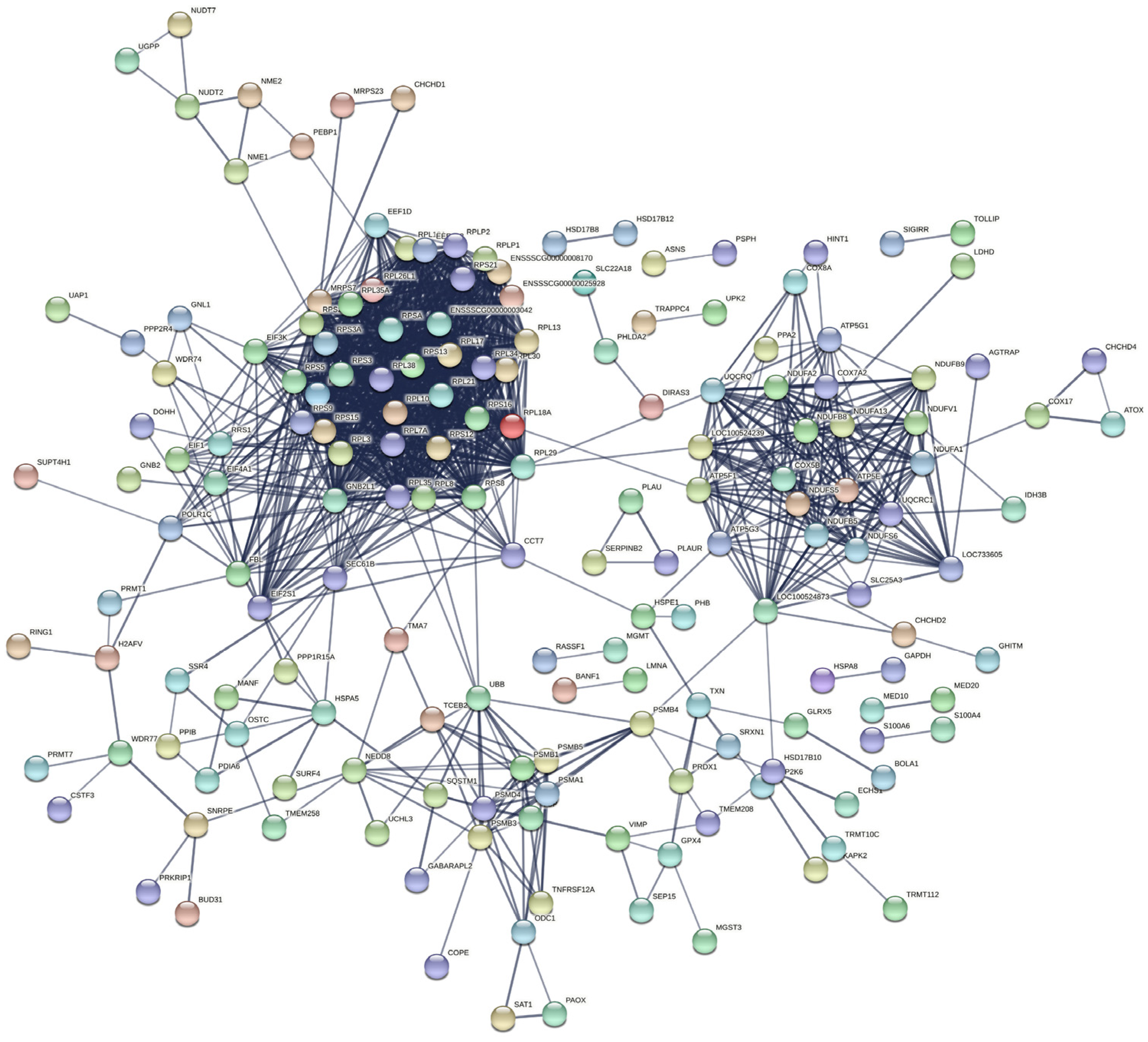
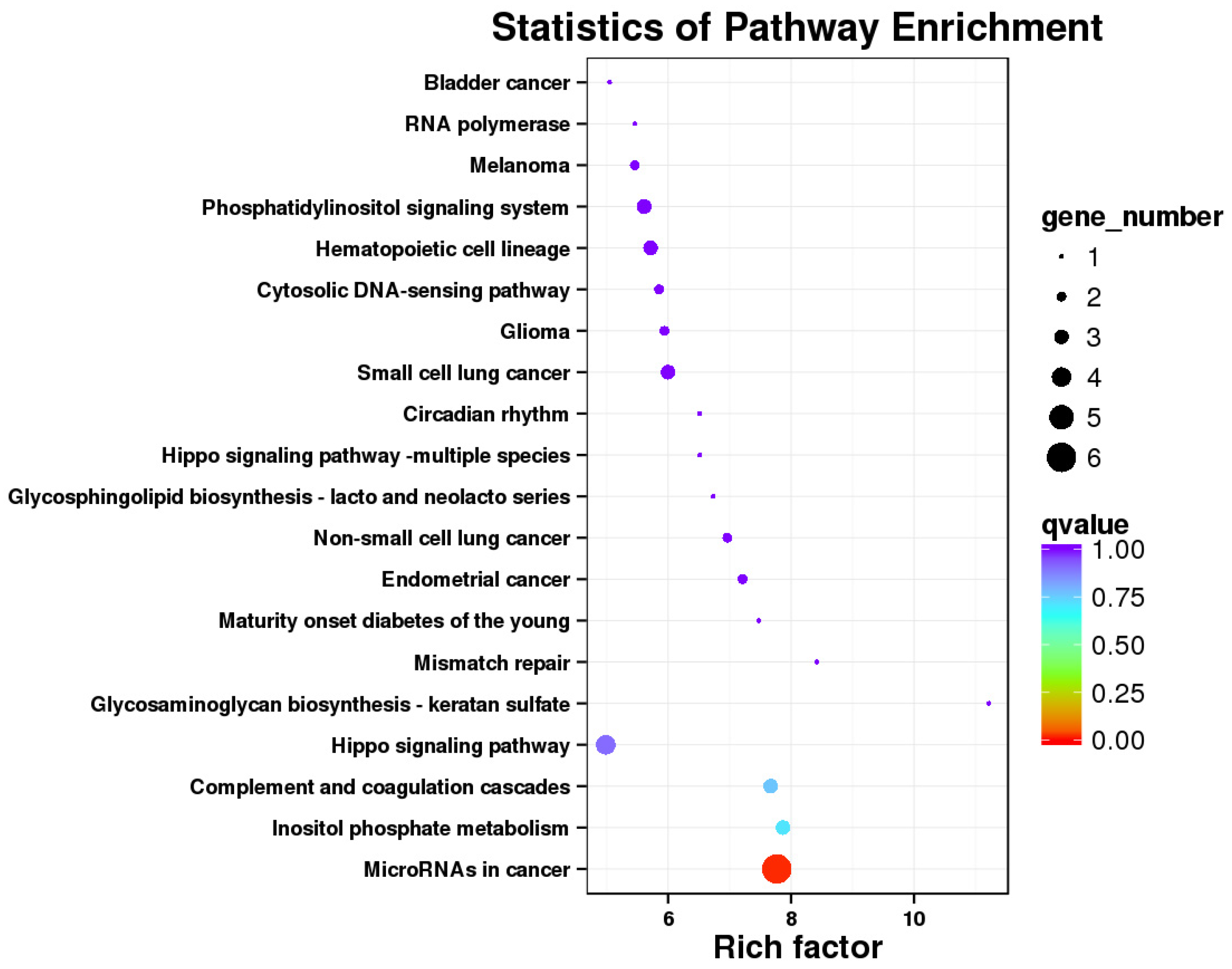
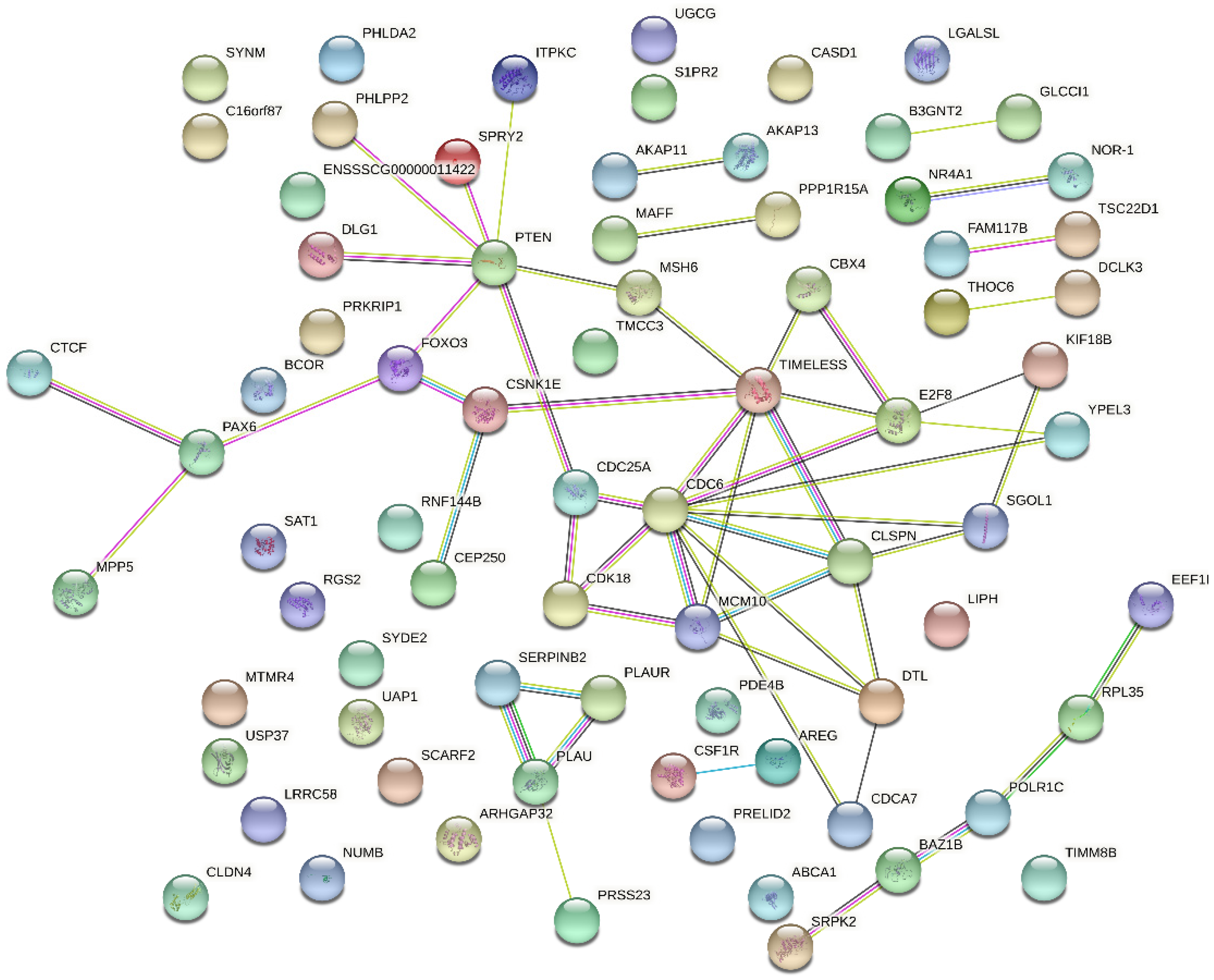
2.5. KEGG Pathway Analysis Revealed That pBD2 Has Multiple Functions in IEPC-J2 Cells against E. coli
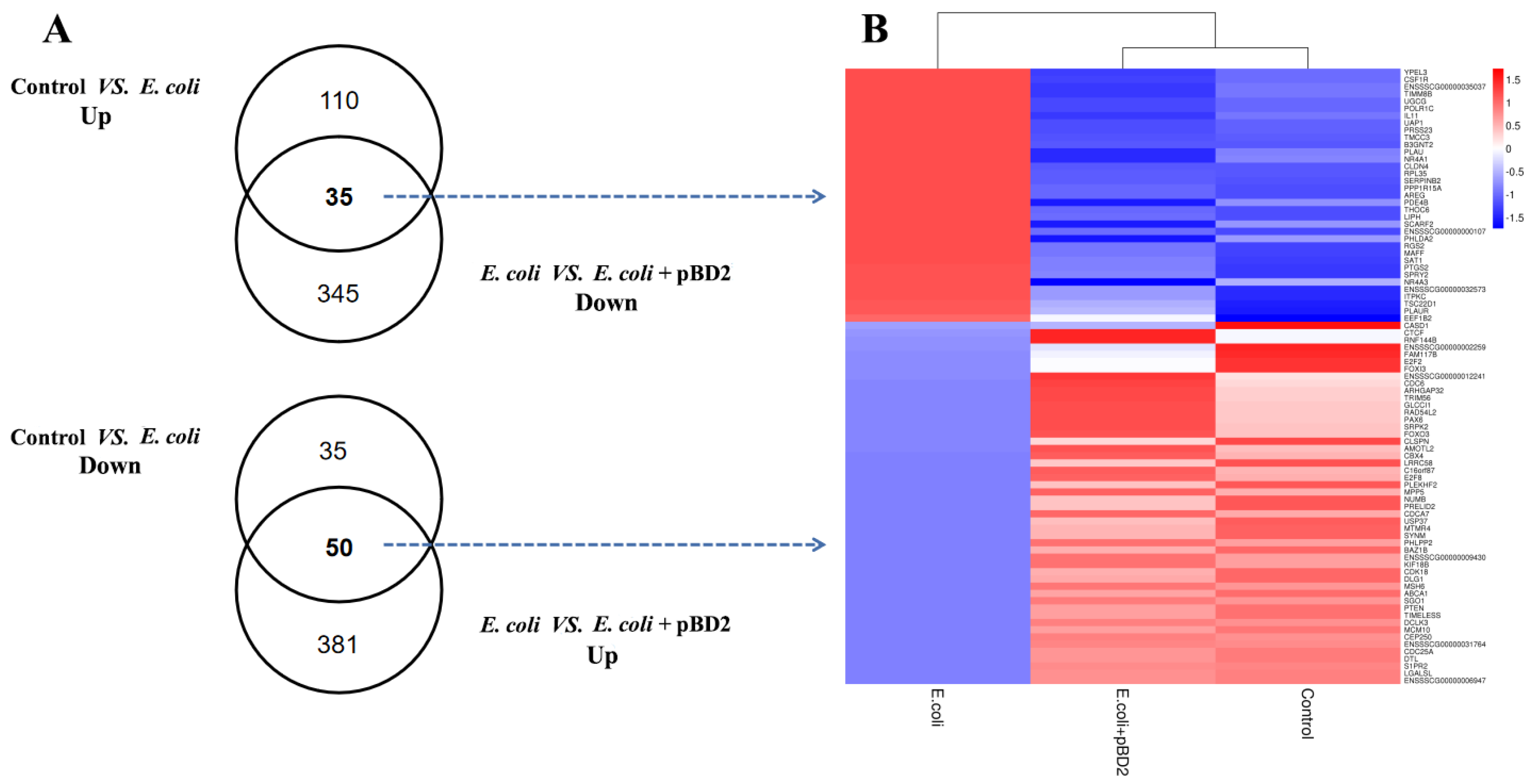
2.6. Comparison of pBD2 Effect with E. coli-Induced Transcriptome Changes
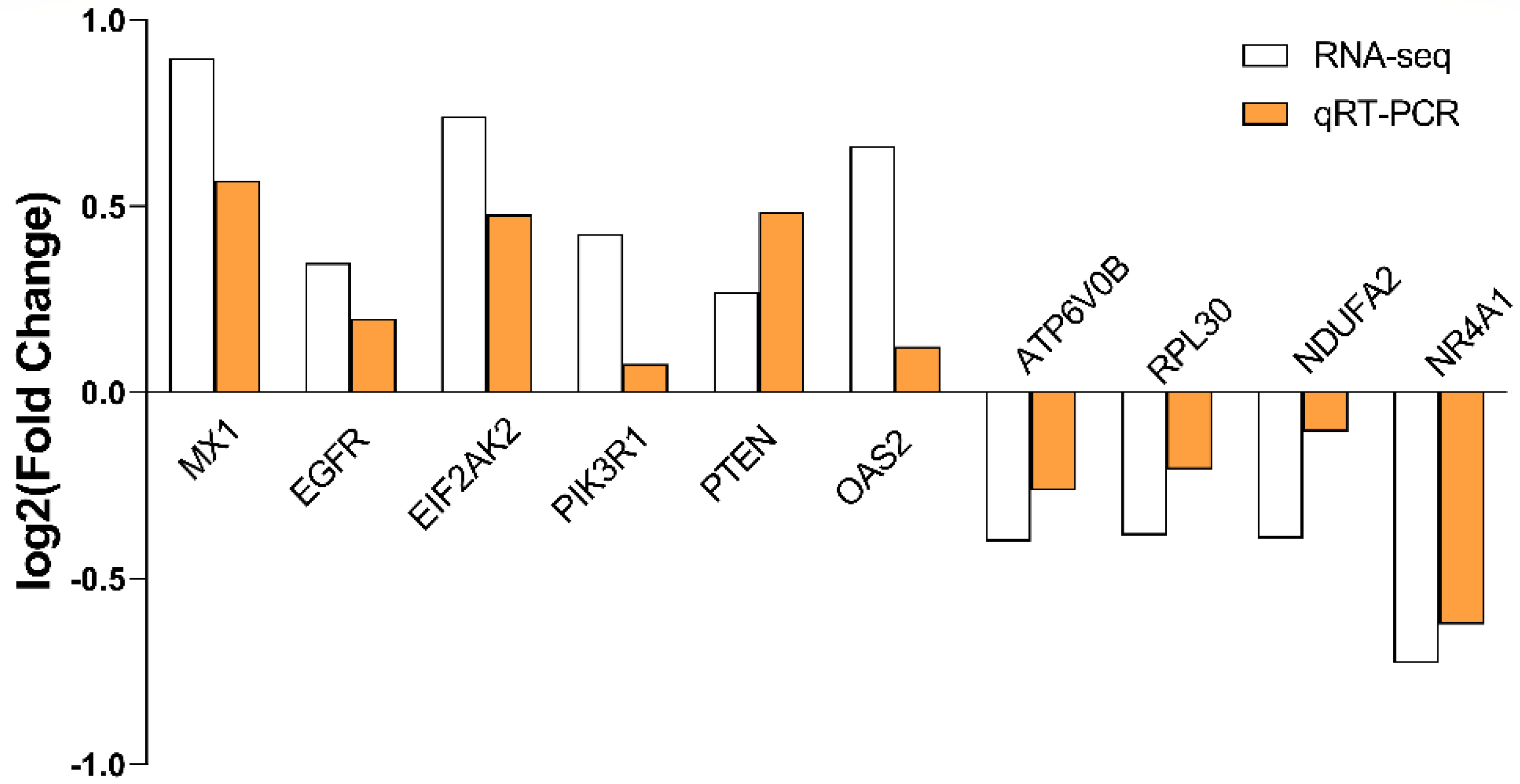
2.7. Validation of RNA-seq Data by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Strains and Cells
4.2. Preparation for pBD2
4.3. Bactericidal Activity
4.4. Cell Culture and Treatment
4.5. Library Preparation and Quality Control
4.6. Analysis of DEGs
4.7. qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shao, Y.; Wang, Y.; Yuan, Y.; Xie, Y. A systematic review on antibiotics misuse in livestock and aquaculture and regulation implications in China. Sci. Total Environ. 2021, 798, 149205. [Google Scholar] [PubMed]
- Blyth, G.A.D.; Connors, L.; Fodor, C.; Cobo, E.R. The network of colonic host defense peptides as an innate immune defense against enteropathogenic bacteria. Front. Immunol. 2020, 11, 965. [Google Scholar] [PubMed]
- Avila, E.E. Functions of antimicrobial peptides in vertebrates. Curr. Protein Pept. Sci. 2017, 18, 1098–1119. [Google Scholar]
- Robinson, K.; Deng, Z.; Hou, Y.; Zhang, G. Regulation of the Intestinal Barrier Function by Host Defense Peptides. Front. Vet. Sci. 2015, 2, 57. [Google Scholar] [PubMed]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar]
- Chen, R.B.; Zhang, K.; Zhang, H.; Gao, C.Y.; Li, C.L. Analysis of the antimicrobial mechanism of porcine beta defensin 2 against E. coli by electron microscopy and differentially expressed genes. Sci. Rep. 2018, 8, 14711. [Google Scholar]
- Zhang, K.; Zhang, H.; Gao, C.; Chen, R.; Li, C. Antimicrobial Mechanism of pBD2 against Staphylococcus aureus. Molecules 2020, 25, 3513. [Google Scholar]
- Han, F.; Zhang, H.; Xia, X.; Xiong, H.; Song, D.; Zong, X.; Wang, Y. Porcine β-defensin 2 attenuates inflammation and mucosal lesions in dextran sodium sulfate-induced colitis. J. Immunol. 2015, 194, 1882–1893. [Google Scholar]
