Integrated Analysis of Transcriptome and Metabolome Reveals Distinct Responses of Pelteobagrus fulvidraco against Aeromonas veronii Infection at Invaded and Recovering Stage
Abstract
1. Introduction
2. Results
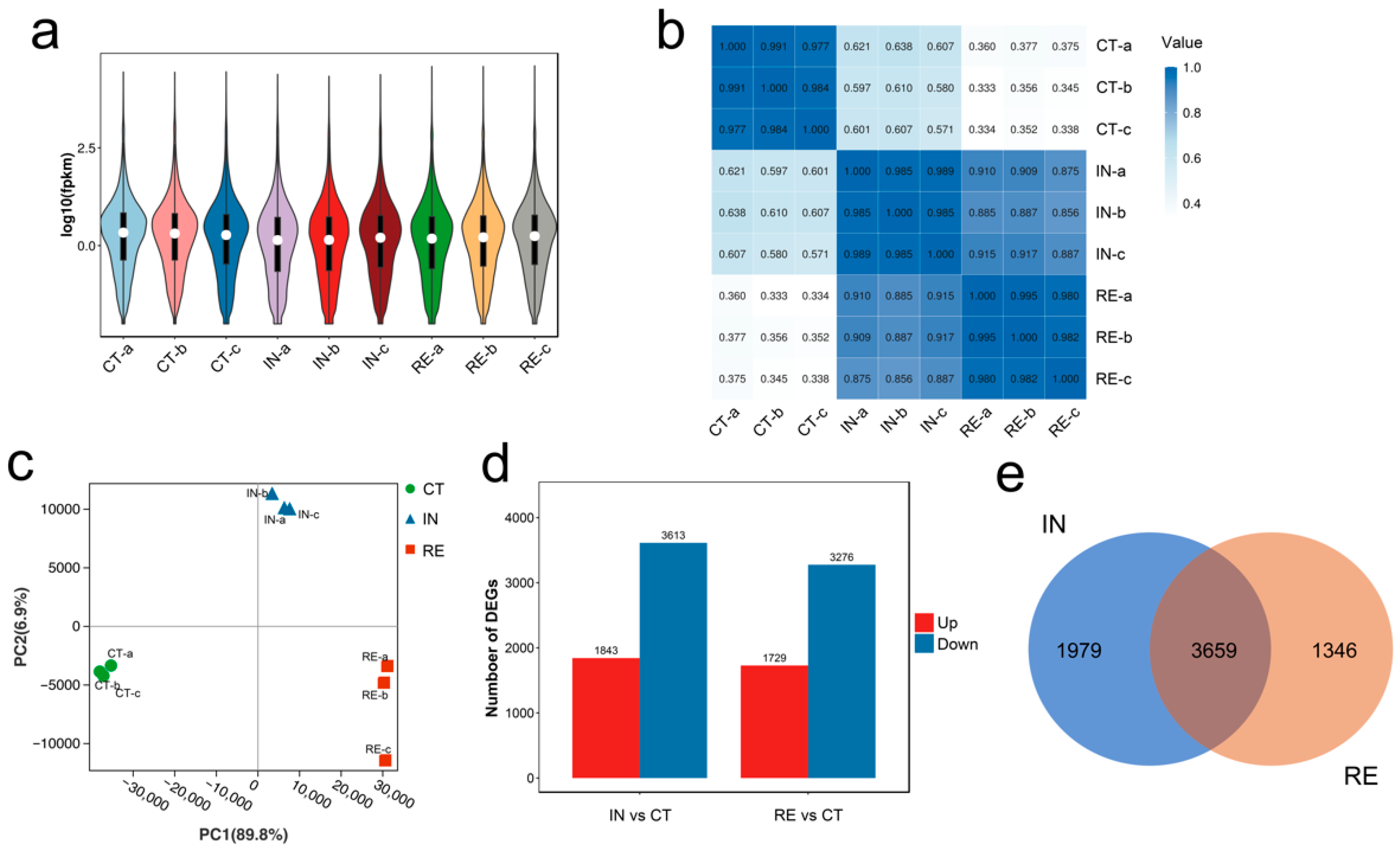
2.1. Transcriptome Data Processing
2.2. Identification of Differentially Expressed Genes (DEGs) at Invaded Stage and Recovering Stage
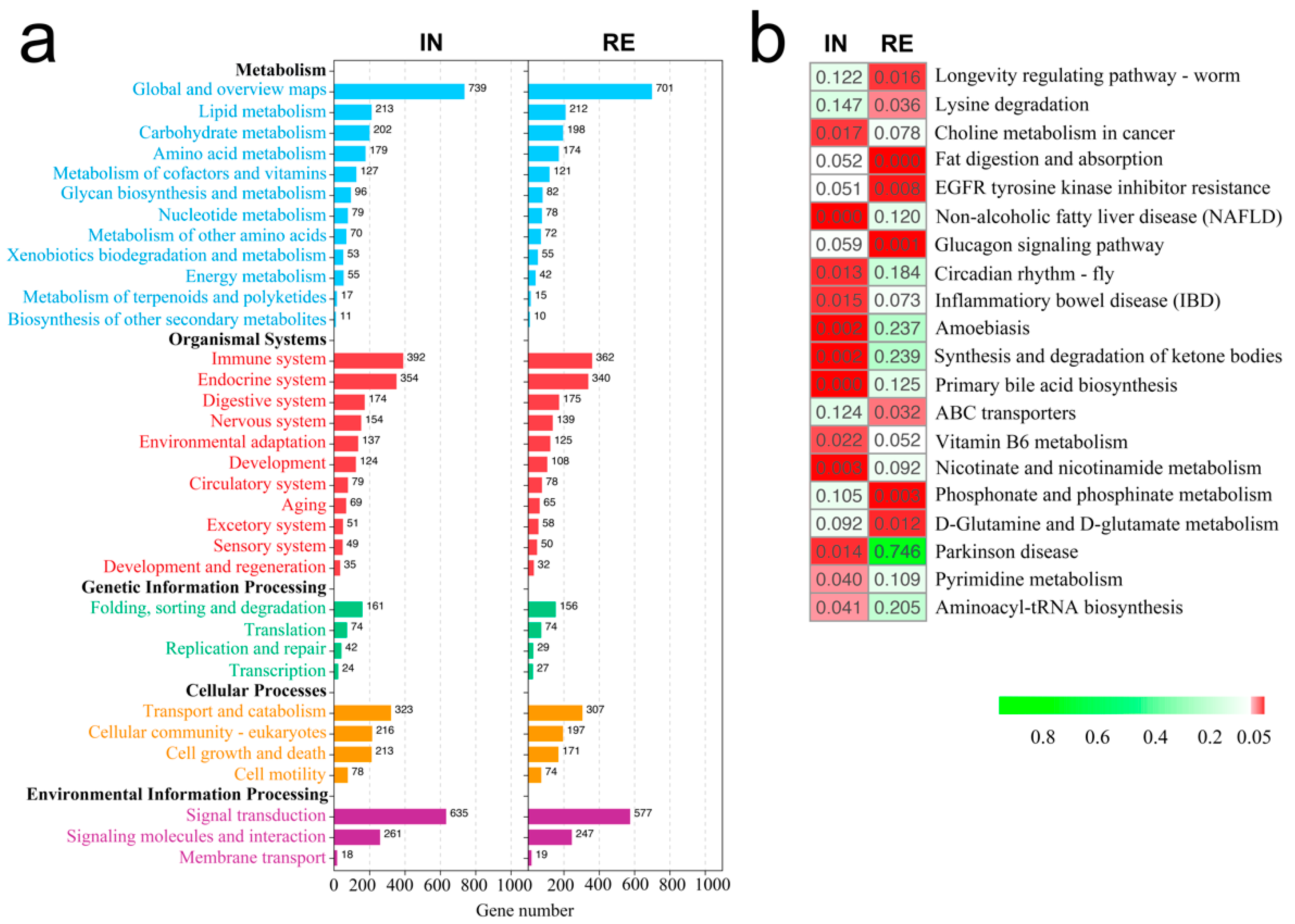
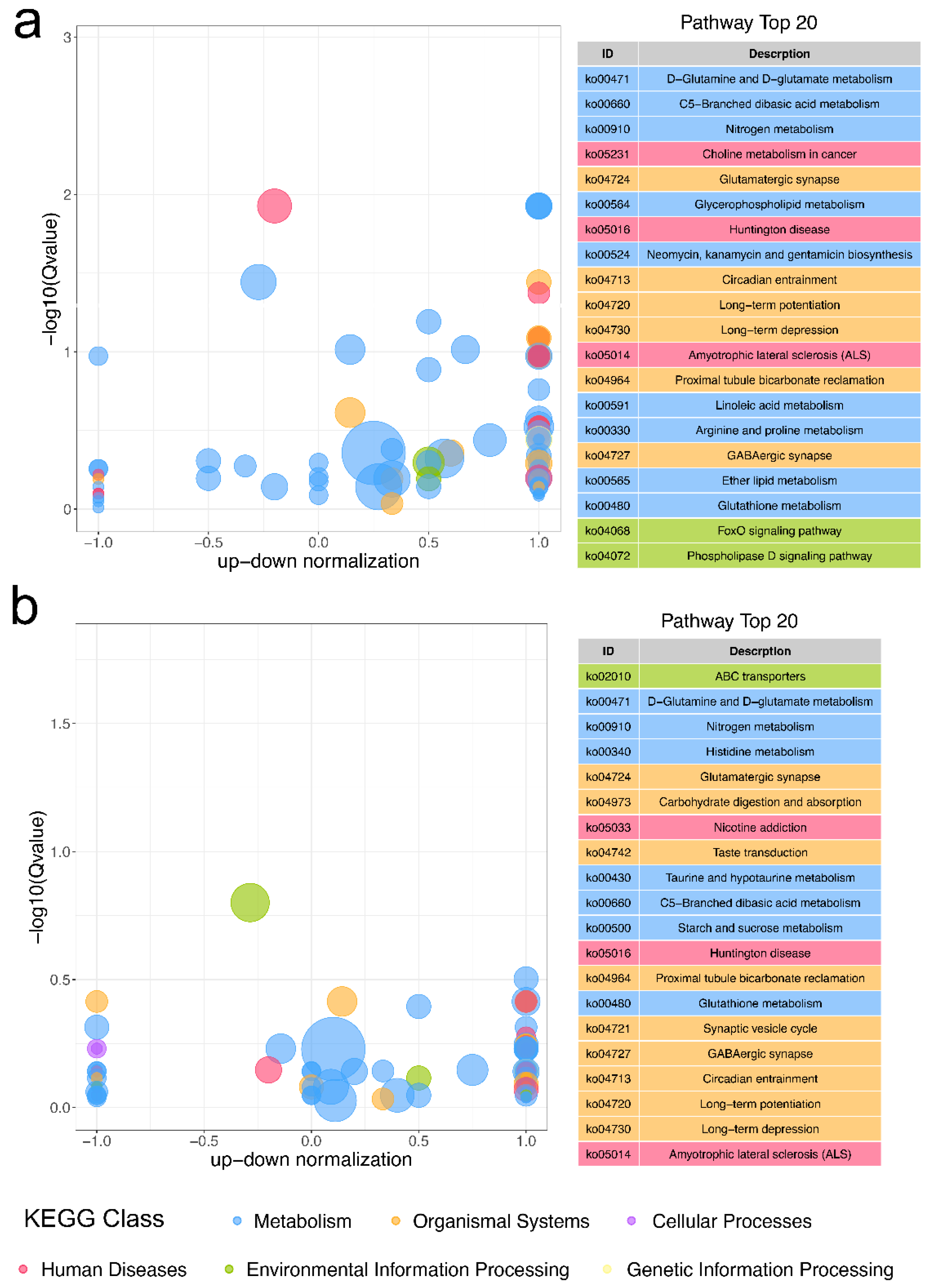
2.3. KEGG Functional Enrichment of DEGs
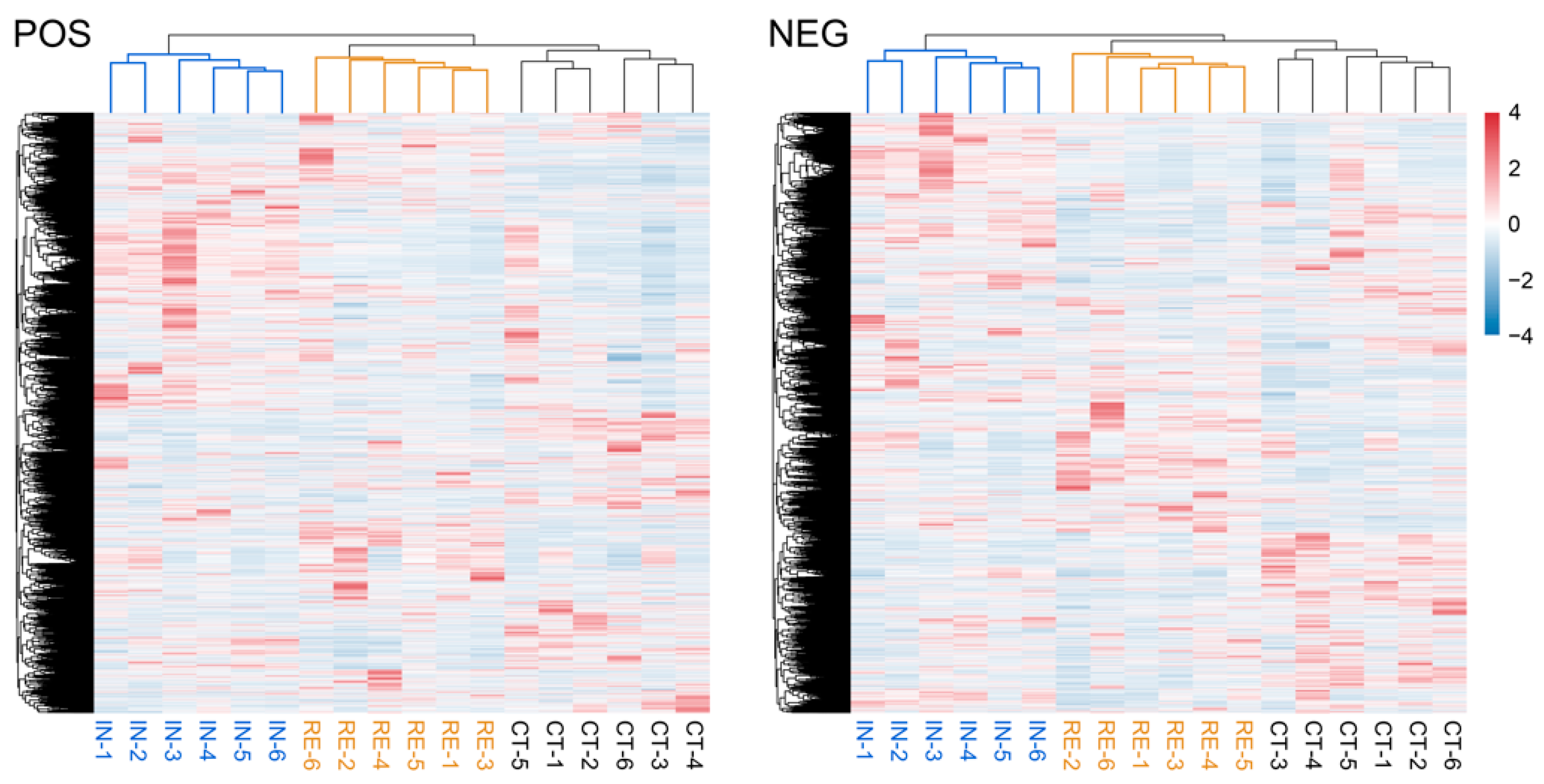
2.4. Characterization of Metabolomic Profiles
2.5. Identification of Differential Abundance Metabolites (DAMs) at Invaded Stage and Recovering Stage
2.6. Functional Enrichment of DAMs
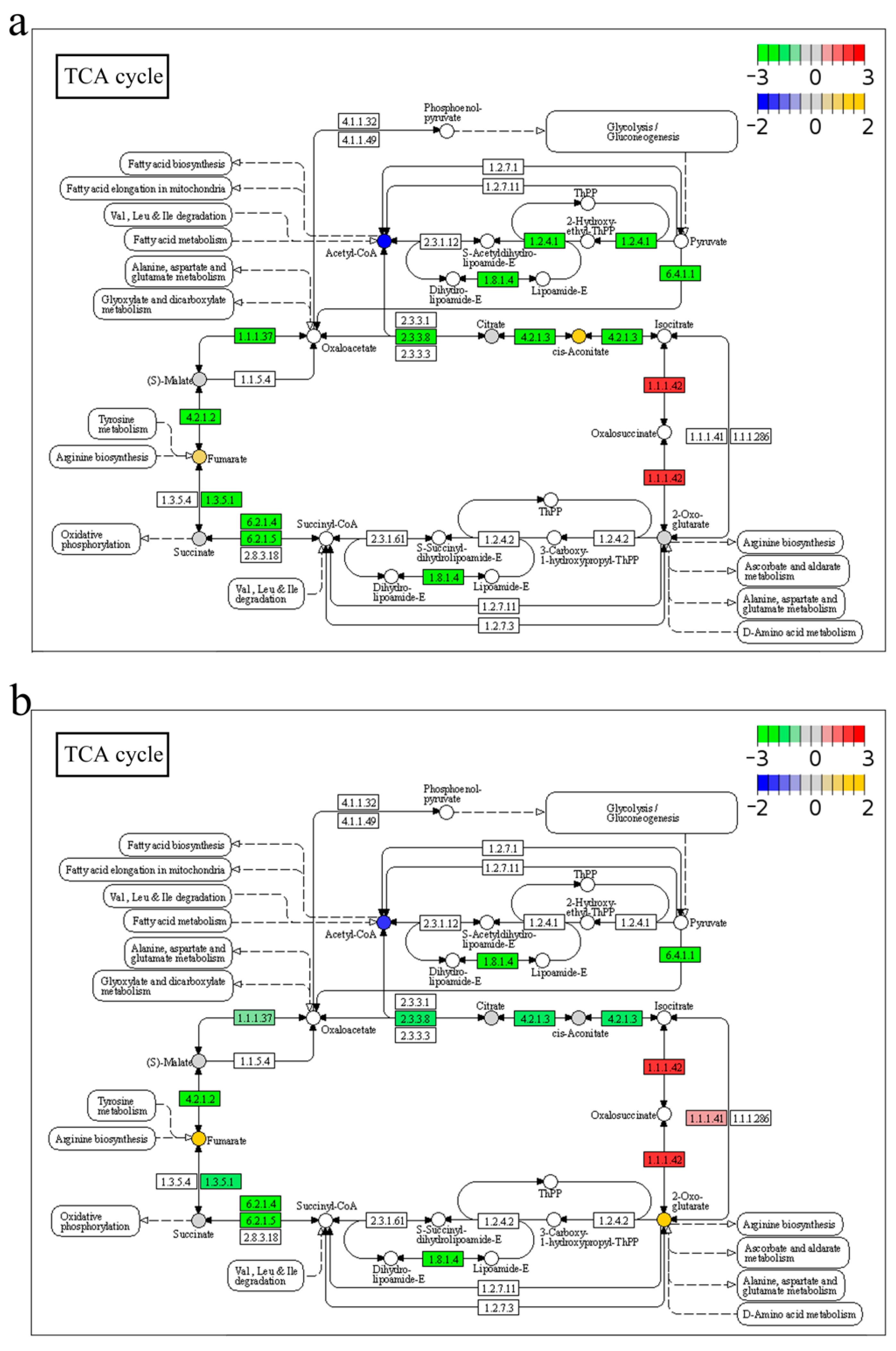
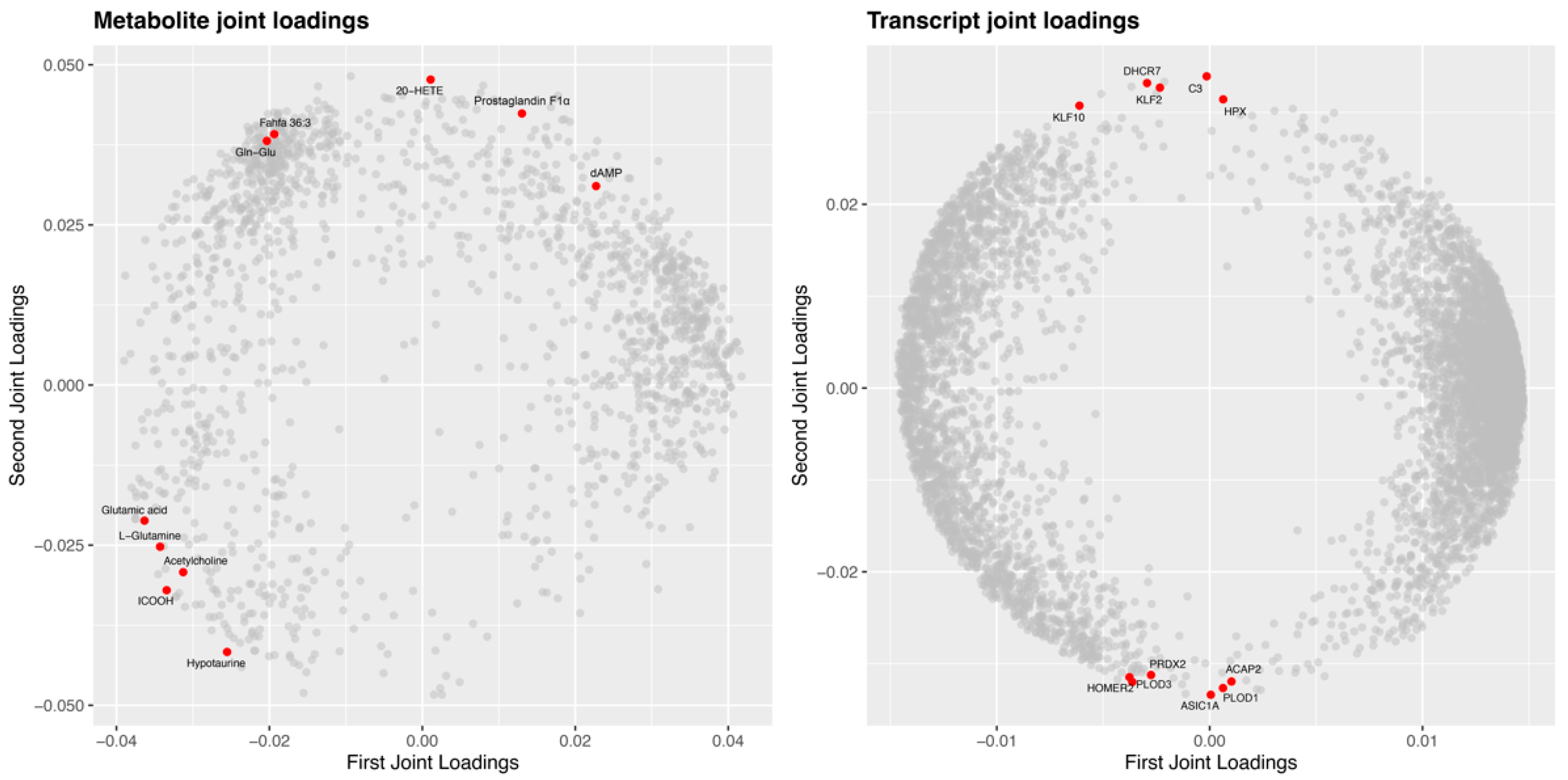
2.7. Integrated Analysis of Transcriptome and Metabolome in Response to A. veronii Infection
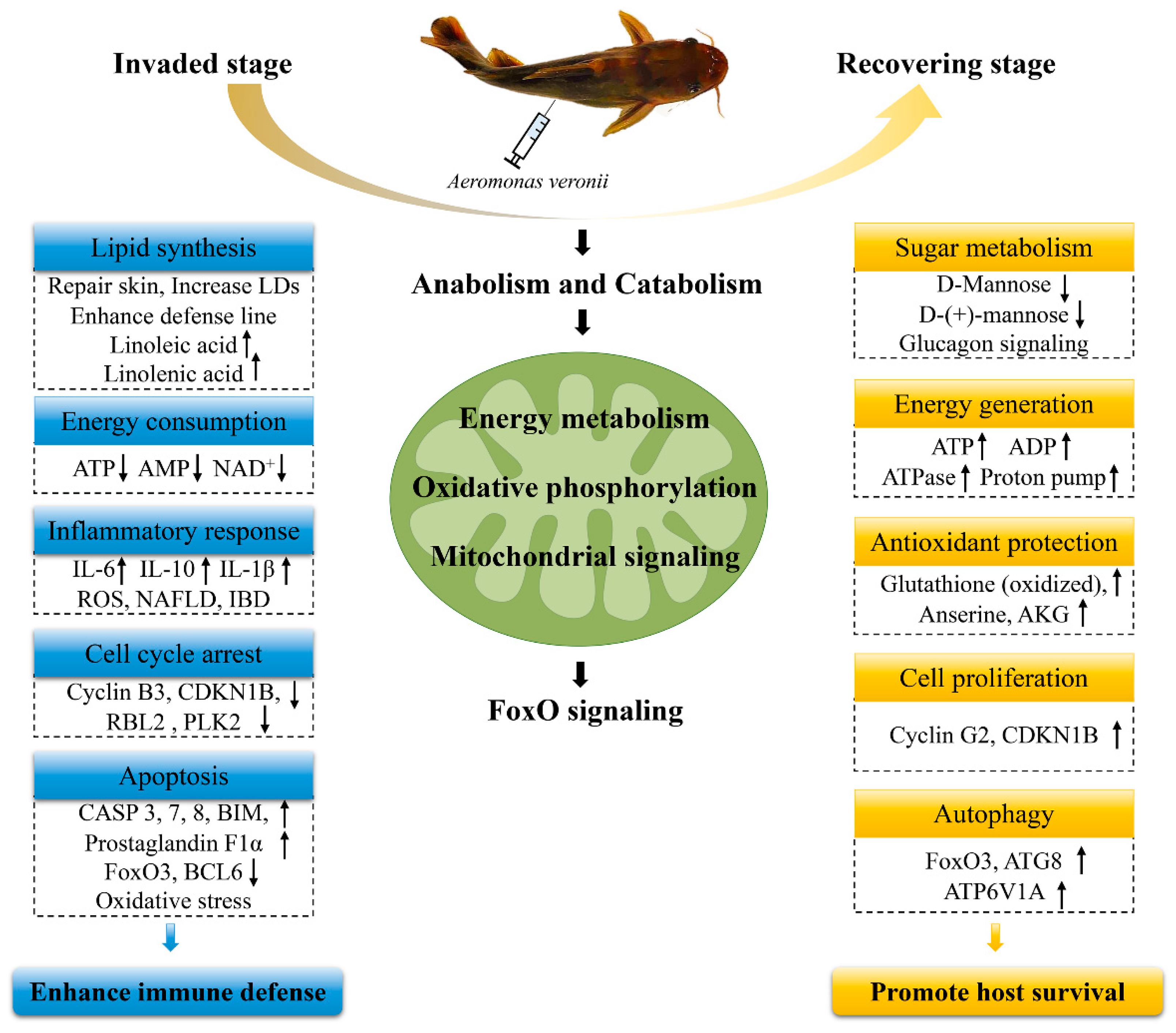
3. Discussion
3.1. Anabolism and Catabolism
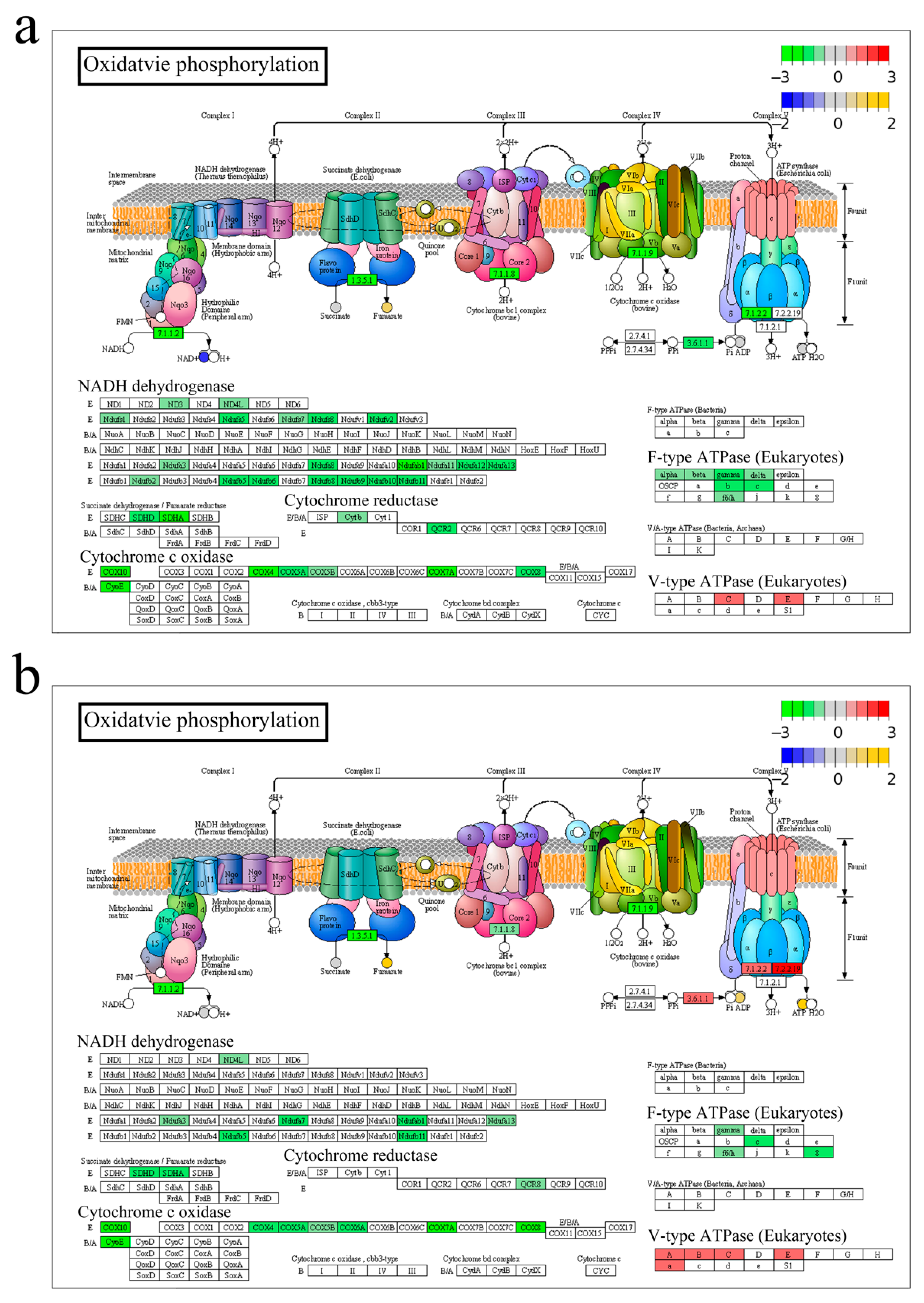
3.2. Energy Metabolism
3.3. FoxO Signaling Pathway
4. Materials and Methods
4.1. Sample Preparation
4.2. Transcriptome Library Construction and Data Processing
4.3. Differential Expression Analysis and Functional Enrichment Analysis
4.4. Metabolites Extraction and LC-MS/MS Analysis
4.5. Metabolome Data Processing and Multivariate Statistical Analysis
4.6. DAMs Identification and KEGG Enrichment Analysis
4.7. Integrated Analysis of Transcriptomic and Metabolomic Data
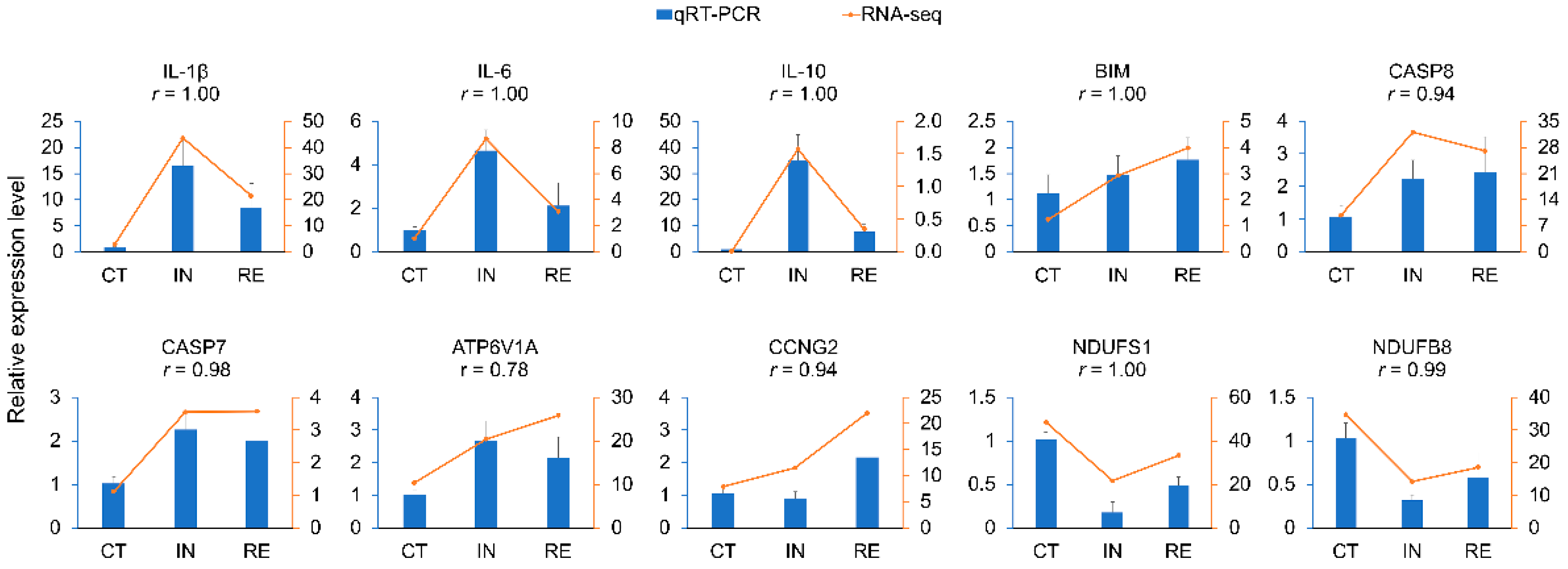
4.8. Experimental Validation of DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A


| Name | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| IL-1β | AGCCTGTGTGTTTGGGGATTGTG | CGCTCCATTCCATCGTTCTCCTTG |
| IL-6 | ACACTCCATTCTGCACAGCTTCAC | CCACTTGTCCACCTTCTTGTCCTC |
| IL-10 | TACTTGGAGACCGTGTTGCC | GCCTTTTCCTCCCATTCGAC |
| BIM | GCTCCTGATTGGTCGGCTGTTAC | CAGTCTGGTGCTACTTGTCTGCTG |
| CASP8 | TTCTGCCCGAGTCTCATCAATAAGC | TCTCCTCTCCTGCATTCTCTTCCAC |
| CASP7 | TTGCGAGACTCTGAGGAGGA | CCTTGTTCTTCTTGCCTCCA |
| ATP6V1A | CCAAGAGTTTACGGGTCGGG | TCCCTCGGTTTTTAGGTGGC |
| CCNG2 | CTGCTGGAGTGAGAGTGAGGA | AGGGAAATAAGGCCCATCTGC |
| NDUFS1 | TCATCGCGCCGGATATTGAA | AACGCAACATGACGCTTGAC |
| NDUFB8 | CCGTATCCTCGCACTGCTGAAG | GTAATCGCCATAGCCCTGTCCATC |
| β-actin | TTCGCTTGGAGATGATGCT | CGTGCTCAATGGGGTACT |
References
- Kim, L.O.; Lee, S.M. Effects of the dietary protein and lipid levels on growth and body composition of bagrid catfish, Pseudobagrus fulvidraco. Aquaculture 2005, 243, 323–329. [Google Scholar] [CrossRef]
