Abstract
The genus Photobacterium is known for its ecophysiological versatility encompassing free-living, symbiotic, and pathogenic lifestyles. Photobacterium sp. CCB-ST2H9 was isolated from estuarine sediment collected at Matang Mangrove, Malaysia. In this study, the genome of CCB-ST2H9 was sequenced, and the pan-genome of 37 Photobacterium strains was analysed. Phylogeny based on core genes showed that CCB-ST2H9 clustered with P. galatheae, forming a distinct clade with P. halotolerans, P. salinisoli, and P. arenosum. The core genome of Photobacterium was conserved in housekeeping functions, while the flexible genome was well represented by environmental genes related to energy production and carbohydrate metabolism. Genomic metrics including 16S rRNA sequence similarity, average nucleotide identity, and digital DNA–DNA hybridization values were below the cut-off for species delineation, implying that CCB-ST2H9 potentially represents a new species. Genome mining revealed that biosynthetic gene clusters (BGCs) involved in producing antimicrobial compounds such as holomycin in CCB-ST2H9 could contribute to the antagonistic potential. Furthermore, the EtOAc extract from the culture broth of CCB-ST2H9 exhibited antagonistic activity against Vibrio spp. Intriguingly, clustering based on BGCs profiles grouped P. galatheae, P. halotolerans, P. salinisoli, P. arenosum, and CCB-ST2H9 together in the heatmap by the presence of a large number of BGCs. These BGCs-rich Photobacterium strains represent great potential for bioactive secondary metabolites production and sources for novel compounds.
1. Introduction
Photobacterium (Vibrionaceae, Gammaproteobacteria) comprises a group of Gram-negative, facultative-aerobic, mostly halophilic, and motile bacteria [1]. The genus currently consists of 37 validly published named species (www.bacterio.net (accessed on 19 May 2022)), with P. phosphoreum as the type species [2]. All species of Photobacterium were originally thought to be luminescent. However, later studies found that only strains of P. angustum, P. aquimaris, P. ganghwense, P. kishitanii, P. leiognathi, P. mandapamensis, and P. phosphoreum display this characteristic [1,3]. Although members of this genus are ubiquitous in marine contexts, isolations of Photobacterium strains from non-marine habitats such as a saline lake, the rhizosphere of a terrestrial weed, and spoiled meat have been reported [4,5,6,7]. Photobacterium exhibits different lifestyles: some occur as free-living in seawater and sediments; others function as symbionts of the light organs of marine fish and squid, decomposers of dead fish, or pathogens for marine animals [1,8,9]. High diversity within Photobacterium has been reported, which could be linked to the lifestyles of the species [10,11]. Photobacterium has been studied for ecophysiological traits such as piezophilic, salt adaptation, bioluminescence, and motility [3,12,13].
Photobacterium also encompasses species that synthesize various secondary metabolites, including those with antibiotic activity [14]. Wietz et al. reported that a pyrrothine antibiotic, holomycin, was produced by P. galatheae S2753 (formerly P. halotolerance) [15]. Holomycin shows a broad antibiotic spectrum on several pathogenic bacteria belonging to the genera Staphylococcus, Enterococcus, Escherichia, and some marine bacteria such as Roseobacter, Pseudoalteromonas, and Vibrio [15,16]. For instance, P. galatheae S2753 also showed antagonism against a bacterial fish pathogen, Vibrio anguillarum, and a human pathogen, Staphylococcus aureus [17]. The mode of action of holomycin is speculated to inhibit RNA polymerase synthesis of the target bacteria [16]. In addition to the production of holomycin, Photobacterium sp. strain S2753 was also capable of producing two novel cyclodepsipeptides, solonamide A and B, which can inhibit the expression of the virulence gene of S. aureus [16]. These findings revealed that some Photobacterium strains have potential as biocontrol agents against pathogenic bacteria. However, the antagonism potential of Photobacterium spp. for use in aquaculture is still not well studied.
Increases in aquaculture production to meet global seafood demand have largely been accomplished through the intensification of current farming systems, resulting in a higher incidence of disease outbreaks. For instance, a shrimp disease, acute hepatopancreatic necrosis disease (AHPND), caused by Vibrio parahaemolyticus (VpAHPND), was reported in China in 2010 [18] and widely spread to South Asia from Latin America [19,20]. A detrimental reduction in shrimp production was caused by VpAHPND due to its high mortality rate, which is 100% on post-larvae of black tiger shrimp (Penaeus monodon) and Pacific white shrimp (P. vannamei), causing severe economic losses [18]. To control VpAHPND, several biocontrol strategies, such as probiotic supplementation, vaccination, and biofloc, have attracted attention in shrimp aquaculture [21,22,23]. However, these methods face several shortcomings, such as the requirement for long-term probiotic supplementation, the low efficacy of vaccination, and the difficulty of producing and maintaining biofloc in the aquaculture field. Therefore, alternative approaches for the control of VpAHPND have been demanded.
A pan-genome is the set of all non-redundant genes present in a given dataset, including the core, which is defined as the homologous genes present in all genomes in the dataset [24]. The status of the core- and pan-genomes is dependent on the number of analysed genomes, the species’ ability to acquire exogenous DNA via horizontal gene transfer, and its ecological niche [25]. A Photobacterium sp. (designated CCB-ST2H9) isolated from sediment collected in a previous metagenomic investigation [26] is the subject of this research. The aim of this study was to sequence the genome of CCB-ST2H9 to provide a glimpse into the genetic basis of antagonism. Core- and pan-genome analysis was performed to provide insights into the genome plasticity within Photobacterium and functional versatility for colonization of different niches. New clustering based on biosynthetic gene clusters (BGCs) profiles separated the Photobacterium into BGCs-rich and BGCs-low groups. Our study yielded a comprehensive understanding of the diversity of the biosynthetic gene clusters in Photobacterium, which will facilitate the use of the bacteria for bioactive metabolites production and as biocontrol agents in aquaculture.
2. Results and Discussion
2.1. Genomic Features
CCB-ST2H9 was isolated from estuarine sediment obtained from the Matang mangrove. Based on a 16S rRNA sequence similarity search performed using the EzTaxon database [27], CCB-ST2H9 is most closely related to P. halotolerans MACL01 (98.03%), P. salinisoli LAM9072 (97.65%), and P. galatheae S2753 (97.56%). These values are slightly below the species demarcation threshold of 98.7% [28].
The complete CCB-ST2H9 genome contained two chromosomes (Chr 1, 1.51 Mb; Chr 2, 1.51 Mb) and a plasmid (79.85Kb), with GC content of 50.3% (Table 1 and Figure S1). The GC content of CCB-ST2H9 was in the upper bound of Photobacterium, which varied between 38.6 and 50.9%. The CCB-ST2H9 genome falls in the Photobacterium genomes size range of 4.38 to 6.53 Mb, with the smallest genome observed for P. malacitanum CECT 9190, associated with diseased redbanded seabream, and the largest for P. alginatilyticum P03D4, isolated from a bottom seawater sample from the East China Sea [29,30]. Concomitantly, P. malacitanum and P. alginatilyticum annotated gene numbers were among the smallest and largest in Photobacterium, respectively. The CCB-ST2H9 genome has 99.2% gene space completeness following assessment with the BUSCO tool (Figure S2). Genome annotation of CCB-ST2H9 resulted in 4915 coding sequences, including 4703 protein-coding genes, 127 tRNA genes, and 37 RNA genes. A total of 3828 CCB-ST2H9 genes were assigned to 22 functional Clusters of Orthologous Groups (COG) classes, with transcription, amino acid transport and metabolism, and inorganic ion transport and metabolism being the most abundant pathways (excluding the unknown function category) (Figure S3a). Gene Ontology (GO) annotation classified 3611 CCB-ST2H9 genes into three functional terms in which the top subgroups were catalytic activity, cellular process, and metabolic process (Figure S3b). From the 2610 CCB-ST2H9 genes mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG), a significant proportion was involved in protein families: genetic information processing and environmental information processing, and protein families: signaling and cellular processes (Figure S3c).