- Li, C.L.; Zhao, Y.C.; Song, X.Y.; Huang, X.X.; Zhao, W.D. Molecular cloning, expression and characterization of the porcine β defensin 2 in E. coli. Protein Pept. Lett. 2013, 20, 715–723. [Google Scholar]
- Li, C.L.; Xu, T.T.; Chen, R.B.; Huang, X.X.; Zhao, Y.C.; Bao, Y.Y.; Zhao, W.D.; Zheng, Z.Y. Cloning, expression and characterization of antimicrobial porcine β defensin 1 in Escherichia coli. Protein Expr. Purif. 2013, 88, 47–53. [Google Scholar]
- Su, G.; Xie, K.; Chen, D.; Yu, B.; Huang, Z.; Luo, Y.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; et al. Differential expression, molecular cloning, and characterization of porcine beta defensin 114. J. Anim. Sci. Biotechnol. 2019, 10, 60. [Google Scholar] [PubMed]
- Xie, K.; Su, G.; Chen, D.; Yu, B.; Huang, Z.; Yu, J.; Zheng, P.; Luo, Y.; Yan, H.; Li, H.; et al. The immunomodulatory function of the porcine β-defensin 129: Alleviate inflammatory response induced by LPS in IPEC-J2 cells. Int. J. Biol. Macromol. 2021, 188, 473–481. [Google Scholar] [PubMed]
- Peng, Z.; Wang, A.; Feng, Q.; Wang, Z.; Ivanova, I.V.; He, X.; Zhang, B.; Song, W. High-level expression, purification and characterisation of porcine β-defensin 2 in Pichia pastoris and its potential as a cost-efficient growth promoter in porcine feed. Appl. Microbiol. Biotechnol. 2014, 98, 5487–5497. [Google Scholar] [PubMed]
- Vermeire, B.; Gonzalez, L.M.; Jansens, R.J.J.; Cox, E.; Devriendt, B. Porcine small intestinal organoids as a model to explore ETEC-host interactions in the gut. Vet. Res. 2021, 52, 94. [Google Scholar]
- Vergauwen, H. The IPEC-J2 cell line. In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models, 1st ed.; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; Volume 12, pp. 125–134. [Google Scholar]
- Holly, M.K.; Diaz, K.; Smith, J.G. Defensins in viral infection and pathogenesis. Annu. Rev. Virol. 2017, 4, 369–391. [Google Scholar]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar]