- Zhang, J.J.; Pei, X.Y.; Wu, Z.W.; Li, J.; Wang, T.; Yin, S.W. A comparative study of immune response between hybrid yellow catfish "Huangyou-1" and its parental populations after challenge with Aeromonas hydrophila or Edwardsiella ictaluri. Aquacult Int. 2019, 27, 859–873. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Figueras, M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013, 36, 371–388. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Figueras, M.J. An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef]
- Liu, D.; Geng, Y.; Wang, K.; Chen, D.; Lai, W. Aeromonas veronii infection in cultured channel catfish, Ictalurus punctatus, in southwest china. Isr. J. Aquacult-Bamid 2016, 68, 1225. [Google Scholar]
- Chen, F.; Sun, J.; Han, Z.; Yang, X.; Xian, J.A.; Lv, A.; Hu, X.; Shi, H. Isolation, identification and characteristics of Aeromonas veronii from diseased crucian carp (Carassius auratus gibelio). Front. Microbiol. 2019, 10, 2742. [Google Scholar] [CrossRef]
- Wang, B.; Hu, J.; Feng, J.; Zhang, Y.; Sun, Y.; Jiang, B.; Li, W.; Liu, C.; Huang, Y.; Su, Y. Acute septicemia and immune response of spotted sea bass (Lateolabrax maculatus) to Aeromonas veronii infection. Fish. Shellfish Immunol. 2022, 124, 47–55. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Johnson, J.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Cheng, Z.X.; Ma, Y.M.; Li, H.; Peng, X.X. N-acetylglucosamine enhances survival ability of tilapias infected by Streptococcus iniae. Fish Shellfish Immunol. 2014, 40, 524–530. [Google Scholar] [CrossRef]
- Maha, I.F.; Xie, X.; Zhou, S.; Yu, Y.; Liu, X.; Zahid, A.; Lei, Y.; Ma, R.; Yin, F.; Qian, D. Skin metabolome reveals immune responses in yellow drum Nibea albiflora to Cryptocaryon irritans infection. Fish Shellfish Immunol. 2019, 94, 661–674. [Google Scholar] [CrossRef]
- Cheng, K.; Tang, Q.; Huang, Y.; Liu, X.; Wang, C. Effect of vitamin D3 on the immunomodulation of head kidney after Edwardsiella ictaluri challenge in yellow catfish (Pelteobagrus fulvidraco). Fish Shellfish Immunol. 2020, 99, 353–361. [Google Scholar] [CrossRef]
- Fu, M.; Kuang, R.; Wang, W.; Yu, Y.; Ai, T.; Liu, X.; Su, J.; Yuan, G. Hepcidin protects yellow catfish (Pelteobagrus fulvidraco) against Aeromonas veronii-induced ascites disease by regulating iron metabolism. Antibiotics 2021, 10, 848. [Google Scholar] [CrossRef]
- Chandel, N. Evolution of mitochondria as signaling organelles. BMC Biol. 2014, 12, 34. [Google Scholar] [CrossRef]
- He, L.D.; Zhu, D.L.; Liang, X.Y.; Li, Y.M.; Liao, L.J.; Yang, C.; Huang, R.; Zhu, Z.; Wang, Y. Multi-omics sequencing provides insights into age-dependent susceptibility of grass carp (Ctenopharyngodon idellus) to reovirus. Front. Immunol. 2021, 12, 694965. [Google Scholar] [CrossRef]
- Liu, K.; Yin, D.; Shu, Y.; Dai, P.; Wu, H. Transcriptome and metabolome analyses of Coilia nasus in response to Anisakidae parasite infection. Fish Shellfish Immunol. 2019, 87, 235–242. [Google Scholar] [CrossRef]