Table 1.
Summary of Photobacterium sp. CCB-ST2H9 genome.
Genomic islands play an important role in bacterial genome evolution and adaptation as they are the probable horizontal origin of genes for pathogenicity, symbiosis, and metabolism [31]. A total of 13 genomic islands ranging in size from 10.2 to 35.8 Kb were detected in the CCB-ST2H9 genome (Table S2). The genomic islands contain a considerable number of hypothetical genes, as well as genes related to energy, cell membrane, and signal transduction (Table S3). In addition, five prophage-like elements were detected in CCB-ST2H9, one of which was classified as intact, three as incomplete, and one as questionable (Table S4).
2.2. Phylogeny of Photobacterium
Although 16S rRNA phylogeny has been widely applied in species classification, the limitations of using this gene as a phylogenetic marker in Vibrionaceae have been noted previously [10,11]. Meanwhile, multigene phylogenies provide better resolution between phylogenetically close strains, and the tree topology is less affected by recombination events [32,33]. In this study, a phylogenomic analysis was performed based on the bacterial core genes from 37 Photobacterium strains, using Vibrio cholerae ATCC 14035 as an outgroup (Figure 1). This tree showed that CCB-ST2H9 clustered with P. galatheae S2753, forming a distinct clade with P. halotolerans DSM18316, P. salinisoli LAM9071, and P. arenosum CAU 1568, distant from the other Photobacterium strains. This grouping appears to be related to environmental sources. With the exception of P. arenosum from marine sediment, this clade consists of Photobacterium isolated from non-marine habitats: CCB-ST2H9 from estuarine sediment, P. halotolerans from a saline lake, and P. salinisoli from saline soil [7,34]. The remaining Photobacterium strains were grouped into the clades Damselae, Leiognathi, Ganghwense, Profundum, Phosphoreum, and Rosenbergii, consistent with prior reports [35,36]. While this clade classification can facilitate the study of large genera by grouping together lines of descent, it is not a standard in nomenclature [8].
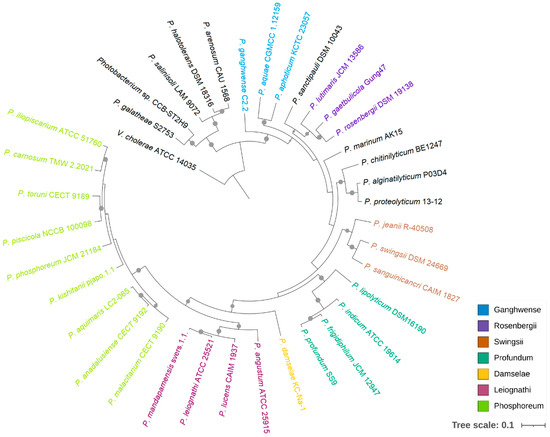
Figure 1.
Phylogenetic relationship of Photobacterium. The maximum likelihood tree was constructed based on 92 bacterial core genes with UBCG. Vibrio cholerae ATCC 14035 was used as an outgroup, and support values of more than 50 are indicated by dots on nodes.
The genome evolutionary relatedness between Photobacterium strains was depicted based on the average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH). In agreement with phylogenomic clustering, the highest ANI was shared between CCB-ST2H9 and P. galatheae (88.95%), P. halotolerans (84.26%), P. arenosum (84.25%), and P. salinisoli (84.25%) (Figure 2). Accordingly, the highest dDDH percentage (32.7%) was between CCB-ST2H9 and P. galatheae (Table S5). Both the sharing ANI and dDDH values between CCB-ST2H9 and other Photobacterium strains were below the proposed 95–96% and 70% cut-off of bacterial species definition [37,38]. Considering the 16S rRNA sequence similarity and ANI and dDDH results, strain CCB-ST2H9 is potentially a new species of the genus Photobacterium.
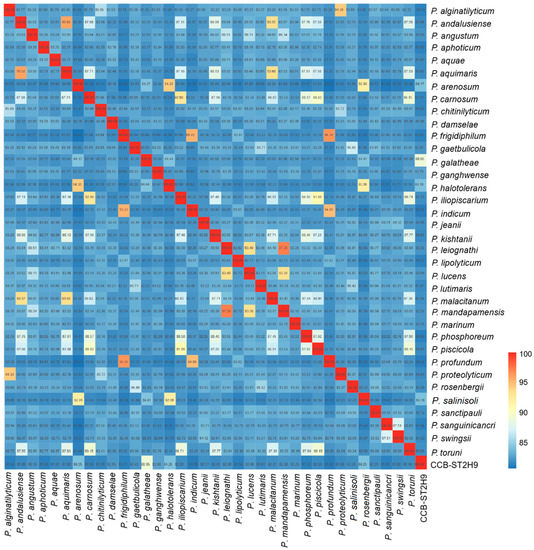
Figure 2.
Average nucleotide identity (ANI) between Photobacterium genomes. The colour bar represents the value of ANI.
2.3. Core and Pan-Genome of Photobacterium
A pan-genome is comprised of core genes found in all strains, accessory genes (shell and cloud) common to two or more strains but not all, and unique genes present only in one strain. The orthologous clustering of protein-coding sequences from the 37 Photobacterium genomes analysed identified a total of 21,786 consensus gene clusters defining the pan-genome (Figure 3a). The size of the Photobacterium pan-genome would continue to expand with the increasing number of genomes incorporated in the analysis and can be considered open (Figure 3b). This pan-genomic feature is typical of taxonomic groups capable of colonizing diverse habitats and hence the opportunity to exchange genetic material with different sources [25,39].
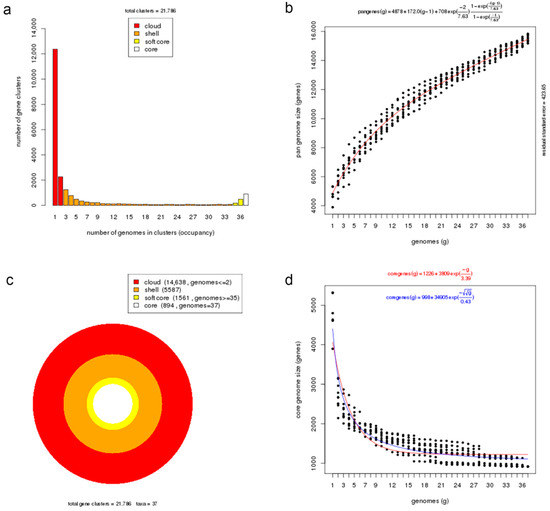
Figure 3.
Core and pan-genome analyses of Photobacterium genus. (a) Barplot of the pan-genome matrix. (b) Plot of the estimated pan-genome size with Tellelin fit. (c) Cloud, shell, soft-core, and core clusters of the pan-genome. (d) Plot of the estimated core genome size with Tellelin and Willenbrock fit.
More than half of the gene sets constituting the pan-genome (14,638 genes, 67%) belonged to the cloud cluster, whereas the shell and the soft-core clusters accounted for 26% (5587 genes) and 7% (1561 genes) of the pan-genome, respectively (Figure 3c). Both the cloud and shell clusters are subsets of the flexible genome that could be applied to infer an organism’s evolutionary trajectory and lifestyle or habitat adaptation [40]. These two clusters are hypothesized to have different rates of gene acquisition and deletion, with the cloud including rapidly gained and lost genes, and the shell containing slowly gained and lost genes [41]. The core genome presented in all examined Photobacterium strains had 894 genes, representing approximately 19% of the total genes in a genome, which indicates significant gene conservation among Photobacterium. The number of core genes decreased rapidly with the addition of genomes but stabilized after the addition of the 20th genome, suggesting that the core genome was closed (Figure 3d). In a previous Photobacterium pan-genome study, a higher number of core genes was observed, likely due to the inclusion of genomes from less diverse species in the analysis and the slightly smaller number of genomes in the dataset [10].
