The Pleiotropy of PAX5 Gene Products and Function
Abstract
1. Introduction
2. The PAX5 Transcription Factor and B-Cell Development
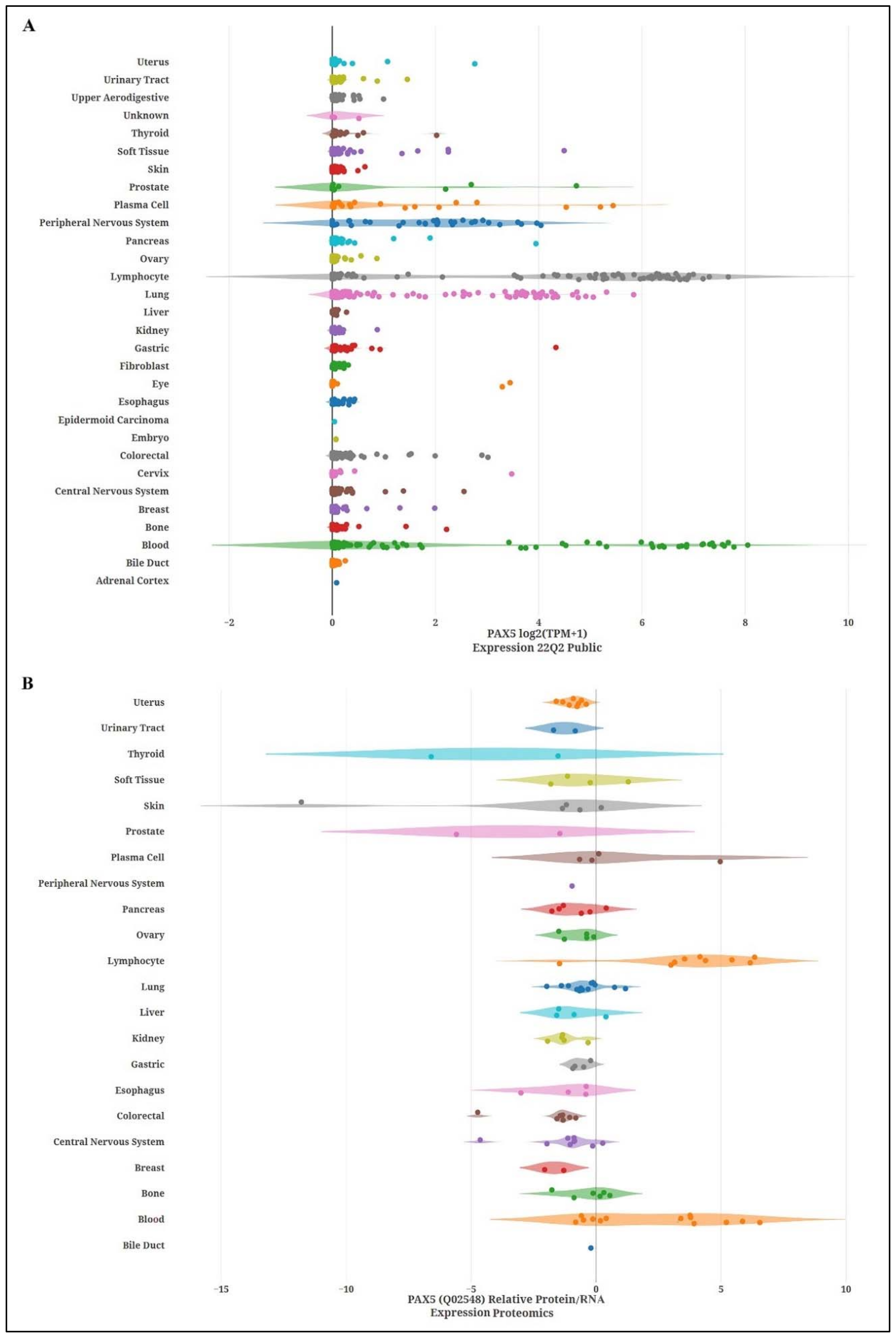
2.1. Expression and Tissue Specificity
2.2. Role of PAX5 in B-Cell Lineage Commitment and Maturation
3. PAX5 and Cancer
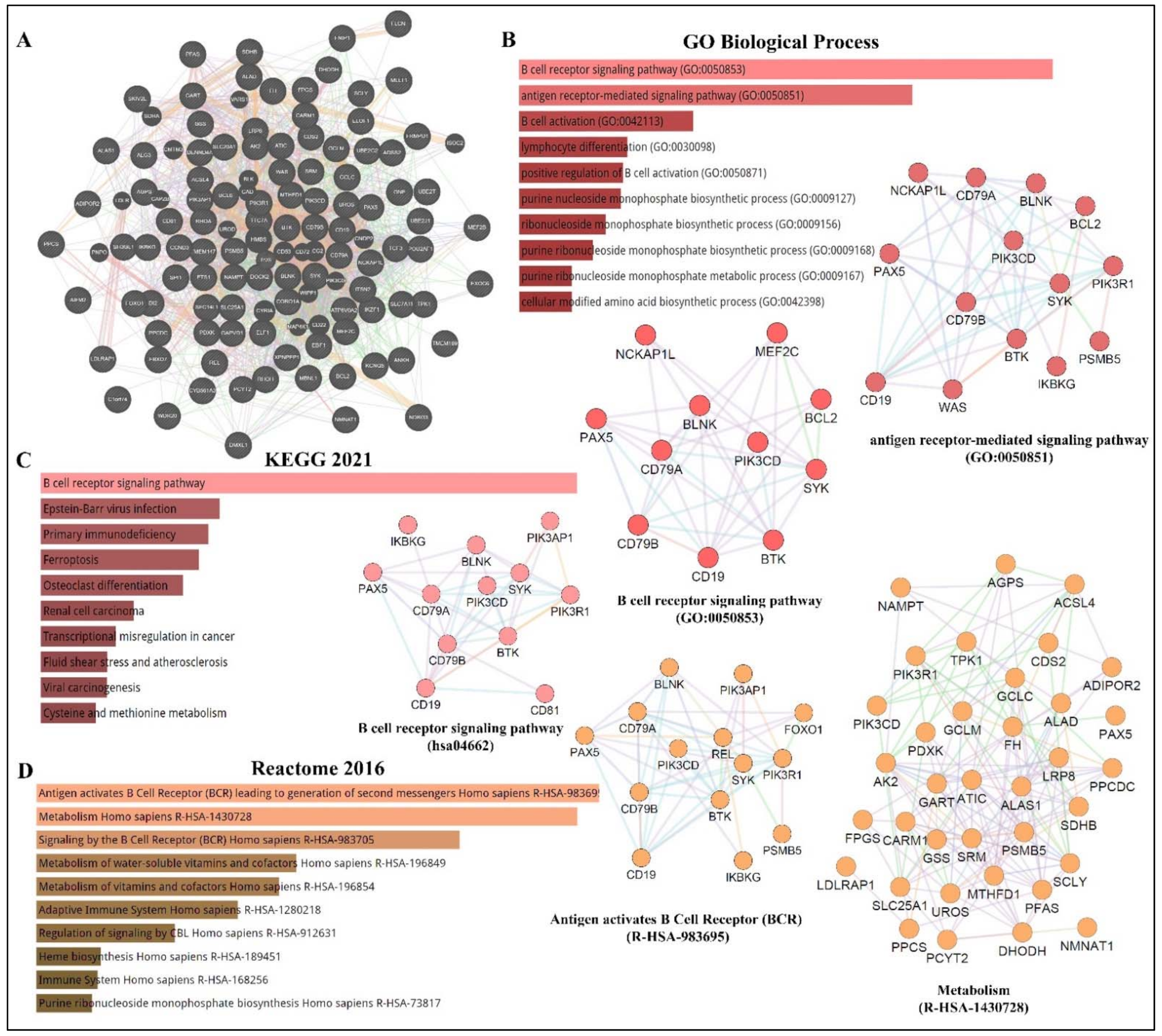
3.1. PAX5 Is a Key Driver of B-Cell Malignancies
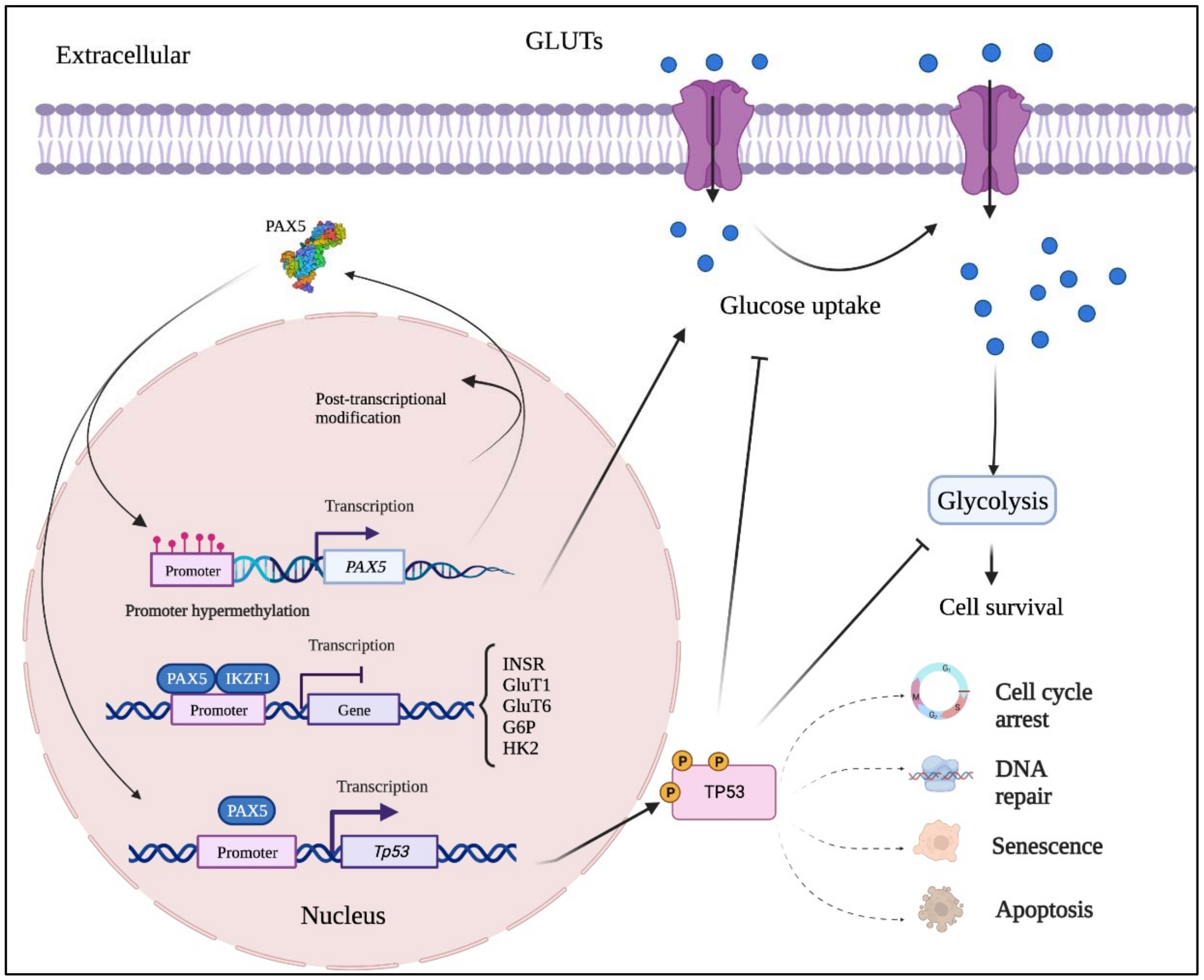
3.2. PAX5 in Non-Hematopoietic Cancers
3.3. PAX5 and Breast Cancer
4. PAX5 Expression and Regulation
4.1. PAX5 Epigenetic Regulation
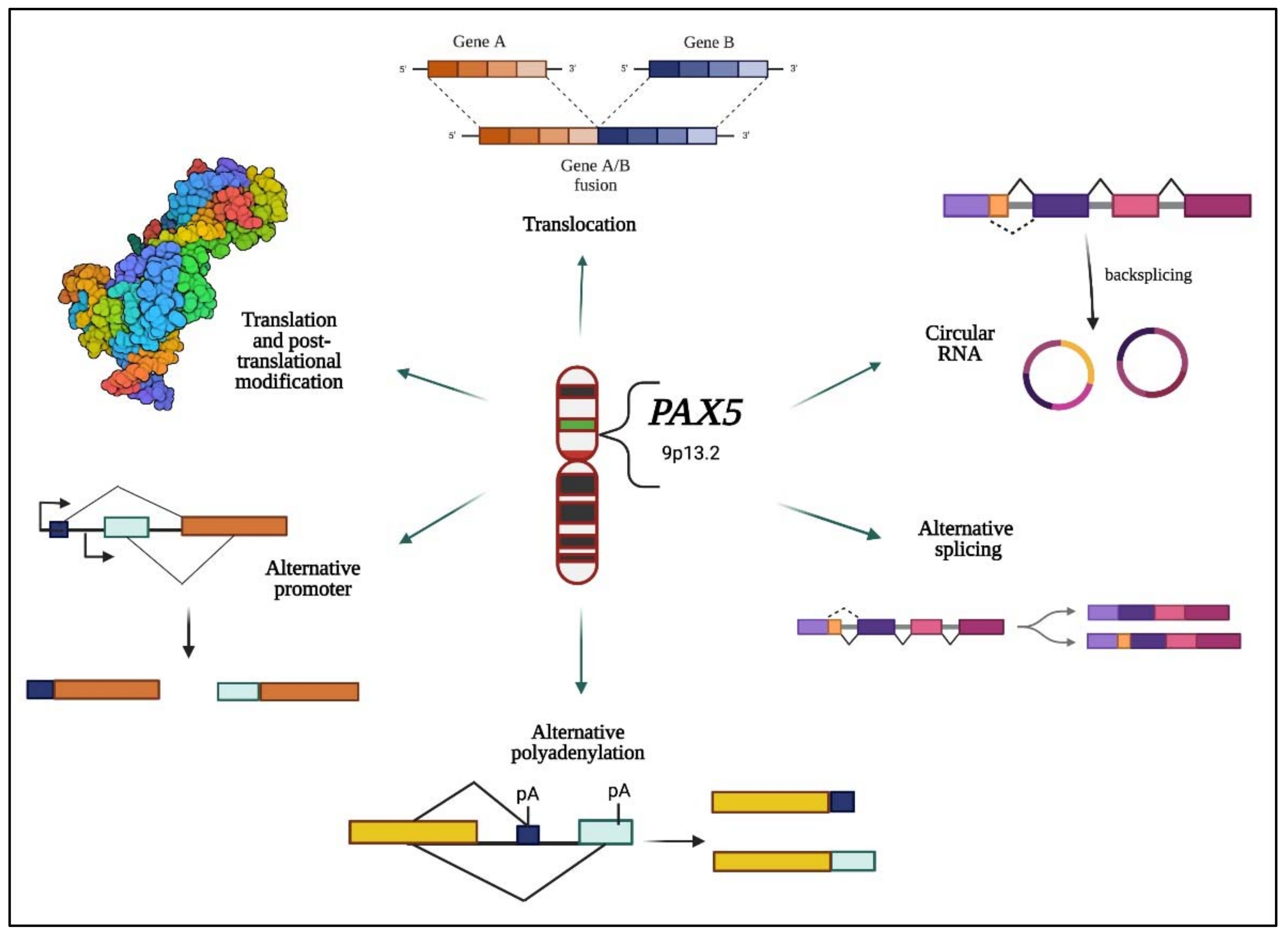
4.2. PAX5 Post-Transcriptional Regulation
4.3. Post-Translational Regulation of PAX5
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, B.; Davidson, E.A.; Liu, W.; Nebert, D.W.; Bruford, E.A.; Zhao, H.; Dermitzakis, E.T.; Thompson, D.C.; Vasiliou, V. Overview of pax gene family: Analysis of human tissue-specific variant expression and involvement in human disease. Hum. Genet. 2021, 140, 381–400. [Google Scholar] [CrossRef]
- Wang, Q.; Fang, W.H.; Krupinski, J.; Kumar, S.; Slevin, M.; Kumar, P. Pax genes in embryogenesis and oncogenesis. J. Cell Mol. Med. 2008, 12, 2281–2294. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.A. Pax proteins and fables of their reconstruction. Crit. Rev. Eukaryot. Gene Expr. 2012, 22, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Robson, E.J.; He, S.J.; Eccles, M.R. A panorama of pax genes in cancer and development. Nat. Rev. Cancer. 2006, 6, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.A. Genetic and biochemical diversity in the pax gene family. Biochem. Cell Biol. 2000, 78, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Dorfler, P.; Aguzzi, A.; Kozmik, Z.; Urbanek, P.; Maurer-Fogy, I.; Busslinger, M. Pax-5 encodes the transcription factor bsap and is expressed in b lymphocytes, the developing cns, and adult testis. Genes Dev. 1992, 6, 1589–1607. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Goorha, S.; Radtke, I.; Miller, C.B.; Coustan-Smith, E.; Dalton, J.D.; Girtman, K.; Mathew, S.; Ma, J.; Pounds, S.B.; et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007, 446, 758–764. [Google Scholar] [CrossRef]
- Bousquet, M.; Broccardo, C.; Quelen, C.; Meggetto, F.; Kuhlein, E.; Delsol, G.; Dastugue, N.; Brousset, P. A novel pax5-eln fusion protein identified in b-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type pax5. Blood 2007, 109, 3417–3423. [Google Scholar] [CrossRef]
- Kurahashi, S.; Hayakawa, F.; Miyata, Y.; Yasuda, T.; Minami, Y.; Tsuzuki, S.; Abe, A.; Naoe, T. Pax5-pml acts as a dual dominant-negative form of both pax5 and pml. Oncogene 2011, 30, 1822–1830. [Google Scholar] [CrossRef]
- Kawamata, N.; Pennella, M.A.; Woo, J.L.; Berk, A.J.; Koeffler, H.P. Dominant-negative mechanism of leukemogenic pax5 fusions. Oncogene 2012, 31, 966–977. [Google Scholar] [CrossRef]
- Busslinger, M.; Klix, N.; Pfeffer, P.; Graninger, P.G.; Kozmik, Z. Deregulation of pax-5 by translocation of the emu enhancer of the igh locus adjacent to two alternative pax-5 promoters in a diffuse large-cell lymphoma. Proc. Natl. Acad. Sci. USA 1996, 93, 6129–6134. [Google Scholar] [CrossRef] [PubMed]