- Xie, X.; Kong, J.; Huang, J.; Zhou, L.; Jiang, Y.; Miao, R.; Yin, F. Integration of metabolomic and transcriptomic analyses to characterize the influence of the gill metabolism of Nibea albiflora on the response to Cryptocaryon irritans infection. Vet. Parasitol. 2021, 298, 109533. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Weng, M.Q.; Zhang, Q.Q.; Li, A.H.; Zhang, J.Y. Transcriptomics analysis of the infected tissue of gibel carp (Carassius auratus gibelio) with liver myxobolosis infers the underlying defense mechanisms from the perspective of immune-metabolic interactions. Aquaculture 2021, 542, 736867. [Google Scholar] [CrossRef]
- Rojo, D.R.; Prough, D.S.; Falduto, M.T.; Boone, D.R.; Micci, M.A.; Kahrig, K.M.; Crookshanks, J.M.; Jimenez, A.; Uchida, T.; Cowart, J.C.; et al. Influence of stochastic gene expression on the cell survival rheostat after traumatic brain injury. PLoS ONE 2011, 6, e23111. [Google Scholar] [CrossRef]
- Kendall, A.C.; Nicolaou, A. Bioactive lipid mediators in skin inflammation and immunity. Prog. Lipid Res. 2013, 52, 141–164. [Google Scholar] [CrossRef]
- Zhang, X.T.; Yu, Y.Y.; Xu, H.Y.; Huang, Z.Y.; Liu, X.; Cao, J.F.; Meng, K.-F.; Wu, Z.-B.; Han, G.-K.; Zhan, M.-T.; et al. Prevailing role of mucosal Igs and B cells in teleost skin immune responses to bacterial infection. J. Immunol. 2021, 206, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Sánchez-Lvarez, M.; Fajardo, A.; Kapetanovic, R.; Steiner, B.; Dutra, F.; Moreira, L.; López, J.A.; Campo, R.; Marí, M.; et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 2020, 370, eaay8085. [Google Scholar] [CrossRef]
- Roder, P.V.; Wu, B.; Liu, Y.; Han, W. Pancreatic regulation of glucose homeostasis. Exp. Mol. Med. 2016, 48, e219. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Huang, X.Y.; Yang, M.J.; Wang, S.; Ren, S.T.; Li, H.; Peng, X.-X. GC/MS-based metabolomics approach to identify biomarkers differentiating survivals from death in crucian carps infected by Edwardsiella tarda. Fish Shellfish Immunol. 2014, 39, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Villadsen, J. Oxidative phosphorylation revisited. Biotechnol. Bioeng. 2015, 112, 429–437. [Google Scholar] [CrossRef]
- Peng, B.; Su, Y.B.; Li, H.; Han, Y.; Guo, C.; Tian, Y.M.; Peng, X.-X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–261. [Google Scholar] [CrossRef]
- Forgac, M. Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 1999, 274, 12951–12954. [Google Scholar] [CrossRef]
- Chung, C.Y.; Shin, H.R.; Berdan, C.A.; Ford, B.; Ward, C.C.; Olzmann, J.A.; Zoncu, R.; Nomura, D.K. Covalent targeting of the vacuolar H(+)-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol. 2019, 15, 776–778. [Google Scholar] [CrossRef]