COG functional assignments revealed that the Photobacterium core genome was enriched in functions involving translation, coenzyme metabolism, and nucleotide metabolism and transport (Figure 4a). These core genes encode housekeeping functions related to fundamental processes in the cell that have been conserved in all species throughout evolution. In addition to translation and coenzyme metabolism, the soft-core genome was also abundant in transcription function. The abundant COG category transcription, which consists of transcriptional regulators, could enable Photobacterium to regulate metabolic processes, contributing to their adaptability to the local environment. Compared to the core genome, the Photobacterium flexible genome was well represented with functions related to energy production and conversion, cell wall/membrane/envelope biogenesis, and carbohydrate metabolism and transport. The flexible genome was enriched with environmental genes, such as those that allow Photobacterium to respond to environmental changes by utilising different types of carbon and energy sources for surviving under various conditions. The flexible genome contributes to functional versatility, which in turn improves the ecological success of the bacteria in diverse environmental niches. Analysis of CCB-ST2H9 specific genes revealed that only 34.8% (201 genes) were assignable to COG functions (Figure 4b). These unique genes that evolved in CCB-ST2H9 likely complement strain-specific activity via unknown mechanisms. Besides function unknown (31%), COGs associated with replication and repair, carbohydrate metabolism and transport, and cell wall/membrane/envelope biogenesis were most abundant among strain-specific genes.
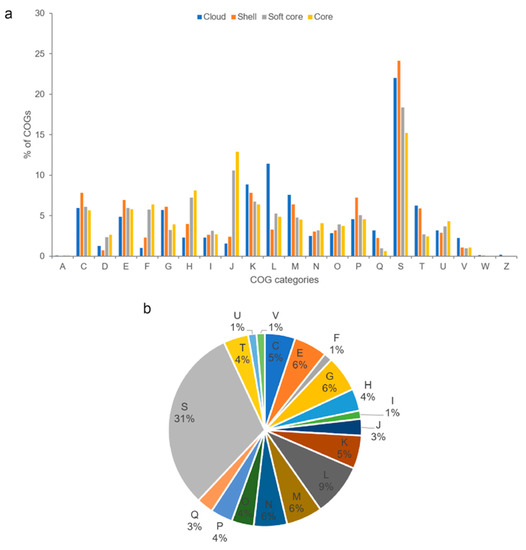
Figure 4.
(a) Distribution of COG functional categories for cloud, shell, soft core, and core Photobacterium pan-genome clusters. (b) COG functional classification of strain CCB-ST2H9 specific genes. A: RNA processing and modification, C: energy production and conversion, D: cell cycle control and mitosis, E: amino acid metabolism and transport, F: nucleotide metabolism and transport, G: carbohydrate metabolism and transport, H: coenzyme metabolism, I: lipid metabolism, J: translation, K: transcription, L: replication and repair, M: cell wall/membrane/envelop biogenesis, N: cell motility, O: post-translational modification, protein turnover, chaperone functions, P: inorganic ion transport and metabolism, Q: secondary structure, S: function unknown, T: signal transduction, U: intracellular trafficking and secretion, V: defense mechanisms, W: extracellular structures, and Z: cytoskeleton.
2.4. Biosynthetic Potential of Photobacterium
To examine the potential of Photobacterium strains to produce bioactive metabolites, the genomes were screened for the presence of biosynthetic gene clusters (BGCs) using antiSMASH software. A total of 238 BGCs were detected amongst the 37 genomes, with beta-lactone (35), ribosomally synthesized and post-translationally modified peptide (RiPP)-like (33), and non-ribosomal peptide synthetase (NRPS) (30) being the most prevalent (Figure 5). Some BGCs co-localized as hybrid clusters, such as NRPS/trans-amino transferase polyketide synthase (transAT-PKS), NRPS/type I polyketide synthase (T1PKS), aryl polyene/ectoine, and NRPS/NRPS-like/T1PKS (Table S6). Ten BGCs were identified in Photobacterium sp. CCB-ST2H9, including clusters potentially encoding beta-lactone, butyrolactone, cyanobactin, ectoine, NRPS, NRPS-like, NRPS/NRPS-like, RiPP-like, and siderophore (Table 2).
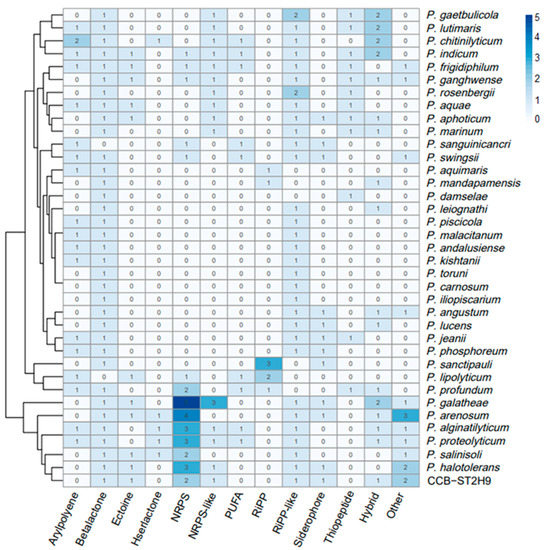
Figure 5.
Heatmap representation of Photobacterium biosynthetic gene clusters (BGCs). The column dendrogram represents hierarchical clustering of bacteria by shared BGCs numbers, and the colour code represents the number of BGCs.

Table 2.
Biosynthetic gene clusters in Photobacterium sp. CCB-ST2H9 predicted using antiSMASH.
All Photobacterium except P. lipolyticum and P. sanguinicancri harbour a beta-lactone cluster. The CCB-ST2H9 beta-lactone cluster contained the biosynthetic gene encoding 2-isopropylmalate synthase, which is involved in the biosynthesis of beta-lactone [42] but shows no correlation with known clusters. CCB-ST2H9 also harbors a BGC for butyrolactone, a quorum sensing molecule related to the regulation of antibiotic production and morphogenesis [43]. The cyanobactin BGC uniquely found in the CCB-ST2H9 genome displayed no match to known clusters. Due to the diversity of gene products and cluster organization in BGCs, some predicted cluster regions do not resemble known database entries. P. indicum, P. frigidiphilum, P. ganghwense, P. lipolyticum, P. galatheae, P. arenosum, P. haloterans, P. salinisoli, and CCB-ST2H9 all have BGCs coding for ectoine that are reported to play a role in osmotolerance [44]. The lack of ectoine in other Photobacterium strains implies that they might utilize different strategies for osmoregulation in hypersaline environments. Although a RiPP-like BGC was detected in CCB-ST2H9, it did not show similarity to known clusters.
Many bacteria have developed the ability to secrete iron-chelating molecules or siderophores that bind iron to form siderophore-iron complexes that are then liberated internally in the cell [45]. Siderophores enable producers to scavenge dissolved iron from the environment and deprive competitors of it. Under iron-limiting conditions, siderophore-producing bacteria have a competitive advantage over other species that lack iron-chelating ability [46]. A siderophore BGC showing high homology (88%) to aerobactin from Aliivibrio fisheri ES114, which is a hydroxamate-type siderophore, was identified in CCB-ST2H9. The presence of the siderophore aerobactin in P. halotolerans MELD1 contributes to the protection of host plants from phytopathogens [47]. Similarly, the siderophore aerobactin in CCB-ST2H9 could contribute to its antagonistic potential. The identified NRPS in CCB-ST2H9 showing 100% similarity to BGC in the database include xenotetrapeptide of Xenorhabdus nematophila ATCC 19061. While X. nematophila has been studied for xenematide activity, the function of xenotetrapeptide in this strain has not been explored [48].