- Gaffo, E.; Boldrin, E.; Dal Molin, A.; Bresolin, S.; Bonizzato, A.; Trentin, L.; Frasson, C.; Debatin, K.-M.; Meyer, L.H.; te Kronnie, G.; et al. Circular rna differential expression in blood cell populations and exploration of circrna deregulation in pediatric acute lymphoblastic leukemia. Sci. Rep. 2019, 9, 14670. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, G.A.; Hannay, B.; Martin, D.; Veilleux, V.; Finn, N. Characterization of new circular rna products from the pax-5 gene in b-cells. J. Immunol. 2020, 204, 223-14. [Google Scholar]
- Hannay, B.; Dumas, P.; Veilleux, V.; LeBlanc, N.; Robichaud, G.A. Discovery of novel non-coding products of the pax-5 gene and their clinical significance in lymphoid cancers. FASEB J. 2018, 32, 677-23. [Google Scholar] [CrossRef]
- Borson, N.D.; Lacy, M.Q.; Wettstein, P.J. Altered mrna expression of pax5 and blimp-1 in b cells in multiple myeloma. Blood 2002, 100, 4629–4639. [Google Scholar] [CrossRef]
- Zwollo, P.; Arrieta, H.; Ede, K.; Molinder, K.; Desiderio, S.; Pollock, R. The pax-5 gene is alternatively spliced during b-cell development. J. Biol. Chem. 1997, 272, 10160–10168. [Google Scholar] [CrossRef]
- Robichaud, G.A.; Nardini, M.; Laflamme, M.; Cuperlovic-Culf, M.; Ouellette, R.J. Human pax-5 c-terminal isoforms possess distinct transactivation properties and are differentially modulated in normal and malignant b cells. J. Biol. Chem. 2004, 279, 49956–49963. [Google Scholar] [CrossRef]
- Beauregard, A.-P.; Hannay, B.; Gharib, E.; Crapoulet, N.; Finn, N.; Guerrette, R.; Ouellet, A.; Robichaud, G.A. Pax-5 protein expression is regulated by transcriptional 3′utr editing. Cells 2021, 11, 76. [Google Scholar] [CrossRef]
- He, T.; Hong, S.Y.; Huang, L.; Xue, W.; Yu, Z.; Kwon, H.; Kirk, M.; Ding, S.-j.; Su, K.; Zhang, Z. Histone acetyltransferase p300 acetylates pax5 and strongly enhances pax5-mediated transcriptional activity. J. Biol. Chem. 2011, 286, 14137–14145. [Google Scholar] [CrossRef]
- Yasuda, T.; Hayakawa, F.; Kurahashi, S.; Sugimoto, K.; Minami, Y.; Tomita, A.; Naoe, T. B cell receptor-erk1/2 signal cancels pax5-dependent repression of blimp1 through pax5 phosphorylation: A mechanism of antigen-triggering plasma cell differentiation. J. Immunol. 2012, 188, 6127–6134. [Google Scholar] [CrossRef]
- Inagaki, Y.; Hayakawa, F.; Hirano, D.; Kojima, Y.; Morishita, T.; Yasuda, T.; Naoe, T.; Kiyoi, H. Pax5 tyrosine phosphorylation by syk co-operatively functions with its serine phosphorylation to cancel the pax5-dependent repression of blimp1: A mechanism for antigen-triggered plasma cell differentiation. Biochem. Biophys Res. Commun. 2016, 475, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Andrieux, J.; Fert-Ferrer, S.; Copin, M.-C.; Huyghe, P.; Pocachard, P.; Lespinasse, J.; Bauters, F.; Laï, J.L.; Quesnel, B. Three new cases of non-hodgkin lymphoma with t (9; 14)(p13; q32). Cancer Genet. Cytogenet. 2003, 145, 65–69. [Google Scholar] [CrossRef]
- Barberis, A.; Widenhorn, K.; Vitelli, L.; Busslinger, M. A novel b-cell lineage-specific transcription factor present at early but not late stages of differentiation. Genes Dev. 1990, 4, 849–859. [Google Scholar] [CrossRef]
- Cresson, C.; Peron, S.; Jamrog, L.; Rouquie, N.; Prade, N.; Dubois, M.; Hebrard, S.; Lagarde, S.; Gerby, B.; Mancini, S.J.C.; et al. Pax5a and pax5b isoforms are both efficient to drive b cell differentiation. Oncotarget 2018, 9, 32841–32854. [Google Scholar] [CrossRef]