- Nunes-Santos, C.J.; Uzel, G.; Rosenzweig, S.D. PI3K pathway defects leading to immunodeficiency and immune dysregulation. J. Allergy Clin. Immunol. 2019, 143, 1676–1687. [Google Scholar]
- Xiao, H.B.; Wang, C.R.; Liu, Z.K.; Wang, J.Y. LPS induces pro-inflammatory response in mastitis mice and mammary epithelial cells: Possible involvement of NF-κB signaling and OPN. Pathol. Biol. 2015, 63, 11–16. [Google Scholar]
- Chen, G.; Goeddel, D.V. TNF-R1 signaling: A beautiful pathway. Science 2002, 296, 1634–1635. [Google Scholar]
- Freche, B.; Reig, N.; van der Goot, F.G. The role of the inflammasome in cellular responses to toxins and bacterial effectors. Semin. Immunopathol. 2007, 29, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Souvannavong, V.; Saidji, N.; Chaby, R. Lipopolysaccharide from Salmonella enterica activates NF-kappaB through both classical and alternative pathways in primary B Lymphocytes. Infect. Immun. 2007, 75, 4998–5003. [Google Scholar] [CrossRef] [PubMed]
- Huxford, T.; Hoffmann, A.; Ghosh, G. Understanding the logic of IκB:NF-κB regulation in structural terms. Curr. Top. Microbiol. Immunol. 2011, 349, 1–24. [Google Scholar] [PubMed]
- Igata, M.; Islam, M.A.; Tada, A.; Takagi, M.; Kober, A.; Albarracin, L.; Aso, H.; Ikeda-Ohtsubo, W.; Miyazawa, K.; Yoda, K.; et al. Transcriptome Modifications in Porcine Adipocytes via Toll-Like Receptors Activation. Front. Immunol. 2019, 10, 1180. [Google Scholar] [CrossRef] [PubMed]
- Schafer, K.A. The cell cycle: A review. Vet. Pathol. 1998, 35, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef]
- Orsolic, I.; Jurada, D.; Pullen, N.; Oren, M.; Eliopoulos, A.G.; Volarevic, S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin. Cancer Biol. 2016, 37–38, 36–50. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- Ganapathy-Kanniappan, S. Molecular intricacies of aerobic glycolysis in cancer: Current insights into the classic metabolic phenotype. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 667–682. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Ashton, T.M.; McKenna, W.G.; Kunz-Schughart, L.A.; Higgins, G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 2482–2490. [Google Scholar] [CrossRef] [PubMed]