Another NRPS with potential biosynthetic novelty shows low homology (38%) with the holomycin gene cluster in Streptomyces clavuligerus ATCC 27064. Holomycin production has been reported in both P. galatheae and P. halotolerans and examined in terms of the biosynthetic gene cluster and physiological role [16,49,50]. Holomycin is a member of the dithiolopyrrolone class that displays antibiotic activity against a broad spectrum of bacteria and inhibits RNA synthesis [51,52]. Further sequence analysis revealed that the CCB-ST2H9 holomycin BGC contains 10 genes homologous to and arranged in the same manner as those in P. galatheae [49]. The cluster is formed by phosphopantothenoylcysteine decarboxylase, HlmF, metallophosphoesterase, HlmY, globin, HlmG, N-acyltransferase, HlmA, acyl-CoA dehydrogenase, HlmB, thioesterase, HlmC, flavin mononucleotide-dependent oxidoreductase, HlmD, MFS transporter, HlmH, transcriptional regulator, HlmX, and core protein NRPS synthetase, HlmE with a characteristic arrangement of cyclization (Cy), adenylation (A), and thiolation (T) domains (Figure 6). The largest block of conserved genes corresponding to hlmA-hlmE is conserved in all holomycin producers, including S. clauvuligerus, Y. ruckeri, Pseudoalteromonas sp., and Photobacterium [49,53,54,55]. The holomycin BGC was also found in the P. arenosum and P. salinosoli genomes, even though antibiotic production and antagonism in these species have not been investigated. The presence of nearly identical holomycin BGCs in P. galatheae, P. halotolerans, P. arenosum, P. salinisoli, and CCB-ST2H9 suggests that this cluster may have originated via a horizontal gene transfer (HGT) event from a donor closely related to these species. Overall, the annotation of CCB-ST2H9 genes involved in the biosynthesis of compounds with antibacterial and iron scavenging functions is in agreement with the strain’s ability to antagonize the growth of pathogenic bacteria.
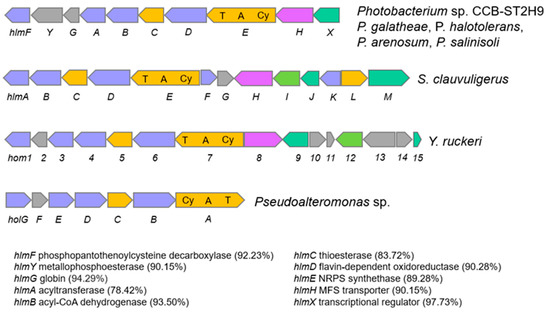
Figure 6.
Comparison of holomycin biosynthetic gene cluster in Photobacterium, S. clauvuligerus, Y. ruckeri, and Pseudoalteromonas sp. Similarity (%) of genes in CCB-ST2H9 to those of P. galatheae are indicated in brackets. Light blue indicates oxidative gene. Orange indicates biosynthesis gene. Magenta indicates MFS transporter genes. Green indicated transcription regulator gene. Grey indicates gene with unknown function.
High variability of the biosynthetic operons in terms of abundance and composition was observed across Photobacterium, with P. galathea carrying the highest number of BGCs (15), while P. carnosum, P. damselae, P. iliopiscarium, and P. toruni have the lowest (2). This is the first study to use clustering based on BGCs profiles to separate the Photobacterium into BGCs-rich and BGCs-low groups. Although no evidence linking environmental sources and BGCs was found, the frequency of BGCs types appears to correlate with genetic proximity. Notably, CCB-ST2H9 displays a similar frequency of BGCs with P. halotolerans, P. arenosum, P. galathea, and P. salinisoli that are phylogenetically close. Except for P. alginatilyticum and P. proteolyticum that are phylogenetically distant, these strains cluster together by the presence of a large number of BGCs in their genomes. Conversely, it could also be claimed that the acquisition of gene clusters leads to ecological diversity and speciation, forming a phylogenetically distinct clade of P. arenosum, P. halotolerans, P. salinisoli, P. galathea, and CCB-ST2H9. The wide distribution of bioactive metabolites in these Photobacterium strains suggests good potential for antibiotic production and as biocontrol agents.
In addition, the variable distribution of BGCs in Photobacterium is indicative of gene loss from the descendants of a cluster-harboring ancestor and HGT or recombination events. Genes encoding traits subjected to weak selection are more likely to be lost, whereas genes that confer positive benefits under certain conditions are likely to be gained. For instance, a polyunsaturated fatty acid (PUFA) biosynthetic cluster that is present in other Photobacterium strains was not detected in CCB-ST2H9. Since the PUFA cluster has been linked to cold temperature adaptation [56], it could have been lost in the evolution of non-psychrophiles Photobacterium. On the other hand, the presence of BGCs for antibiotics in Photobacterium such as P. arenosum, P. halotolerans, P. salinisoli, P. galatheae, and CCB-ST2H9 raises the intriguing question of the ecological significance of the BGCs to the producers. In habitats characterized by dense multispecies communities, bacteria compete with their neighbours for scarce resources [57]. Antibiotic compound productions are proposed as weapons that provide fitness advantage over other occupants of the same ecological niche [58]. Compared to free-living Photobacterium, densely colonized sediment and soil could be the factor driving the evolution of antibiotic-producing traits in strains such as P. arenosum, P. salinisoli, and CCB-ST2H9. This view is in line with results from pelagic ocean samples, which showed increased antagonism of particle-attached bacteria compared to free-living bacteria [59]. Overall, the distinct BGC profiles displayed by Photobacterium from diverse habitats provide clues that metabolite production capacity could be the result of habitat-specific adaptation.
2.5. Antimicrobial Activity of EtOAc Extract against Vibrio spp.
As analysed above, CCB-ST2H9 possesses the holomycin biosynthetic gene cluster in its genome. To confirm the production of antagonistic compounds, EtOAc extraction was performed from the culture supernatant. Disc diffusion assay of the crude extract resuspended in methanol was conducted against seven Vibrio spp., including VpAHPND, and two bacterial strains, E. coli DH5α and Bacillus sp. CCB-MMP212, as positive controls. Oliver and co-researchers reported that holomycin exhibited antimicrobial activity against both Gram-positive and negative bacteria such as Staphylococcus and E. coli [51]. Besides that, holomycin isolated from P. galarheae S2753 exhibited the antagonistic activity against several marine bacteria, including the genus Vibrio [49]. As shown in Figure 7, the crude extract also showed antimicrobial activity on Bacillus sp. CCB-MMP212 (Gram-positive) and E. coli DH5α (Gram-negative). All Vibrio spp. tested were susceptible to the crude extract (Figure 7). It was reported that AHPND could be caused not only by V. parahaemolyticus but also other non-pathogenic Vibrio species because of the introduction of the pVA1 plasmid carrying the pirAB gene encoding homologues of Photorhabdus insect-related (Pir) toxins [60]. This finding suggests that CCB-ST2H9 might have the potential as a novel biocontrol agent due to its broad host range of antagonistic compounds.
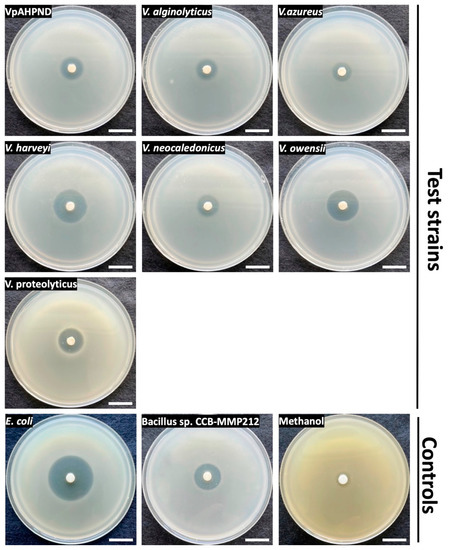
Figure 7.