- Kikuchi, H.; Nakayama, M.; Kuribayashi, F.; Mimuro, H.; Imajoh-Ohmi, S.; Nishitoh, H.; Takami, Y.; Nakayama, T. Paired box gene 5 isoforms a and b have different functions in transcriptional regulation of b cell development-related genes in immature b cells. Microbiol. Immunol. 2015, 59, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, G.A.; Perreault, J.-P.; Ouellette, R.J. Development of an isoform-specific gene suppression system: The study of the human pax-5b transcriptional element. Nucleic Acids Res. 2008, 36, 4609–4620. [Google Scholar] [CrossRef][Green Version]
- Horcher, M.; Souabni, A.; Busslinger, M. Pax5/bsap maintains the identity of b cells in late b lymphopoiesis. Immunity 2001, 14, 779–790. [Google Scholar] [CrossRef]
- Cobaleda, C.; Schebesta, A.; Delogu, A.; Busslinger, M. Pax5: The guardian of b cell identity and function. Nat. Immunol. 2007, 8, 463–470. [Google Scholar] [CrossRef]
- Zriwil, A.; Böiers, C.; Kristiansen, T.A.; Wittmann, L.; Yuan, J.; Nerlov, C.; Sitnicka, E.; Jacobsen, S.E.W. Direct role of flt 3 in regulation of early lymphoid progenitors. Br. J. Haematol. 2018, 183, 588–600. [Google Scholar] [CrossRef]
- Namen, A.E.; Lupton, S.; Hjerrild, K.; Wignall, J.; Mochizuki, D.Y.; Schmierer, A.; Mosley, B.; March, C.J.; Urdal, D.; Gillis, S. Stimulation of b-cell progenitors by cloned murine interleukin-7. Nature 1988, 333, 571–573. [Google Scholar] [CrossRef]
- Matsuuchi, L.; Gold, M.R. New views of bcr structure and organization. Curr. Opin. Immunol. 2001, 13, 270–277. [Google Scholar] [CrossRef]
- Kozmik, Z.; Wang, S.; Dorfler, P.; Adams, B.; Busslinger, M. The promoter of the cd19 gene is a target for the b-cell-specific transcription factor bsap. Mol. Cell. Biol. 1992, 12, 2662–2672. [Google Scholar] [PubMed]
- Maier, H.; Colbert, J.; Fitzsimmons, D.; Clark, D.R.; Hagman, J. Activation of the early b-cell-specific mb-1 (ig-alpha) gene by pax-5 is dependent on an unmethylated ets binding site. Mol. Cell. Biol. 2003, 23, 1946–1960. [Google Scholar] [CrossRef]
- Hirokawa, S.; Sato, H.; Kato, I.; Kudo, A. Ebf-regulating pax5 transcription is enhanced by stat5 in the early stage of b cells. Eur. J. Immunol. 2003, 33, 1824–1829. [Google Scholar] [CrossRef] [PubMed]
- Pridans, C.; Holmes, M.L.; Polli, M.; Wettenhall, J.M.; Dakic, A.; Corcoran, L.M.; Smyth, G.K.; Nutt, S.L. Identification of pax5 target genes in early b cell differentiation. J. Immunol. 2008, 180, 1719–1728. [Google Scholar] [CrossRef]
- Pang, S.H.M.; de Graaf, C.A.; Hilton, D.J.; Huntington, N.D.; Carotta, S.; Wu, L.; Nutt, S.L. Pu.1 is required for the developmental progression of multipotent progenitors to common lymphoid progenitors. Front. Immunol. 2018, 9, 1264. [Google Scholar] [CrossRef]
- Carotta, S.; Holmes, M.; Pridans, C.; Nutt, S.L. Pax5 maintains cellular identity by repressing gene expression throughout b cell differentiation. Cell Cycle 2006, 5, 2452–2456. [Google Scholar] [CrossRef]
- Schebesta, M.; Pfeffer, P.L.; Busslinger, M. Control of pre-bcr signaling by pax5-dependent activation of the blnk gene. Immunity 2002, 17, 473–485. [Google Scholar] [CrossRef]
- Reth, M.; Nielsen, P. Signaling circuits in early b-cell development. Adv. Immunol. 2014, 122, 129–175. [Google Scholar]