- Huang, T.; Huang, X.; Yao, M. miR-143 inhibits intracellular salmonella growth by targeting ATP6V1A in macrophage cells in pig. Res. Vet. Sci. 2018, 117, 138–143. [Google Scholar] [CrossRef]
- Silwal, P.; Kim, J.K.; Yuk, J.M.; Jo, E.K. AMP-activated protein kinase and host defense against infection. Int. J. Mol. Sci. 2018, 19, 3495. [Google Scholar] [CrossRef]
- Martinez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Peng, M.; Yin, N.; Chhangawala, S.; Xu, K.; Leslie, C.S.; Li, M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 2016, 354, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Zaslona, Z.; O’Neill, L.J. Cytokine-like roles for metabolites in immunity. Mol. Cell 2020, 78, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Angajala, A.; Lim, S.; Phillips, J.B.; Jim, J.H.; Yates, C.; You, Z.B.; Tan, M. Diverse roles of mitochondria in immune responses: Novel insights into immuno-metabolism. Front. Immunol. 2018, 9, 1605. [Google Scholar] [CrossRef] [PubMed]
- West, A.P.; Brodsky, I.E.; Rahner, C.; Woo, D.K.; Erdjument-Bromage, H.; Tempst, P.; Walsh, M.C.; Choi, Y.; Shadel, G.S.; Ghosh, S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 2011, 472, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Ralf, W.; Sabine, W.; Frank, T. Organ and tissue fibrosis: Molecular signals, cellular mechanisms and translational implications. Mol. Aspects Med. 2018, 65, 2–15. [Google Scholar]
- Souvannavong, V.; Lemaire, C.; Chaby, R. Lipopolysaccharide protects primary B lymphocytes from apoptosis by preventing mitochondrial dysfunction and bax translocation to mitochondria. Infect. Immun 2004, 72, 3260–3266. [Google Scholar] [CrossRef]
- Ma, C.; Wickham, M.E.; Guttman, J.A.; Deng, W.; Vallance, B.A. Citrobacter rodentium infection causes both mitochondrial dysfunction and intestinal epithelial barrier disruption in vivo: Role of mitochondrial associated protein (Map). Cell. Microbiol. 2010, 8, 1669–1686. [Google Scholar] [CrossRef]
- Pellegrino, M.W.; Haynes, C.M. Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection. BMC Biol. 2015, 13, 22. [Google Scholar] [CrossRef]
- Ran, K.Y.; Eun, L.S.; Kang, I.C.; Il-Chol, K.; Choy, H.E.; Haeng, R.J. A bacterial RTX toxin causes programmed necrotic cell death through calcium-mediated mitochondrial dysfunction. J. Infect. Dis. 2013, 207, 1406–1415. [Google Scholar]
- Lu, X.Y.; Xuan, W.T.; Li, J.J.; Yao, H.W.; Huang, C.; Li, J. AMPK protects against alcohol-induced liver injury through UQCRC2 to up-regulate mitophagy. Autophagy 2021, 17, 3622–3643. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; He, L.; Zhao, Y.; Zhang, L.; He, Y.; Li, J.; Li, C.; Zhang, B.; Huang, Q.; Xing, J.; et al. Akap1 deficiency exacerbates diabetic cardiomyopathy in mice by NDUFS1-mediated mitochondrial dysfunction and apoptosis. Diabetologia 2020, 63, 1072–1087. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Luo, J.; Tian, W.; Yan, W.; Ge, S.; Zhang, Y.; Sun, W. gamma-Tocotrienol inhibits oxidative phosphorylation and triggers apoptosis by inhibiting mitochondrial complex I subunit NDUFB8 and complex II subunit SDHB. Toxicology 2019, 417, 42–53. [Google Scholar] [CrossRef]