Disk diffusion assay of EtOAc extract on seven Vibrio species and control strains. Scale bars indicate 2 cm.
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
Photobacterium sp. strain CCB-ST2H9 was isolated from estuarine sediment collected from the Matang Mangrove Forest in Malaysia (4°12′48.06″ N, 100°38′49.5204″ E) [26]. The isolation was performed on a high-nutrient artificial seawater agar medium (H-ASWM) (0.5% tryptone, 2.4% artificial seawater, 10 mM HEPES, pH 7.6) [61] following the method previously described [62]. The strain was deposited at the Microbial Biodiversity Library of the Centre for Chemical Biology (CCB-MBL), University Sains Malaysia. CCB-ST2H9 was cultured aerobically in H-ASWM at 30 °C. For antagonistic analysis of crude extracts from cell culture of ST2H9, the AHPND-causing V. parahaemolyticus-like strain (VpAHPND) was obtained from Universiti Malaysia Terengganu [63]. V. harveyi JCM 33361T, V. owensii JCM 16517T, and V. proteolyticus JCM 21193T were obtained from the Japan Collection of Microorganisms, whereas V. alginolyticus CCB-CTB329, V. azureus CCB-ST2H38, and V. neocaledonicus CCB-ST4H6 were from CCB-MBL. These strains were incubated on H-ASWM agar plates at 30 °C. The cells growing on the agar plate were inoculated into 10 mL of H-ASWM broth and incubated for 16 h at 30 °C with shaking (200 rpm). After the incubation period, 200 µL of the suspension was transferred to 10 mL of H-ASWM broth and incubated for 2 h at 30 °C with 200 rpm of shaking to obtain exponential phase cells. Escherichia coli DH5α (Invitrogen, Carlsabd, CA, USA) and Bacillus sp. CCB-MMP212 from CCB-MBL were used as positive controls and cultured on Lysogeny-Broth (LB) agar plate at 37 °C and Marine Agar (MB) plate at 30 °C, respectively.
3.2. Genome Sequencing
Genomic DNA was extracted from mid-logarithmic phase culture following the method of Sokolov et al. [64] with some modifications: sample lysis was performed with a buffer containing 50 mM NaCl, 50 mM Tris-HCl pH8, 50 mM EDTA, 2% SDS, and isopropanol precipitation was replaced by SPRI bead purification. The quality and quantity of isolated DNA were assayed using 1% (w/v) agarose gel electrophoresis and Qubit fluorimetry (Thermo Scientific, Waltham, MA, USA), respectively. The Illumina sequencing library was constructed using the NEBNext Ultra DNA kit (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions. The genome of CCB-ST2H9 was sequenced on a NovaSeq 6000 (Illumina, San Diego, CA, USA) at 150 bp paired-end, generating 10.26 million reads totaling 1.53 Gb. The DNA library for Nanopore sequencing was prepared using the Ligation sequencing kit (SQK-LSK109, Oxford Nanopore, UK). The library was sequenced with a PromethION and yielded 182,476 reads with a total of 1.92 Gb.
3.3. Genome Assembly and Annotation
Raw Illumina reads were trimmed with fastp (v0.23.2) (https://github.com/OpenGene/fastp, accessed on 12 August 2022) to remove low-quality bases and adaptor sequences. Nanopore sequences were preprocessed to remove reads with qscore below 7. Hybrid assembly of Illumina and Nanopore reads was performed using Unicycler (v0.5.0) (https://github.com/rrwick/Unicycler, accessed on 12 August 2022) [65] with default settings. Genome assembly metrics were computed with QUAST (v5.0.2) (http://quast.sourceforge.net, accessed on 12 August 2022). Assembly completeness was evaluated using BUSCO (v5.3.2) (https://busco.ezlab.org, accessed on 12 August 2022) against the bacteria_odb10 dataset. Genome annotation was carried out through Prokka pipeline (v1.14.6) (https://github.com/tseemann/prokka, accessed on 12 August 2022) [66] and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Coding sequences (CDS) were functionally annotated via the eggNOG-mapper (v2.1.6) (http://eggnog-mapper.embl.de, accessed on 12 August 2022) and BlastKoala server (https://www.kegg.jp/blastkoala, accessed on 12 August 2022). Genomic Islands were detected by IslandViewer4 (https://www.pathogenomics.sfu.ca/islandviewer, accessed on 12 August 2022) with IslandPath-DIMOB and SIGI-HMM predictions, and prophage was screened using PHASTER (https://phaster.ca, accessed on 12 August 2022). Secondary metabolite gene clusters were predicted using antiSMASH (v6.0.1) (https://antismash.secondarymetabolites.org, accessed on 12 August 2022) [67] with detection strictness set to ‘relaxed’. The KnownClusterBlast feature of antiSMASH was used to compare identified clusters against known gene clusters from the Minimum Information about a Biosynthetic Gene Cluster (MIBiG) database.
3.4. Comparative Genomics
The genome sequences of Photobacterium strains from different habitats were retrieved from the GenBank database (Table S1). Genome-based phylogeny was carried out using the UBCG (v3.0) tool (https://www.ezbiocloud.net/tools/ubcg ) [68] based on 92 core genes. A maximum-likelihood tree was inferred with RAxML (v8.2.12) (https://github.com/stamatak/standard-RAxML, accessed on 12 August 2022) under the GTR + GAMMA model and 100 bootstrap replications. The phylogenetic tree was visualized using the Interactive Tree of Life (http://itol.embl.de (accessed on 12 August 2022)). The average nucleotide identity value was calculated using GET_HOMOLOGUES (v3.4.3) (https://github.com/eead-csic-compbio/get_homologues, accessed on 12 August 2022) [69] with parameters ‘-a CDS -A -t 0 -M’. The DNA–DNA hybridization was estimated in silico using the Genome-to-Genome Distance Calculator (v3.0) (https://ggdc.dsmz.de/ggdc.php, accessed on 12 August 2022) [70] with formula 2.
Gene clustering was performed with GET_HOMOLOGIES based on the bidirectional best-hit (BDBH), COGtriangles, and OrthoMCL algorithms. BLASTp searches were done with a 75% minimum alignment coverage and E-value set at 1×10−5. The core-genome was estimated from the consensus clusters defined by BDBH, COGtriangles, and OrthoMCL, whereas the pan-genome was computed from COGtriangles and OrthoMCL clusters.
3.5. Extraction of Antagonistic Compounds from CCB-ST2H9 and Its Disk Diffusion Assay
The extraction of antagonistic compounds from CCB-ST2H9 was conducted according to the method of Mansson [16] with slight modifications. CCB-ST2H9 was cultured in 300 mL of Sigma Sea Salt medium (4% Sigma Sea Salt, 0.4% glucose, and 0.3% casamino acids with 10 mM HEPES, pH 7.6) for 72 h at 30 °C with shaking (200 rpm). After the incubation, the cell suspension was centrifuged at 8000× g for 20 min to remove the cells. An equal volume of ethyl acetate (EtOAc) was added and mixed into the supernatant. The EtOAc fraction was evaporated using a rotary evaporator (Heidolph, Germany). The crude extract was resuspended in methanol (200 mg/200 µL), and the solution was subjected to a test for its antagonistic activity on Vibrio spp.
Double-layer agar plates containing test strains were prepared to test the antagonistic activity of crude extract. Four mL of H-ASWM soft agar (0.3%) containing 200 µL of the exponential phase cells of each Vibrio species were overlayed on H-ASWM agar plates. The disk soaked in 20 µL of the extract was placed on the center of the double-layer agar plate, and the plate was incubated for 1 day at 30 °C. The double-layer agar plates containing E. coli DH5α or Bacillus sp. CCB-MMP212 were prepared by LB agar and MB, respectively. The disk with soaked methanol was used as a negative control.