- Shahjahani, M.; Norozi, F.; Ahmadzadeh, A.; Shahrabi, S.; Tavakoli, F.; Asnafi, A.A.; Saki, N. The role of pax5 in leukemia: Diagnosis and prognosis significance. Med. Oncol. 2015, 32, 360. [Google Scholar] [CrossRef]
- Gonda, H.; Sugai, M.; Nambu, Y.; Katakai, T.; Agata, Y.; Mori, K.J.; Yokota, Y.; Shimizu, A. The balance between pax5 and id2 activities is the key to aid gene expression. J. Exp. Med. 2003, 198, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Nutt, S.L.; Morrison, A.M.; Dorfler, P.; Rolink, A.; Busslinger, M. Identification of bsap (pax-5) target genes in early b-cell development by loss- and gain-of-function experiments. EMBO J. 1998, 17, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Nera, K.-P.; Kohonen, P.; Narvi, E.; Peippo, A.; Mustonen, L.; Terho, P.; Koskela, K.; Buerstedde, J.-M.; Lassila, O. Loss of pax5 promotes plasma cell differentiation. Immunity 2006, 24, 283–293. [Google Scholar] [CrossRef]
- Souabni, A.; Cobaleda, C.; Schebesta, M.; Busslinger, M. Pax5 promotes b lymphopoiesis and blocks t cell development by repressing notch1. Immunity 2002, 17, 781–793. [Google Scholar] [CrossRef]
- Lin, K.-I.; Angelin-Duclos, C.; Kuo, T.C.; Calame, K. Blimp-1-dependent repression of pax-5 is required for differentiation of b cells to immunoglobulin m-secreting plasma cells. Mol. Cell. Biol. 2002, 22, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Kallies, A.; Nutt, S.L. Terminal differentiation of lymphocytes depends on blimp-1. Curr. Opin. Immunol. 2007, 19, 156–162. [Google Scholar] [CrossRef]
- Delogu, A.; Schebesta, A.; Sun, Q.; Aschenbrenner, K.; Perlot, T.; Busslinger, M. Gene repression by pax5 in b cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity 2006, 24, 269–281. [Google Scholar] [CrossRef]
- Liu, G.J.; Jaritz, M.; Wohner, M.; Agerer, B.; Bergthaler, A.; Malin, S.G.; Busslinger, M. Repression of the b cell identity factor pax5 is not required for plasma cell development. J. Exp. Med. 2020, 217, e20200147. [Google Scholar] [CrossRef]
- Cobaleda, C.; Jochum, W.; Busslinger, M. Conversion of mature b cells into t cells by dedifferentiation to uncommitted progenitors. Nature 2007, 449, 473–477. [Google Scholar] [CrossRef]
- Nutt, S.L.; Heavey, B.; Rolink, A.G.; Busslinger, M. Commitment to the b-lymphoid lineage depends on the transcription factor pax5. Nature 1999, 401, 556–562. [Google Scholar] [CrossRef]
- Proulx, M.; Cayer, M.-P.; Drouin, M.; Laroche, A.; Jung, D. Overexpression of pax5 induces apoptosis in multiple myeloma cells. Int. J. Hematol. 2010, 92, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Cozma, D.; Yu, D.; Hodawadekar, S.; Azvolinsky, A.; Grande, S.; Tobias, J.W.; Metzgar, M.H.; Paterson, J.; Erikson, J.; Marafioti, T.; et al. B cell activator pax5 promotes lymphomagenesis through stimulation of b cell receptor signaling. J. Clin. Investig. 2007, 117, 2602–2610. [Google Scholar] [CrossRef] [PubMed]
- Torlakovic, E.; Slipicevic, A.; Robinson, C.; DeCoteau, J.F.; Alfsen, G.C.; Vyberg, M.; Chibbar, R.; Flørenes, V.A. Pax-5 expression in nonhematopoietic tissues. Am. J. Clin. Pathol. 2006, 126, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Poppe, B.; De Paepe, P.; Michaux, L.; Dastugue, N.; Bastard, C.; Herens, C.; Moreau, E.; Cavazzini, F.; Yigit, N.; Van Limbergen, H.; et al. Pax5/igh rearrangement is a recurrent finding in a subset of aggressive b-nhl with complex chromosomal rearrangements. Genes Chromosomes Cancer 2005, 44, 218–223. [Google Scholar] [CrossRef]