- Shima, A.; Fukushima, K.; Arai, T.; Terada, H. Dual inhibitory effects of the peptide antibiotics leucinostatins on oxidative phosphorylation in mitochondria. Cell Struct. Funct. 1990, 15, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lu, L.; Li, C.; Shao, G.; Chen, X. Transcriptome analysis revealed multiple immune processes and energy metabolism pathways involved in the defense response of the large yellow croaker Larimichthys crocea against Pseudomonas plecoglossicida. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100886. [Google Scholar] [CrossRef]
- Narra, H.P.; Sahni, A.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Global transcriptomic profiling of pulmonary gene expression in an experimental murine model of Rickettsia conorii infection. Genes 2019, 10, 204. [Google Scholar] [CrossRef]
- Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 2005, 30, 283–286. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Academy. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Wang, J.; Maldonado, M.A. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell. Mol. Immunol. 2006, 3, 255–261. [Google Scholar]
- Rechsteiner, M.; Hill, C.P. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005, 15, 27–33. [Google Scholar] [CrossRef]
- Anbanandam, A.; Albarado, D.C.; Tirziu, D.C.; Simons, M.; Veeraraghavan, S. Molecular basis for proline- and arginine-rich peptide inhibition of proteasome. J. Mol. Biol. 2008, 384, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, H.; Xu, Y.; Ning, W.; Hu, S.; Wei, S.; Song, H.; Sun, J.; Ziebolz, D.; Schmalz, G.; et al. Implications of human antimicrobial peptide defensin beta-1 in clinical oral squamous cell carcinoma patients via an integrated bioinformatics approach. Comput. Math. Methods Med. 2022, 2022, 2203615. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.J.A.; Kvansakul, M.; Lay, F.T.; Phan, T.K.; Hulett, M.D. Defensin-lipid interactions in membrane targeting: Mechanisms of action and opportunities for the development of antimicrobial and anticancer therapeutics. Biochem. Soc. Trans. 2022, 50, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Ibusuki, R.; Uto, H.; Oda, K.; Ohshige, A.; Tabu, K.; Mawatari, S.; Kumagai, K.; Kanmura, S.; Tamai, T.; Moriuchi, A.; et al. Human neutrophil peptide-1 promotes alcohol-induced hepatic fibrosis and hepatocyte apoptosis. PLoS ONE 2017, 12, e0174913. [Google Scholar] [CrossRef] [PubMed]
- Carnero, A.; Blanco-Aparicio, C.; Renner, O.; Link, W.; Leal, J.F. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets 2008, 8, 187–198. [Google Scholar] [CrossRef]
- Park, J.W.; Wollmann, G.; Urbiola, C.; Fogli, B.; Florio, T.; Geley, S.; Klimaschewski, L. Sprouty2 enhances the tumorigenic potential of glioblastoma cells. Neuro-Oncol. 2018, 20, 1044–1054. [Google Scholar] [CrossRef]
- Liao, P.H.; Wang, Y.Y.; Wang, W.C.; Chen, C.H.; Kao, Y.H.; Hsu, J.W.; Chen, C.Y.; Chen, P.H.; Yuan, S.S.; Chen, Y.K. Overexpression of sprouty2 in human oral squamous cell carcinogenesis. Arch. Oral Biol. 2018, 87, 131–142. [Google Scholar] [CrossRef]
- Chen, G.; Sun, J.; Xie, M.; Yu, S.; Tang, Q.; Chen, L. PLAU promotes cell proliferation and epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Front. Genet. 2021, 12, 651882. [Google Scholar] [CrossRef]
- Shen, T.; Huang, S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anti-Cancer Agents Med. Chem. 2012, 12, 631–639. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, F.; He, Z.; Zuo, M.Z. E2F2/5/8 serve as potential prognostic biomarkers and targets for human ovarian cancer. Front. Oncol. 2019, 9, 161. [Google Scholar] [CrossRef]
- Hashemi Goradel, N.; Najafi, M.; Salehi, E.; Farhood, B.; Mortezaee, K. Cyclooxygenase-2 in cancer: A review. J. Cell. Physiol. 2019, 234, 5683–5699. [Google Scholar] [CrossRef] [PubMed]