4. Conclusions
This study provides insights into the genomic characteristics of Photobacterium sp. CCB-ST2H9 and its antagonistic potential against Vibrio spp. Photobacterium has an open pan-genome encompassing a core genome encoding essential functions and a flexible genome containing environmental genes contributing to adaptation to diverse habitats. While phylogenomic analysis showed that CCB-ST2H9 was closely related to P. galatheae, P. halotolerans, P. salinisoli, and P. arenosum, genomic metrics including 16S rRNA sequence similarity, ANI, and dDDH values between CCB-ST2H9 and these species were below the boundary of a bacterial species. AntiSMASH prediction revealed that CCB-ST2H9 harbours 10 BGCs, including three NRPSs predicted to be involved in the production of antibiotics such as holomycin and xenotetrapeptide. These NRPSs, along with the BGCs for butyrolactone and siderophore, could contribute to the antagonistic potential of CCB-ST2H9. The BGCs profile revealed that CCB-ST2H9, along with the other six Photobacterium spp., formed a BGCs-rich group. In addition, the production of antagonistic compounds against the causative agent of shrimp disease was also experimentally confirmed. Photobacterium spp. are promising resources for the genetic exploration and exploitation of bioactive secondary metabolites.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23179712/s1. References [4,5,7,17,29,30,34,35,36,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96] are cited in the supplementary materials.
Author Contributions
Conceptualization, N.-S.L. and G.F.; methodology, N.-S.L., G.F. and W.L.H.; formal analysis, N.-S.L.; resources, N.M. and N.A.A.; manuscript writing, N.-S.L. and G.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a Universiti Sains Malaysia (USM) short-term research grant (304.PCCB.6315625).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Photobacterium sp. CCB-ST2H9 whole genome project is available in the NCBI database under the accession number CP100425-CP100427, SAMN25271433 (BioSample) and PRJNA800629 (BioProject).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Urbanczyk, H.; Ast, J.C.; Dunlap, P.V. Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol. Rev. 2011, 35, 324–342. [Google Scholar] [CrossRef] [PubMed]
- Beijerinck, M.W. Le Photobacterium luminosum, bactérie lumineuse de la Mer du Nord. Arch. Neerl. Sci. Exactes Nat. 1889, 23, 401–427. [Google Scholar]
- Ast, J.C.; Dunlap, P.V. Phylogenetic resolution and habitat specificity of members of the Photobacterium phosphoreum species group. Environ. Microbiol. 2005, 7, 1641–1654. [Google Scholar] [PubMed]
- Lucena, T.; Ruvira, M.A.; Pascual, J.; Garay, E.; Macián, M.C.; Arahal, D.R.; Pujalte, M.J. Photobacterium aphoticum sp. nov., isolated from coastal water. Int. J. Syst. Evol. Microbiol. 2011, 61, 1579–1584. [Google Scholar] [CrossRef]
- Yoshizawa, S.; Wada, M.; Kita-Tsukamoto, K.; Yokota, A.; Kogure, K. Photobacterium aquimaris sp. nov., a luminous marine bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 2009, 59, 1438–1442. [Google Scholar] [CrossRef][Green Version]
- Mathew, D.C.; Ho, Y.N.; Gicana, R.G.; Mathew, G.M.; Chien, M.C.; Huang, C.C. A rhizosphere-associated symbiont, Photobacterium spp. strain MELD1, and its targeted synergistic activity for phytoprotection against mercury. PLoS ONE 2015, 10, e0121178. [Google Scholar] [CrossRef]
- Rivas, R.; García-Fraile, P.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, E. Photobacterium halotolerans sp. nov., isolated from Lake Martel in Spain. Int. J. Syst. Evol. Microbiol. 2006, 56, 1067–1071. [Google Scholar] [CrossRef]
- Labella, A.M.; Arahal, D.R.; Castro, D.; Lemos, M.L.; Borrego, J.J. Revisiting the genus Photobacterium: Taxonomy, ecology and pathogenesis. Int. Microbiol. 2017, 20, 1–10. [Google Scholar]
- Rivas, A.J.; Lemos, M.L.; Osorio, C.R. Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front. Microbiol. 2013, 4, 283. [Google Scholar] [CrossRef]
- Machado, H.; Gram, L. Comparative genomics reveals high genomic diversity in the genus Photobacterium. Front. Microbiol. 2017, 8, 1204. [Google Scholar] [CrossRef]
- Fuertes-Perez, S.; Hauschild, P.; Hilgarth, M.; Vogel, R.F. Biodiversity of Photobacterium spp. isolated from meats. Front. Microbiol. 2019, 10, 2399. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, P.; Hilgarth, M.; Vogel, R.F. Hydrostatic pressure- and halotolerance of Photobacterium phosphoreum and P. carnosum isolated from spoiled meat and salmon. Food Microbiol. 2021, 99, 103679. [Google Scholar] [CrossRef] [PubMed]
- Eloe, E.A.; Lauro, F.M.; Vogel, R.F.; Bartlett, D.H. The deep-sea bacterium Photobacterium profundum SS9 utilizes separate flagellar systems for swimming and swarming under high-pressure conditions. Appl. Environ. Microbiol. 2008, 74, 6298–6305. [Google Scholar] [CrossRef] [PubMed]
- Fuertes-Perez, S.; Vogel, R.F.; Hilgarth, M. Comparative genomics of Photobacterium species from terrestrial and marine habitats. Curr. Res. Microb. Sci. 2021, 2, 100087. [Google Scholar] [CrossRef]
- Wietz, M.; Mansson, M.; Gotfredsen, C.H.; Larsen, T.O.; Gram, L. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs 2010, 8, 2946–2960. [Google Scholar] [CrossRef]
- Mansson, M.; Gram, L.; Larsen, T.O. Production of bioactive secondary metabolites by marine vibrionaceae. Mar. Drugs 2011, 9, 1440–1468. [Google Scholar] [CrossRef]
- Machado, H.; Giubergia, S.; Mateiu, R.V.; Gram, L. Photobacterium galatheae sp. nov., a bioactive bacterium isolated from a mussel in the Solomon Sea. Int. J. Syst. Evol. Microbiol. 2015, 65, 4503–4507. [Google Scholar] [CrossRef]
- Lightner, D.V.; Redman, R.M.; Pantoja, C.; Noble, B.L.; Tran, L. Early mortality syndrome affects shrimp in Asia. Glob. Aquacult. Advocate 2012, 15, 40. [Google Scholar]
- Amatul-Samahah, M.; Muthukrishnan, S.; Omar, W.; Ikhsan, N.; Md Yasin, I. Vibrio spp. associated with acute hepatopancreatic necrosis disease (AHPND) found in penaeid shrimp pond from the east coast of peninsular Malaysia. J. Environ. Biol. 2020, 41, 1160–1170. [Google Scholar] [CrossRef]
- Yu, L.H.; Teh, C.S.J.; Yap, K.P.; Thong, K.L. Diagnostic approaches and contribution of next-generation sequencing technologies in genomic investigation of Vibrio parahaemolyticus that caused acute hepatopancreatic necrosis disease (AHPND). Aquac. Int. 2020, 28, 2547–2559. [Google Scholar] [CrossRef]
- Truc, L.N.T.; Ngoc, A.T.; Hong, T.T.T.; Thanh, T.N.; Kim, H.H.; Kim, L.P.; Truong, G.H.; Quoc, P.T.; Ngoc, T.N.T. Selection of lactic acid bacteria (LAB) antagonizing Vibrio Parahaemolyticus: The pathogen of acute hepatopancreatic necrosis disease (AHPND) in whiteleg shrimp (Penaeus Vannamei). Biology 2019, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Kewcharoen, W.; Srisapoome, P. Probiotic effects of Bacillus spp. from Pacific white shrimp (Litopenaeus vannamei) on water quality and shrimp growth, immune responses, and resistance to Vibrio parahaemolyticus (AHPND strains). Fish Shellfish Immunol. 2019, 94, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Bossier, P.; Norouzitallab, P.; Vanrompay, D. Trained immunity and perspectives for shrimp aquaculture. Rev. Aquac. 2020, 12, 2351–2370. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [PubMed]