- Krenacs, L.; Himmelmann, A.; Quintanilla-Martinez, L.; Fest, T.; Riva, A.; Wellmann, A.; Bagdi, E.; Kehrl, J.; Jaffe, E.; Raffeld, M. Transcription factor b-cell-specific activator protein (bsap) is differentially expressed in b cells and in subsets of b-cell lymphomas. Blood 1998, 92, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.; Morin, P.; Ouellette, R.; Robichaud, G. The pax-5 gene: A pluripotent regulator of b-cell differentiation and cancer disease. Cancer Res. 2011, 71, 7345–7350. [Google Scholar] [CrossRef] [PubMed]
- Arthur, S.E.; Jiang, A.; Grande, B.M.; Alcaide, M.; Cojocaru, R.; Rushton, C.K.; Mottok, A.; Hilton, L.K.; Lat, P.K.; Zhao, E.Y.; et al. Genome-wide discovery of somatic regulatory variants in diffuse large b-cell lymphoma. Nat. Commun. 2018, 9, 4001. [Google Scholar] [CrossRef]
- Puente, X.S.; Bea, S.; Valdes-Mas, R.; Villamor, N.; Gutierrez-Abril, J.; Martin-Subero, J.I.; Munar, M.; Rubio-Perez, C.; Jares, P.; Aymerich, M.; et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic burkitt lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef]
- Cazzaniga, G.; Daniotti, M.; Tosi, S.; Giudici, G.; Aloisi, A.; Pogliani, E.; Kearney, L.; Biondi, A. The paired box domain gene pax5 is fused to etv6/tel in an acute lymphoblastic leukemia case. Cancer Res. 2001, 61, 4666–4670. [Google Scholar]
- Nebral, K.; Konig, M.; Harder, L.; Siebert, R.; Haas, O.A.; Strehl, S. Identification of pml as novel pax5 fusion partner in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2007, 139, 269–274. [Google Scholar] [CrossRef]
- Familiades, J.; Bousquet, M.; Lafage-Pochitaloff, M.; Béné, M.C.; Beldjord, K.; De Vos, J.; Dastugue, N.; Coyaud, E.; Struski, S.; Quelen, C.; et al. Pax5 mutations occur frequently in adult b-cell progenitor acute lymphoblastic leukemia and pax5 haploinsufficiency is associated with bcr-abl1 and tcf3-pbx1 fusion genes: A graall study. Leukemia 2009, 23, 1989–1998. [Google Scholar] [CrossRef] [PubMed]
- Medvedovic, J.; Ebert, A.; Tagoh, H.; Busslinger, M. Pax5: A master regulator of b cell development and leukemogenesis. Adv. Immunol. 2011, 111, 179–206. [Google Scholar]
- Jamrog, L.; Chemin, G.; Fregona, V.; Coster, L.; Pasquet, M.; Oudinet, C.; Rouquié, N.; Prade, N.; Lagarde, S.; Cresson, C.; et al. Pax5-eln oncoprotein promotes multistep b-cell acute lymphoblastic leukemia in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 10357–10362. [Google Scholar] [CrossRef]
- Medina, K.L. Assembling a gene regulatory network for specification of the b cell fate. Dev. Cell 2004, 7, 607–617. [Google Scholar] [CrossRef]
- Okuyama, K.; Strid, T.; Kuruvilla, J.; Somasundaram, R.; Cristobal, S.; Smith, E.; Prasad, M.; Fioretos, T.; Lilljebjörn, H.; Soneji, S.; et al. Pax5 is part of a functional transcription factor network targeted in lymphoid leukemia. PLoS Genet. 2019, 15, e1008280. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Miller, C.B.; Radtke, I.; Phillips, L.A.; Dalton, J.; Ma, J.; White, D.; Hughes, T.P.; Le Beau, M.M.; Pui, C.-H. Bcr–abl1 lymphoblastic leukaemia is characterized by the deletion of ikaros. Nature 2008, 453, 110–114. [Google Scholar] [CrossRef]