- Akagi, S.; Nakamura, K.; Matsubara, H.; Kusano, K.F.; Kataoka, N.; Oto, T.; Miyaji, K.; Miura, A.; Ogawa, A.; Yoshida, M.; et al. Prostaglandin I2 induces apoptosis via upregulation of Fas ligand in pulmonary artery smooth muscle cells from patients with idiopathic pulmonary arterial hypertension. Int. J. Cardiol. 2013, 165, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, G.; Lu, Z.; Geurts, A.M.; Usa, K.; Jacob, H.J.; Cowley, A.W.; Wang, N.; Liang, M. Antithrombin III/SerpinC1 insufficiency exacerbates renal ischemia/reperfusion injury. Kidney Int. 2015, 88, 796–803. [Google Scholar] [CrossRef]
- Häcker, G. Apoptosis in infection. Microbes Infect. 2017, 20, 552–559. [Google Scholar] [CrossRef]
- Ning, X.H.; Sun, L. Identification and characterization of immune-related lncRNAs and lncRNA-miRNA-mRNA networks of Paralichthys olivaceus involved in Vibrio anguillarum infection. BMC Genom. 2021, 22, 447. [Google Scholar] [CrossRef]
- Wu, R.; Sheng, X.; Tang, X.; Xing, J.; Zhan, W. Transcriptome analysis of flounder (Paralichthys olivaceus) gill in response to lymphocystis disease virus (LCDV) infection: Novel insights into fish defense mechanisms. Int. J. Mol. Sci. 2018, 19, 160. [Google Scholar] [CrossRef]
- Gong, Q.Y.; Yang, M.J.; Yang, L.F.; Chen, Z.G.; Jiang, M.; Peng, B. Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb. Biotechnol. 2020, 13, 796–812. [Google Scholar] [CrossRef]
- Liu, S.; He, L.; Yao, K. The antioxidative function of alpha-ketoglutarate and its applications. Biomed. Res. Int. 2018, 2018, 3408467. [Google Scholar] [CrossRef]
- Fraternale, A.; Paoletti, M.F.; Casabianca, A.; Nencioni, L.; Garaci, E.; Palamara, A.T.; Magnani, M. GSH and analogs in antiviral therapy. Mol. Aspects Med. 2009, 30, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Godos, J.; Castellano, S.; Micek, A.; Murabito, P.; Galvano, F.; Ferri, R.; Grosso, G.; Caraci, F. The therapeutic potential of carnosine/anserine supplementation against cognitive decline: A systematic review with meta-analysis. Biomedicines 2021, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hou, Y.; Yi, D.; Li, Y.; Ding, B.; Zhu, H.; Liu, J.; Xiao, H.; Wu, G. Dietary supplementation with glutamate precursor α-ketoglutarate attenuates lipopolysaccharide-induced liver injury in young pigs. Amino Acids 2015, 47, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Wang, H.T.; Gaur, U.; Little, P.J.; Xu, J.P.; Zheng, W.H. FOXO signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar] [CrossRef]