- Uraki, S.; Sugimoto, K.; Shiraki, K.; Tameda, M.; Inagaki, Y.; Ogura, S.; Kasai, C.; Nojiri, K.; Yoneda, M.; Yamamoto, N.; et al. Human β-defensin-3 inhibits migration of colon cancer cells via downregulation of metastasis-associated 1 family, member 2 expression. Int. J. Oncol. 2014, 45, 1059–1064. [Google Scholar] [CrossRef]
- Droin, N.; Hendra, J.B.; Ducoroy, P.; Solary, E. Human defensins as cancer biomarkers and antitumour molecules. J. Proteom. 2009, 72, 918–927. [Google Scholar] [CrossRef]
- Ghavami, S.; Asoodeh, A.; Klonisch, T.; Halayko, A.J.; Kadkhoda, K.; Kroczak, T.J.; Gibson, S.B.; Booy, E.P.; Naderi-Manesh, H.; Los, M. Brevinin-2R(1) semi-selectively kills cancer cells by a distinct mechanism, which involves the lysosomal-mitochondrial death pathway. J. Cell. Mol. Med. 2008, 12, 1005–1022. [Google Scholar] [CrossRef] [PubMed]
- Arimura, Y.; Ashitani, J.; Yanagi, S.; Tokojima, M.; Abe, K.; Mukae, H.; Nakazato, M. Elevated serum beta-defensins concentrations in patients with lung cancer. Anticancer Res. 2004, 24, 4051–4057. [Google Scholar] [PubMed]
- Wong, C.C.; Zhang, L.; Ren, S.X.; Shen, J.; Chan, R.L.; Cho, C.H. Antibacterial peptides and gastrointestinal diseases. Curr. Pharm. Des. 2011, 17, 1583–1586. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Pan, B.; Yang, L.; Wang, B.; Li, J. Beta defensin 3 enhances ovarian granulosa cell proliferation and migration via ERK1/2 pathway in vitro†. Biol. Reprod. 2019, 100, 1057–1065. [Google Scholar] [CrossRef]
- Kiatsurayanon, C.; Niyonsaba, F.; Smithrithee, R.; Akiyama, T.; Ushio, H.; Hara, M.; Okumura, K.; Ikeda, S.; Ogawa, H. Host defense (Antimicrobial) peptide, human β-defensin-3, improves the function of the epithelial tight-junction barrier in human keratinocytes. J. Investig. Dermatol. 2014, 134, 2163–2173. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Samples | Clean Reads | Clean Bases | GC Content | %≥Q30 |
|---|---|---|---|---|
| Control-1 | 35,369,943 | 10,533,069,694 | 52.27% | 94.10% |
| Control-2 | 32,617,012 | 9,703,386,646 | 52.26% | 94.14% |
| Control-3 | 31,534,327 | 9,375,721,120 | 52.35% | 94.31% |