- Chan, L.N.; Chen, Z.; Braas, D.; Lee, J.-W.; Xiao, G.; Geng, H.; Cosgun, K.N.; Hurtz, C.; Shojaee, S.; Cazzaniga, V.; et al. Metabolic gatekeeper function of b-lymphoid transcription factors. Nature 2017, 542, 479–483. [Google Scholar] [CrossRef]
- Szablewski, L. Expression of glucose transporters in cancers. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2013, 1835, 164–169. [Google Scholar] [CrossRef]
- Sadras, T.; Chan, L.N.; Xiao, G.; Muschen, M. Metabolic gatekeepers of pathological b cell activation. Annu. Rev. Pathol. 2021, 16, 323–349. [Google Scholar] [CrossRef]
- Li, S.-J.; Yang, X.-N.; Qian, H.-Y. Antitumor effects of wnt2b silencing in glut1 overexpressing cisplatin resistant head and neck squamous cell carcinoma. Am. J. Cancer Res. 2015, 5, 300. [Google Scholar] [PubMed]
- Kurimoto, K.; Hayashi, M.; Guerrero-Preston, R.; Koike, M.; Kanda, M.; Hirabayashi, S.; Tanabe, H.; Takano, N.; Iwata, N.; Niwa, Y.; et al. Pax5 gene as a novel methylation marker that predicts both clinical outcome and cisplatin sensitivity in esophageal squamous cell carcinoma. Epigenetics 2017, 12, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, X.; Chu, E.; Go, M.Y.; Xu, L.; Zhao, G.; Li, L.; Dai, N.; Si, J.; Tao, Q.; et al. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology 2011, 53, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cheung, K.F.; Ma, X.; Tian, L.; Zhao, J.; Go, M.Y.; Shen, B.; Cheng, A.S.; Ying, J.; Tao, Q.; et al. Epigenetic inactivation of paired box gene 5, a novel tumor suppressor gene, through direct upregulation of p53 is associated with prognosis in gastric cancer patients. Oncogene 2012, 31, 3419–3430. [Google Scholar] [CrossRef] [PubMed]
- Vidal, L.; Perry, J.; Vouyovitch, C.; Pandey, V.; Brunet-Dunand, S.; Mertani, H.; Liu, D.-X.; Lobie, P. Pax5alpha enhances the epithelial behavior of human mammary carcinoma cells. Mol. Cancer Res. MCR 2010, 8, 444–456. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Michailidi, C.; Marchionni, L.; Pickering, C.R.; Frederick, M.J.; Myers, J.N.; Yegnasubramanian, S.; Hadar, T.; Noordhuis, M.G.; Zizkova, V.; et al. Key tumor suppressor genes inactivated by “greater promoter” methylation and somatic mutations in head and neck cancer. Epigenetics 2014, 9, 1031–1046. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Downing, J.R. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: Recent insights and future directions. Leukemia 2009, 23, 1209–1218. [Google Scholar] [CrossRef]
- Shah, S.; Schrader, K.A.; Waanders, E.; Timms, A.E.; Vijai, J.; Miething, C.; Wechsler, J.; Yang, J.; Hayes, J.; Klein, R.J.; et al. A recurrent germline pax5 mutation confers susceptibility to pre-b cell acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 1226–1231. [Google Scholar] [CrossRef]
- Heltemes-Harris, L.; Willette, M.; Ramsey, L.; Qiu, Y.; Neeley, E.; Zhang, N.; Thomas, D.; Koeuth, T.; Baechler, E.; Kornblau, S.; et al. Ebf1 or pax5 haploinsufficiency synergizes with stat5 activation to initiate acute lymphoblastic leukemia. J. Exp. Med. 2011, 208, 1135–1149. [Google Scholar] [CrossRef]
- Roberts, K.G.; Morin, R.D.; Zhang, J.; Hirst, M.; Zhao, Y.; Su, X.; Chen, S.C.; Payne-Turner, D.; Churchman, M.L.; Harvey, R.C.; et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 2012, 22, 153–166. [Google Scholar] [CrossRef]
- Pui, C.-H.; Robison, L.L.; Look, A.T. Acute lymphoblastic leukaemia. Lancet 2008, 371, 1030–1043. [Google Scholar] [CrossRef]