- Van der Vos, K.E.; Eliasson, P.; Proikas-Cezanne, T.; Vervoort, S.J.; van Boxtel, R.; Putker, M.; van Zutphen, I.J.; Mauthe, M.; Zellmer, S.; Pals, C.; et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat. Cell Biol. 2012, 14, 829–837. [Google Scholar] [CrossRef]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef]
- Ning, X.H.; Sun, L. Gene network analysis reveals a core set of genes involved in the immune response of Japanese flounder (Paralichthys olivaceus) against Vibrio anguillarum infection. Fish Shellfish Immunol. 2020, 98, 800–809. [Google Scholar] [CrossRef]
- Ning, X.H.; Sun, L. Micro-transcriptome analysis reveals immune-related microrna regulatory networks of Paralichthys olivaceus induced by Vibrio anguillarum infection. Int. J. Mol. Sci. 2020, 21, 4252. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, H.; Zhang, L.; Li, Q.; Huang, Z.; Zhao, Z.; Ke, H.; Xiao, Y.; Su, X.; Liu, Q.; et al. MiRNA-seq analysis of spleen and head kidney tissue from aquacultured largemouth bass (Micropterus salmoides) in response to Aeromonas hydrophila infection. Funct. Integr. Genom. 2021, 21, 101–111. [Google Scholar] [CrossRef]
- Fei, Y.Y.; Feng, Z.Z.; Wu, K.X.; Luo, Y.; Yu, L.; Zhang, Y.; Lu, L.; Xu, D.; Lv, L. MicroRNA expression profiling of caudal fin cell of C. auratus gibelio upon cyprinid herpesvirus 2 infection. Dev. Comp. Immunol. 2020, 107, 103637. [Google Scholar] [CrossRef]
- Li, X.; Zheng, S.; Wu, G. Nutrition and metabolism of glutamate and glutamine in fish. Amino Acids 2020, 52, 671–691. [Google Scholar] [CrossRef] [PubMed]
- Graciano, T.S.; Michelato, M.; Neu, D.H.; Vitor, L.; Vidal, O.; Xavier, T.O.; de Moura, L.B.; Furuya, W.M. Performance and body composition of Nile tilapia fed diets supplemented with AminoGut during sex reversal period. Semin. Cienc. Agrar. 2014, 35, 2779–2789. [Google Scholar] [CrossRef]
- Pereira, R.T.; Rosa, P.V.; Gatlin, D.M. Glutamine and arginine in diets for Nile tilapia: Effects on growth, innate immune responses, plasma amino acid profiles and whole-body composition. Aquaculture 2017, 473, 135–144. [Google Scholar] [CrossRef]
- Liu, X.; Cai, X.; Zhang, D.; Xu, C.; Xiao, W. Zebrafish foxo3b negatively regulates antiviral response through suppressing the transactivity of irf3 and irf7. J. Immunol. 2016, 197, 4736–4749. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Liu, J.; Huang, D.; Guo, Y.; Luo, K.; Yang, M.; Gao, W.; Xu, Q.; Zhang, W.; Mai, K. FoxO3 modulates LPS-activated hepatic inflammation in turbot (Scophthalmus maximus L.). Front. Immunol. 2021, 12, 679704. [Google Scholar] [CrossRef] [PubMed]