| E. coli-1 | 24,055,348 | 7,170,857,754 | 52.64% | 93.23% |
| E. coli-2 | 25,929,754 | 7,718,143,996 | 52.37% | 93.55% |
| E. coli-3 | 22,754,029 | 6,773,668,654 | 52.28% | 93.90% |
| pBD2-1 | 20,721,673 | 6,176,881,268 | 51.76% | 93.46% |
| pBD2-2 | 23,894,463 | 7,107,703,946 | 52.28% | 94.16% |
| pBD2-3 | 21,228,755 | 6,327,976,062 | 51.94% | 93.67% |
| Samples | Mapped Reads | Unique Mapped Reads | Multiple Map Reads |
|---|---|---|---|
| Control-1 | 67,898,715 (95.98%) | 66,065,506 (93.39%) | 1,833,209 (2.59%) |
| Control-2 | 62,849,152 (96.34%) | 61,097,600 (93.66%) | 1,751,552 (2.69%) |
| Control-3 | 60,714,671 (96.27%) | 59,081,017 (93.68%) | 1,633,654 (2.59%) |
| E. coli-1 | 46,168,875 (95.96%) | 44,891,836 (93.31%) | 1,277,039 (2.65%) |
| E. coli-2 | 49,843,598 (96.11%) | 48,508,765 (93.54%) | 1,334,833 (2.57%) |
| E. coli-3 | 43,776,016 (96.19%) | 42,624,022 (93.66%) | 1,151,994 (2.53%) |
| pBD2-1 | 39,790,781 (96.01%) | 38,760,294 (93.53%) | 1,030,487 (2.49%) |
| pBD2-2 | 46,057,704 (96.38%) | 44,834,945 (93.82%) | 1,222,759 (2.56%) |
| pBD2-3 | 40,692,378 (95.84%) | 39,483,201 (92.99%) | 1,209,177 (2.85%) |
| KEGG Pathway | ID | Gene Name | Corrected p-Value |
|---|---|---|---|
| NF-kappa B signaling pathway | ko04064 | TNFAIP3, BIRC3, CXCL8, TICAM1, NFKBIA, ICAM1, PTGS2, BCL10, PLAU, NFKB1, RELB, NFKB2 | 0.00000127 |
| TNF signaling pathway | ko04668 | TNFAIP3, MAP3K8, BIRC3, EDN1, NFKBIA, ICAM1, PTGS2, ENSSSCG00000008954, CXCL2, NFKB1, CCL20, BCL3 | 0.00000233 |
| NOD-like receptor signaling pathway | ko04621 | TNFAIP3, BIRC3, CXCL8, NFKBIA, NFKBIB, NFKB1 | 0.014302364 |
| Chemokine signaling pathway | ko04062 | FOXO3, RAC1, CXCL8, NFKBIA, NFKBIB, ENSSSCG00000008954, CXCL2, NFKB1, GNAI1, CCL20 | 0.014397446 |
| Genes | Sequence (5′ → 3′) | Size (bp) | GenBank Number | |
|---|---|---|---|---|
| MX1 | Forward | GTTACCGGGACAGCGAGATT | 105 | NM_214061.2 |
| Reverse | CATGACTGATTCCCACGCCT | |||
| EGFR | Forward | AGGACGAAGCAACATGGTCA | 132 | NM_214007.1 |
| Reverse | TGCATAGCACAGGTTTCGGT | |||