- Bosi, E.; Monk, J.M.; Aziz, R.K.; Fondi, M.; Nizet, V.; Palsson, B. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Natl. Acad. Sci. USA 2016, 113, E3801–E3809. [Google Scholar] [CrossRef]
- Priya, G.; Lau, N.-S.; Furusawa, G.; Dinesh, B.; Foong, S.Y.; Amirul, A.-A.A. Metagenomic insights into the phylogenetic and functional profiles of soil microbiome from a managed mangrove in Malaysia. Agri Gene 2018, 9, 5–15. [Google Scholar] [CrossRef]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Labella, A.M.; Castro, M.D.; Manchado, M.; Lucena, T.; Arahal, D.R.; Borrego, J.J. Photobacterium malacitanum sp. nov., and Photobacterium andalusiense sp. nov., two new bacteria isolated from diseased farmed fish in Southern Spain. Syst. Appl. Microbiol. 2018, 41, 444–451. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Yang, X.; Sun, H.; Li, B.; Zhang, X.H. Photobacterium alginatilyticum sp. nov., a marine bacterium isolated from bottom seawater. Int. J. Syst. Evol. Microbiol. 2017, 67, 1912–1917. [Google Scholar] [CrossRef]
- Dobrindt, U.; Hochhut, B.; Hentschel, U.; Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2004, 2, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Gontcharov, A.A.; Marin, B.; Melkonian, M. Are combined analyses better than single gene phylogenies? A case study using SSU rDNA and rbcL sequence comparisons in the Zygnematophyceae (Streptophyta). Mol. Biol. Evol. 2004, 21, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Kita-Tsukamoto, K.; Thompson, F.L. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 2007, 189, 7932–7936. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Kong, D.; Wang, Y.; Ma, Q.; Han, X.; Zhou, Y.; Jiang, X.; Zhang, Y.; Ruan, Z.; Zhang, Q. Photobacterium salinisoli sp. nov., isolated from a sulfonylurea herbicide-degrading consortium enriched with saline soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 3910–3916. [Google Scholar] [CrossRef]
- Enciso-Ibarra, J.; González-Castillo, A.; Soto-Rodriguez, S.A.; Enciso-Ibarra, K.; Bolán-Mejia, C.; Gomez-Gil, B. Photobacterium lucens sp. nov., isolated from a cultured shrimp Penaeus Vannamei. Curr Microbiol 2020, 77, 1111–1116. [Google Scholar] [CrossRef]
- Gomez-Gil, B.; Roque, A.; Rotllant, G.; Peinado, L.; Romalde, J.L.; Doce, A.; Cabanillas-Beltrán, H.; Chimetto, L.A.; Thompson, F.L. Photobacterium swingsii sp. nov., isolated from marine organisms. Int. J. Syst. Evol. Microbiol. 2011, 61, 315–319. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef]
- Nelson, W.C.; Stegen, J.C. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 2015, 6, 713. [Google Scholar] [CrossRef]
- Collins, R.E.; Higgs, P.G. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol. Biol. Evol. 2012, 29, 3413–3425. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.L.; Christenson, J.K.; Wackett, L.P. Biosynthesis and chemical diversity of β-lactone natural products. Nat. Prod. Rep. 2019, 36, 458–475. [Google Scholar] [CrossRef] [PubMed]
- Takano, E. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 2006, 9, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Empadinhas, N.; da Costa, M.S. Osmoadaptation mechanisms in prokaryotes: Distribution of compatible solutes. Int. Microbiol. 2008, 11, 151–161. [Google Scholar] [PubMed]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Mathew, D.C.; Lo, S.C.; Mathew, G.M.; Chang, K.H.; Huang, C.C. Genomic sequence analysis of a plant-associated Photobacterium halotolerans MELD1: From marine to terrestrial environment? Stand. Genomic. Sci. 2016, 11, 56. [Google Scholar] [CrossRef]
- Lang, G.; Kalvelage, T.; Peters, A.; Wiese, J.; Imhoff, J.F. Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J. Nat. Prod. 2008, 71, 1074–1077. [Google Scholar] [CrossRef]
- Zhang, S.D.; Isbrandt, T.; Lindqvist, L.L.; Larsen, T.O.; Gram, L. Holomycin, an antibiotic secondary metabolite, is required for biofilm formation by the native producer Photobacterium galatheae S2753. Appl. Environ. Microbiol. 2021, 87, e00169-21. [Google Scholar] [CrossRef]
- Machado, H.; Sonnenschein, E.C.; Melchiorsen, J.; Gram, L. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genom. 2015, 16, 158. [Google Scholar] [CrossRef]
- Oliva, B.; O’Neill, A.; Wilson, J.M.; O’Hanlon, P.J.; Chopra, I. Antimicrobial properties and mode of action of the pyrrothine holomycin. Antimicrob. Agents Chemother. 2001, 45, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.; Tipper, D.J.; Davies, J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 1973, 3, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Baker, A.T.; Raab, A.; Huang, S.; Wang, T.; Yu, Y.; Jaspars, M.; Secombes, C.J.; Deng, H. The fish pathogen Yersinia ruckeri produces holomycin and uses an RNA methyltransferase for self-resistance. J. Biol. Chem. 2013, 288, 14688–14697. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, D.; Haines, A.S.; Song, Z.; Murphy, A.C.; Hothersall, J.; Stephens, E.R.; Gurney, R.; Cox, R.J.; Crosby, J.; Willis, C.L.; et al. A natural plasmid uniquely encodes two biosynthetic pathways creating a potent anti-MRSA antibiotic. PLoS ONE 2011, 6, e18031. [Google Scholar] [CrossRef]
- Li, B.; Walsh, C.T. Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc. Natl. Acad. Sci. USA 2010, 107, 19731–19735. [Google Scholar] [CrossRef]
- Yoshida, K.; Hashimoto, M.; Hori, R.; Adachi, T.; Okuyama, H.; Orikasa, Y.; Nagamine, T.; Shimizu, S.; Ueno, A.; Morita, N. Bacterial long-chain polyunsaturated fatty acids: Their biosynthetic genes, functions, and practical use. Mar. Drugs 2016, 14, 94. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Granato, E.T.; Meiller-Legrand, T.A.; Foster, K.R. The evolution and ecology of bacterial warfare. Curr. Biol. 2019, 29, R521–R537. [Google Scholar] [CrossRef]
- Long, R.A.; Azam, F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 2001, 67, 4975–4983. [Google Scholar] [CrossRef]
- Dong, X.; Song, J.; Chen, J.; Bi, D.; Wang, W.; Ren, Y.; Wang, H.; Wang, G.; Tang, K.F.J.; Wang, X.; et al. Conjugative transfer of the pVA1-type plasmid carrying the pirABvp genes results in the formation of new AHPND-causing Vibrio. Front. Cell. Infect. Microbiol. 2019, 9, 195. [Google Scholar] [CrossRef]