- English, L.; Chemali, M.; Duron, J.; Rondeau, C.; Laplante, A.; Gingras, D.; Alexander, D.; Leib, D.; Norbury, C.; Lippe, R.; et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009, 10, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Lv, M.; Qiu, X.; Wang, X.; Zhang, A.; Yang, K.; Zhou, H. IFN-gamma manipulates NOD1-mediated interaction of autophagy and Edwardsiella piscicida to augment intracellular clearance in Fish. J. Immunol. 2021, 207, 1087–1098. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Novak, B.; Vinod, P.K.; Freire, P.; Kapuy, O. Systems-level feedback in cell-cycle control. Biochem. Soc. Trans. 2010, 38, 1242–1246. [Google Scholar] [CrossRef]
- Whitcomb, E.A.; Tsai, Y.C.; Basappa, J.; Liu, K.; Le Feuvre, A.K.; Weissman, A.M.; Taylor, A. Stabilization of p27(Kip1)/CDKN1B by UBCH7/UBE2L3 catalyzed ubiquitinylation: A new paradigm in cell-cycle control. FASEB J. 2019, 33, 1235–1247. [Google Scholar] [CrossRef]
- Enrico, T.P.; Stallaert, W.; Wick, E.T.; Ngoi, P.; Emanuele, M.J. Cyclin F drives proliferation through SCF-dependent degradation of the retinoblastoma-like tumor suppressor p130/RBL2. Elife 2021, 10, e70691. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Jo, M.; Curry Jr, T.E.; Liu, J. Hormonal induction of polo-like kinases (Plks) and impact of Plk2 on cell cycle progression in the rat ovary. PLoS ONE 2012, 7, e41844. [Google Scholar]
- Zhao, Y.N.; Guo, X.; Ma, Z.G.; Gu, L.; Ge, J.; Li, Q. Pro-apoptotic protein BIM in apoptosis of glucocorticoid-sensitive and -resistant acute lymphoblastic leukemia CEM cells. Med. Oncol. 2011, 28, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Zhang, R.; Li, C.; Jiang, L.; Xiang, M.; Ye, Z.; Kita, H.; Melnick, A.M.; Dent, A.L.; Sun, J. BCL6 modulates tissue neutrophil survival and exacerbates pulmonary inflammation following influenza virus infection. Proc. Natl. Acad. Sci. USA 2019, 116, 11888–11893. [Google Scholar] [CrossRef]
- Wesch, N.; Kirkin, V.; Rogov, V.V. Atg8-family proteins-structural features and molecular interactions in autophagy and beyond. Cells 2020, 9, 2008. [Google Scholar] [CrossRef]
- Zhang, G.; Li, J.; Zhang, J.; Liang, X.; Yin, S. Integrated analysis of transcriptomic, miRNA and proteomic changes of a novel hybrid yellow catfish uncovers key roles for miRNAs in heterosis. Mol. Cell Proteom. 2019, 18, 1437–1453. [Google Scholar] [CrossRef]
- Zhang, P.; Yu, C.; Sun, L. Japanese flounder (Paralichthys olivaceus) Bmal1 is involved in the regulation of inflammatory response and bacterial infection. Aquaculture 2020, 525, 735330. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Bouhaddani, S.E.; Houwing-Duistermaat, J.; Salo, P.; Perola, M.; Jongbloed, G.; Uh, H.W. Evaluation of O2PLS in Omics data integration. BMC Bioinform. 2016, 17 (Suppl. S2), 11. [Google Scholar] [CrossRef]