| EIF2AK2 | Forward | CCCTGCACTTCTAGCCATCT | 121 | NM_214319.1 |
| Reverse | CGACCACTGGCCATTTCTTTC | |||
| PIK3R1 | Forward | CTTGAGTCGGGTGCTGGAAC | 164 | XM_021076847.1 |
| Reverse | AACGCGTCCCTAACCGATTC | |||
| PTEN | Forward | TGCAATCCTCAGTTTGTGGT | 224 | NM_001143696.1 |
| Reverse | TCCTCTGGTCCTGGTATGAAG | |||
| OAS2 | Forward | AGCCAGAGCAATGGGAAACT | 228 | NM_001031796.1 |
| Reverse | GAGTTGCCCCTCAAGACTGT | |||
| ATP6VOB | Forward | AACCCCAGCCTCTTCGTAAA | 100 | XM_021096834.1 |
| Reverse | TCACTCTGGAGGTCTGAAGG | |||
| RPL30 | Forward | GACAAGGTCCAATGTTCCCA | 110 | NM_001190178.1 |
| Reverse | CCAACCTCTTTTGTAGCCGT | |||
| NDUFA2 | Forward | TGCTAAGTGGCAAAGCCTG | 167 | XM_003124046.4 |
| Reverse | GGTAGAGGGTGGAACAAGGAA | |||
| NR4A1 | Forward | TGAGAAGGTTCCCGGCTTTG | 196 | ENSSSCG00000031321 |
| Reverse | GATGCTGTCGATCCAGTCCC | |||
| TUBA1B | Forward | TACTCACCTCGACTCTTAGC | 103 | NM_001044544.1 |
| Reverse | GATGCACTCACGCATGG | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, S.; Lin, X.; Zhan, F.; Shen, X.; Liang, Y.; Li, C. Transcriptome Analysis Reveals the Multiple Functions of pBD2 in IPEC-J2 Cells against E. coli. Int. J. Mol. Sci. 2022, 23, 9754. https://doi.org/10.3390/ijms23179754
Lian S, Lin X, Zhan F, Shen X, Liang Y, Li C. Transcriptome Analysis Reveals the Multiple Functions of pBD2 in IPEC-J2 Cells against E. coli. International Journal of Molecular Sciences. 2022; 23(17):9754. https://doi.org/10.3390/ijms23179754
Chicago/Turabian StyleLian, Shaoqiang, Xiaqing Lin, Fengting Zhan, Xiaoyang Shen, Yu Liang, and Chunli Li. 2022. "Transcriptome Analysis Reveals the Multiple Functions of pBD2 in IPEC-J2 Cells against E. coli" International Journal of Molecular Sciences 23, no. 17: 9754. https://doi.org/10.3390/ijms23179754
APA StyleLian, S., Lin, X., Zhan, F., Shen, X., Liang, Y., & Li, C. (2022). Transcriptome Analysis Reveals the Multiple Functions of pBD2 in IPEC-J2 Cells against E. coli. International Journal of Molecular Sciences, 23(17), 9754. https://doi.org/10.3390/ijms23179754