- Furusawa, G.; Lau, N.S.; Shu-Chien, A.C.; Jaya-Ram, A.; Amirul, A.A. Identification of polyunsaturated fatty acid and diterpenoid biosynthesis pathways from draft genome of Aureispira sp. CCB-QB1. Mar. Genom. 2015, 19, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, B.; Lau, N.S.; Furusawa, G.; Kim, S.W.; Taylor, T.D.; Foong, S.Y.; Shu-Chien, A.C. Comparative genome analyses of novel Mangrovimonas-like strains isolated from estuarine mangrove sediments reveal xylan and arabinan utilization genes. Mar. Genom. 2016, 25, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Deris, Z.M.; Iehata, S.; Ikhwanuddin, M.; Sahimi, M.B.M.K.; Dinh Do, T.; Sorgeloos, P.; Sung, Y.Y.; Wong, L.L. Immune and bacterial toxin genes expression in different giant tiger prawn, Penaeus monodon post-larvae stages following AHPND-causing strain of Vibrio parahaemolyticus challenge. Aquac. Rep. 2020, 16, 100248. [Google Scholar] [CrossRef]
- Sokolov, E.P. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J. Molluscan Stud. 2000, 66, 573–575. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Reichelt, J.L.; Baumann, P.; Baumann, L. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch. Microbiol. 1976, 110, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, L.Z.; Song, L.; Zhou, Y.G.; Qi, F.J.; Liu, Z.P. Photobacterium aquae sp. nov., isolated from a recirculating mariculture system. Int. J. Syst. Evol. Microbiol. 2014, 64, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Weerawongwiwat, V.; Yoon, S.; Kim, J.-H.; Yoon, J.-H.; Lee, J.S.; Sukhoom, A.; Kim, W. Photobacterium arenosum sp. nov., isolated from marine sediment sand. Int. J. Syst. Evol. Microbiol. 2021, 71, 005034. [Google Scholar] [CrossRef] [PubMed]
- Hilgarth, M.; Fuertes, S.; Ehrmann, M.; Vogel, R.F. Photobacterium carnosum sp. nov., isolated from spoiled modified atmosphere packaged poultry meat. Syst. Appl. Microbiol. 2018, 41, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Xue, C.X.; Li, B.; Zhou, S.; Liu, L.; Zhang, X.H. Photobacterium chitinilyticum sp. nov., a marine bacterium isolated from seawater at the bottom of the East China Sea. Int. J. Syst. Evol. Microbiol. 2019, 69, 1477–1483. [Google Scholar] [CrossRef]
- Lee, K.; Kim, H.K.; Sohn, H.; Cho, Y.; Choi, Y.M.; Jeong, D.G.; Kim, J.H. Genomic insights into Photobacterium damselae subsp. damselae strain KC-Na-1, isolated from the finless porpoise (Neophocaena asiaeorientalis). Mar. Genomics 2018, 37, 26–30. [Google Scholar] [CrossRef]
- Seo, H.J.; Bae, S.S.; Lee, J.H.; Kim, S.J. Photobacterium frigidiphilum sp. nov., a psychrophilic, lipolytic bacterium isolated from deep-sea sediments of Edison Seamount. Int. J. Syst. Evol. Microbiol. 2005, 55, 1661–1666. [Google Scholar] [CrossRef]
- Kim, Y.O.; Kim, K.K.; Park, S.; Kang, S.J.; Lee, J.H.; Lee, S.J.; Oh, T.K.; Yoon, J.H. Photobacterium gaetbulicola sp. nov., a lipolytic bacterium isolated from a tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2010, 60, 2587–2591. [Google Scholar] [CrossRef]
- Lascu, I.; Mereuță, I.; Chiciudean, I.; Hansen, H.; Avramescu, S.M.; Tănase, A.M.; Stoica, I. Complete genome sequence of Photobacterium ganghwense C2.2: A new polyhydroxyalkanoate production candidate. MicrobiologyOpen 2021, 10, e1182. [Google Scholar] [CrossRef]
- Urakawa, H.; Kita-Tsukamoto, K.; Ohwada, K. Reassessment of the taxonomic position of Vibrio iliopiscarius (Onarheim et al. 1994) and proposal for Photobacterium iliopiscarium comb. nov. Int. J. Syst. Bacteriol. 1999, 49, 257–260. [Google Scholar] [CrossRef][Green Version]
- Xie, C.H.; Yokota, A. Transfer of Hyphomicrobium indicum to the genus Photobacterium as Photobacterium indicum comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 2113–2116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chimetto, L.A.; Cleenwerck, I.; Thompson, C.C.; Brocchi, M.; Willems, A.; De Vos, P.; Thompson, F.L. Photobacterium jeanii sp. nov., isolated from corals and zoanthids. Int. J. Syst. Evol. Microbiol. 2010, 60, 2843–2848. [Google Scholar] [CrossRef] [PubMed]
- Ast, J.C.; Cleenwerck, I.; Engelbeen, K.; Urbanczyk, H.; Thompson, F.L.; De Vos, P.; Dunlap, P.V. Photobacterium kishitanii sp. nov., a luminous marine bacterium symbiotic with deep-sea fishes. Int. J. Syst. Evol. Microbiol. 2007, 57, 2073–2078. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk, H.; Urbanczyk, Y.; Hayashi, T.; Ogura, Y. Diversification of two lineages of symbiotic Photobacterium. PLoS ONE 2013, 8, e82917. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Lee, J.K.; Kim, Y.O.; Oh, T.K. Photobacterium lipolyticum sp. nov., a bacterium with lipolytic activity isolated from the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 2005, 55, 335–339. [Google Scholar] [CrossRef][Green Version]
- Jung, S.Y.; Jung, Y.T.; Oh, T.K.; Yoon, J.H. Photobacterium lutimaris sp. nov., isolated from a tidal flat sediment in Korea. Int. J. Syst. Evol. Microbiol. 2007, 57, 332–336. [Google Scholar] [CrossRef]
- Srinivas, T.N.; Vijaya Bhaskar, Y.; Bhumika, V.; Anil Kumar, P. Photobacterium marinum sp. nov., a marine bacterium isolated from a sediment sample from Palk Bay, India. Syst. Appl. Microbiol. 2013, 36, 160–165. [Google Scholar] [CrossRef]
- Figge, M.J.; Cleenwerck, I.; van Uijen, A.; De Vos, P.; Huys, G.; Robertson, L. Photobacterium piscicola sp. nov., isolated from marine fish and spoiled packed cod. Syst. Appl. Microbiol. 2014, 37, 329–335. [Google Scholar] [CrossRef]
- DeLong, E.F.; Franks, D.G.; Yayanos, A.A. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl. Environ. Microbiol. 1997, 63, 2105–2108. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, M.; Wang, F.; Wang, E.T.; Du, Z.; Wu, C.; Zhang, Z.; Liu, W.; Xie, Z. Photobacterium proteolyticum sp. nov., a protease-producing bacterium isolated from ocean sediments of Laizhou Bay. Int. J. Syst. Evol. Microbiol. 2017, 67, 1835–1840. [Google Scholar] [CrossRef]
- Thompson, F.L.; Thompson, C.C.; Naser, S.; Hoste, B.; Vandemeulebroecke, K.; Munn, C.; Bourne, D.; Swings, J. Photobacterium rosenbergii sp. nov. and Enterovibrio coralii sp. nov., vibrios associated with coral bleaching. Int. J. Syst. Evol. Microbiol. 2005, 55, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.P.B.; Duytschaever, G.; Chimetto Tonon, L.A.; Fróes, A.M.; de Oliveira, L.S.; Amado-Filho, G.M.; Francini-Filho, R.B.; De Vos, P.; Swings, J.; Thompson, C.C.; et al. Photobacterium sanctipauli sp. nov. isolated from bleached Madracis decactis (Scleractinia) in the St Peter & St Paul Archipelago, Mid-Atlantic Ridge, Brazil. PeerJ 2014, 2, e427. [Google Scholar] [PubMed]
- Gomez-Gil, B.; Roque, A.; Rotllant, G.; Romalde, J.L.; Doce, A.; Eggermont, M.; Defoirdt, T. Photobacterium sanguinicancri sp. nov. isolated from marine animals. Antonie Van Leeuwenhoek 2016, 109, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Labella, A.M.; Arahal, D.R.; Lucena, T.; Manchado, M.; Castro, D.; Borrego, J.J. Photobacterium toruni sp. nov., a bacterium isolated from diseased farmed fish. Int. J. Syst. Evol. Microbiol. 2017, 67, 4518–4525. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).