- Li, Q.; Guo, S.; Jiang, X.; Bryk, J.; Naumann, R.; Enard, W.; Tomita, M.; Sugimoto, M.; Khaitovich, P.; Pääbo, S. Mice carrying a human GLUD2 gene recapitulate aspects of human transcriptome and metabolome development. Proc. Natl. Acad. Sci. USA 2016, 113, 5358–5363. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G., Jr.; Brouwer, C. Pathview web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed]
- Pavlidis, P. Using ANOVA for gene selection from microarray studies of the nervous system. Methods 2003, 31, 282–289. [Google Scholar] [CrossRef]


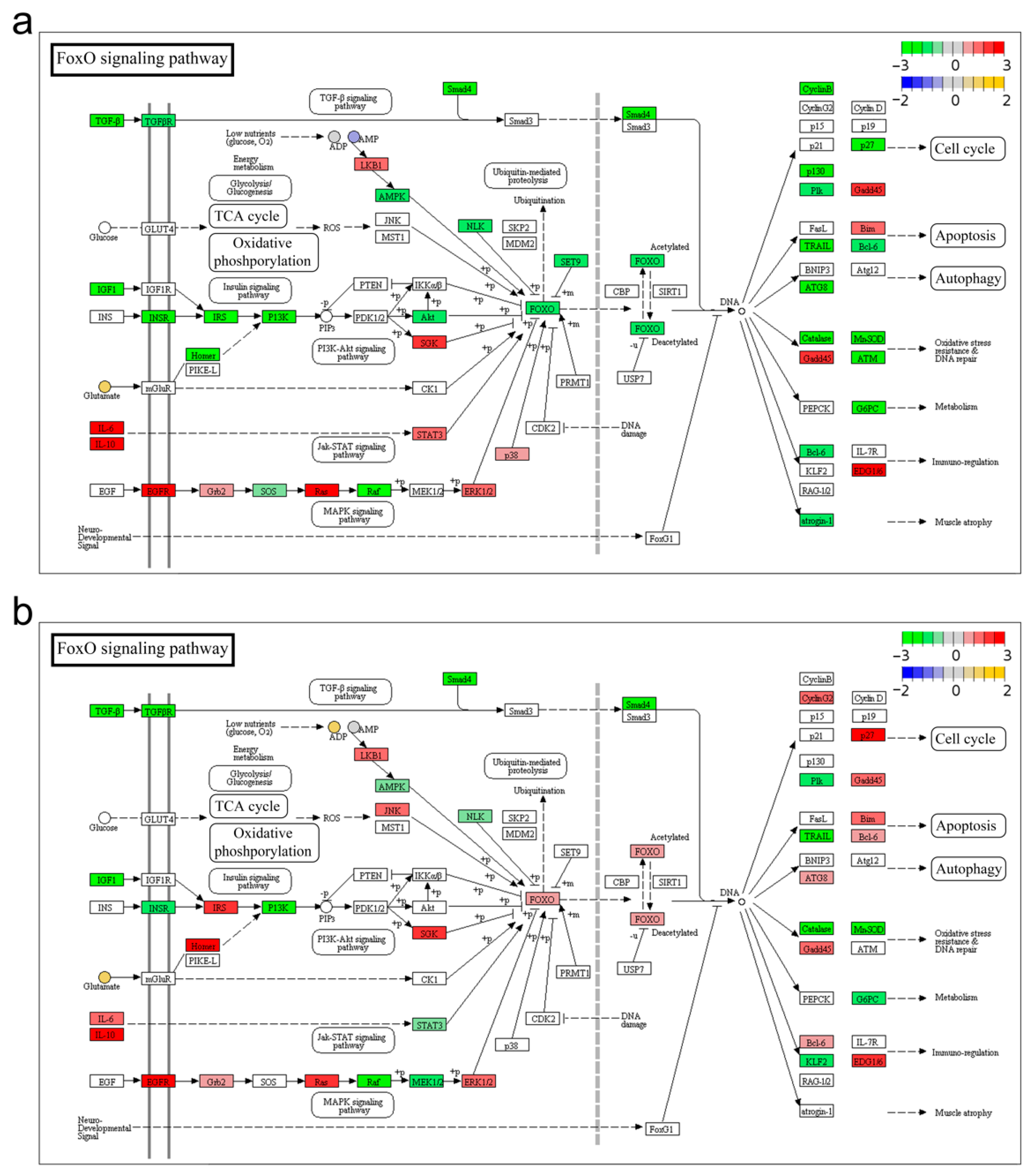
| Sample | Raw Reads | HQ Reads | HQ Ratio (%) | RHQ Reads | Mapped Reads | Mapping Ratio (%) |
|---|---|---|---|---|---|---|
| CT-a | 78,771,364 | 78,499,694 | 99.66 | 45,734,330 | 43,105,538 | 94.25 |
| CT-b | 76,322,050 | 76,044,322 | 99.64 | 40,809,162 | 38,653,690 | 94.72 |
| CT-c | 79,193,264 | 78,939,460 | 99.68 | 57,590,950 | 54,559,150 | 94.74 |
| IN-a | 89,965,810 | 89,748,254 | 99.76 | 66,765,870 | 62,919,273 | 94.24 |
| IN-b | 99,306,580 | 99,037,870 | 99.73 | 73,854,352 | 69,466,476 | 94.06 |
| IN-c | 91,103,442 | 90,859,558 | 99.73 | 52,051,328 | 48,980,490 | 94.10 |
| RE-a | 80,482,820 | 80,287,406 | 99.76 | 51,564,106 | 49,152,222 | 95.32 |
| RE-b | 85,969,140 | 85,755,306 | 99.75 | 52,182,432 | 49,328,966 | 94.53 |
| RE-c | 76,547,964 | 76,350,666 | 99.74 | 47,252,734 | 44,118,118 | 93.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, X.; Peng, Y.; Tang, P.; Zhang, Y.; Wang, L.; Zhang, W.; Zhang, K.; Ji, J.; Yin, S. Integrated Analysis of Transcriptome and Metabolome Reveals Distinct Responses of Pelteobagrus fulvidraco against Aeromonas veronii Infection at Invaded and Recovering Stage. Int. J. Mol. Sci. 2022, 23, 10121. https://doi.org/10.3390/ijms231710121
Ning X, Peng Y, Tang P, Zhang Y, Wang L, Zhang W, Zhang K, Ji J, Yin S. Integrated Analysis of Transcriptome and Metabolome Reveals Distinct Responses of Pelteobagrus fulvidraco against Aeromonas veronii Infection at Invaded and Recovering Stage. International Journal of Molecular Sciences. 2022; 23(17):10121. https://doi.org/10.3390/ijms231710121
Chicago/Turabian StyleNing, Xianhui, Ye Peng, Peng Tang, Yiran Zhang, Lingling Wang, Wenwen Zhang, Kai Zhang, Jie Ji, and Shaowu Yin. 2022. "Integrated Analysis of Transcriptome and Metabolome Reveals Distinct Responses of Pelteobagrus fulvidraco against Aeromonas veronii Infection at Invaded and Recovering Stage" International Journal of Molecular Sciences 23, no. 17: 10121. https://doi.org/10.3390/ijms231710121
APA StyleNing, X., Peng, Y., Tang, P., Zhang, Y., Wang, L., Zhang, W., Zhang, K., Ji, J., & Yin, S. (2022). Integrated Analysis of Transcriptome and Metabolome Reveals Distinct Responses of Pelteobagrus fulvidraco against Aeromonas veronii Infection at Invaded and Recovering Stage. International Journal of Molecular Sciences, 23(17), 10121. https://doi.org/10.3390/ijms231710121