- Kawamata, N.; Ogawa, S.; Zimmermann, M.; Niebuhr, B.; Stocking, C.; Sanada, M.; Hemminki, K.; Yamatomo, G.; Nannya, Y.; Koehler, R.; et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc. Natl. Acad. Sci. USA 2008, 105, 11921–11926. [Google Scholar] [CrossRef] [PubMed]
- Coyaud, E.; Struski, S.; Prade, N.; Familiades, J.; Eichner, R.; Quelen, C.; Bousquet, M.; Mugneret, F.; Talmant, P.; Pages, M.P.; et al. Wide diversity of pax5 alterations in b-all: A groupe francophone de cytogenetique hematologique study. Blood 2010, 115, 3089–3097. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Wright, S.L.; Konn, Z.J.; Matheson, E.; Minto, L.; Moorman, A.V.; Parker, H.; Griffiths, M.; Ross, F.M.; Davies, T.; et al. Variable breakpoints target pax5 in patients with dicentric chromosomes: A model for the basis of unbalanced translocations in cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 17050–17054. [Google Scholar] [CrossRef]
- Nebral, K.; Denk, D.; Attarbaschi, A.; König, M.; Mann, G.; Haas, O.A.; Strehl, S. Incidence and diversity of pax5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 2009, 23, 134–143. [Google Scholar] [CrossRef]
- Fazio, G.; Daniele, G.; Cazzaniga, V.; Impera, L.; Severgnini, M.; Iacobucci, I.; Galbiati, M.; Leszl, A.; Cifola, I.; De Bellis, G.; et al. Three novel fusion transcripts of the paired box 5 gene in b-cell precursor acute lymphoblastic leukemia. Haematologica 2015, 100, e14–e17. [Google Scholar] [CrossRef]
- Fazio, G.; Cazzaniga, V.; Palmi, C.; Galbiati, M.; Giordan, M.; te Kronnie, G.; Rolink, A.; Biondi, A.; Cazzaniga, G. Pax5/etv6 alters the gene expression profile of precursor b cells with opposite dominant effect on endogenous pax5. Leukemia 2013, 27, 992–995. [Google Scholar] [CrossRef]
- Smeenk, L.; Fischer, M.; Jurado, S.; Jaritz, M.; Azaryan, A.; Werner, B.; Roth, M.; Zuber, J.; Stanulla, M.; den Boer, M.L.; et al. Molecular role of the pax5-etv6 oncoprotein in promoting b-cell acute lymphoblastic leukemia. EMBO J. 2017, 36, 718–735. [Google Scholar] [CrossRef]
- Revilla, I.D.R.; Bilic, I.; Vilagos, B.; Tagoh, H.; Ebert, A.; Tamir, I.M.; Smeenk, L.; Trupke, J.; Sommer, A.; Jaritz, M.; et al. The b-cell identity factor pax5 regulates distinct transcriptional programmes in early and late b lymphopoiesis. EMBO J. 2012, 31, 3130–3146. [Google Scholar] [CrossRef]
- Iida, S.; Rao, P.H.; Nallasivam, P.; Hibshoosh, H.; Butler, M.; Louie, D.C.; Dyomin, V.; Ohno, H.; Chaganti, R.S.; Dalla-Favera, R. The t(9;14)(p13;q32) chromosomal translocation associated with lymphoplasmacytoid lymphoma involves the pax-5 gene. Blood 1996, 88, 4110–4117. [Google Scholar] [CrossRef]
- Souabni, A.; Jochum, W.; Busslinger, M. Oncogenic role of pax5 in the t-lymphoid lineage upon ectopic expression from the immunoglobulin heavy-chain locus. Blood 2007, 109, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Baumann Kubetzko, F.; Di Paolo, C.; Maag, C.; Meier, R.; Schäfer, B.; Betts, D.; Stahel, R.; Himmelmann, A. The pax5 oncogene is expressed in n-type neuroblastoma cells and increases tumorigenicity of a s-type cell line. Carcinogenesis 2004, 25, 1839–1846. [Google Scholar] [CrossRef]
- Kozmik, Z.; Sure, U.; Rüedi, D.; Busslinger, M.; Aguzzi, A. Deregulated expression of pax5 in medulloblastoma. Proc. Natl. Acad. Sci. USA 1995, 92, 5709–5713. [Google Scholar] [CrossRef]
- Kanteti, R.; Nallasura, V.; Loganathan, S.; Tretiakova, M.; Kroll, T.; Krishnaswamy, S.; Faoro, L.; Cagle, P.; Husain, A.N.; Vokes, E.E.; et al. Pax5 is expressed in small-cell lung cancer and positively regulates c-met transcription. Lab. Investig. 2009, 89, 301–314. [Google Scholar] [CrossRef]
- Dong, B.W.; Zhang, W.B.; Qi, S.M.; Yan, C.Y.; Gao, J. Transactivation of ptgs2 by pax5 signaling potentiates cisplatin resistance in muscle-invasive bladder cancer cells. Biochem. Biophys. Res. Commun. 2018, 503, 2293–2300. [Google Scholar] [CrossRef]
- Stuart, E.T.; Haffner, R.; Oren, M.; Gruss, P. Loss of p53 function through pax-mediated transcriptional repression. EMBO J. 1995, 14, 5638–5645. [Google Scholar] [CrossRef] [PubMed]
- Stuart, E.T.; Kioussi, C.; Aguzzi, A.; Gruss, P. Pax5 expression correlates with increasing malignancy in human astrocytomas. Clin. Cancer Res. 1995, 1, 207–214. [Google Scholar]
- Yang, R.; Klimentova, J.; Gockel-Krzikalla, E.; Ly, R.; Gmelin, N.; Hotz-Wagenblatt, A.; Rehulkova, H.; Stulik, J.; Rosl, F.; Niebler, M. Combined transcriptome and proteome analysis of immortalized human keratinocytes expressing human papillomavirus 16 (hpv16) oncogenes reveals novel key factors and networks in hpv-induced carcinogenesis. mSphere 2019, 4, e00129-19. [Google Scholar] [CrossRef]
- Norhany, S.; Kouzu, Y.; Uzawa, K.; Hayama, M.; Higo, M.; Koike, H.; Kasamatu, A.; Tanzawa, H. Overexpression of pax5 in oral carcinogenesis. Oncol. Rep. 2006, 16, 1003–1008. [Google Scholar] [CrossRef][Green Version]
- Mžik, M.; Chmelařová, M.; John, S.; Laco, J.; Slabý, O.; Kiss, I.; Bohovicová, L.; Palička, V.; Nekvindová, J. Aberrant methylation of tumour suppressor genes wt1, gata5 and pax5 in hepatocellular carcinoma. Clin. Chem. Lab. Med. (CCLM) 2016, 54, 1971–1980. [Google Scholar] [CrossRef]
- Benzina, S.; Beauregard, A.-P.; Guerrette, R.; Jean, S.; Faye, M.D.; Laflamme, M.; Maïcas, E.; Crapoulet, N.; Ouellette, R.J.; Robichaud, G.A. Pax-5 is a potent regulator of e-cadherin and breast cancer malignant processes. Oncotarget 2017, 8, 12052–12066. [Google Scholar] [PubMed]
- Crapoulet, N.; O’Brien, P.; Ouellette, R.; Robichaud, G. Coordinated expression of pax-5 and fak1 in metastasis. Anti-Cancer Agents Med. Chem. 2011, 11, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Benzina, S.; Harquail, J.; Guerrette, R.; O’Brien, P.; Jean, S.; Crapoulet, N.; Robichaud, G.A. Breast cancer malignant processes are regulated by pax-5 through the disruption of fak signaling pathways. J. Cancer 2016, 7, 2035–2044. [Google Scholar] [CrossRef]
- Livide, G.; Epistolato, M.C.; Amenduni, M.; Disciglio, V.; Marozza, A.; Mencarelli, M.A.; Toti, P.; Lazzi, S.; Hadjistilianou, T.; De Francesco, S.; et al. Epigenetic and copy number variation analysis in retinoblastoma by ms-mlpa. Pathol. Oncol. Res. 2012, 18, 703–712. [Google Scholar]
- Deng, J.; Liang, H.; Zhang, R.; Dong, Q.; Hou, Y.; Yu, J.; Fan, D.; Hao, X. Applicability of the methylated cpg sites of paired box 5 (pax5) promoter for prediction the prognosis of gastric cancer. Oncotarget 2014, 5, 7420–7430. [Google Scholar]
- Kolhe, R.; Reid, M.D.; Lee, J.R.; Cohen, C.; Ramalingam, P. Immunohistochemical expression of pax5 and tdt by merkel cell carcinoma and pulmonary small cell carcinoma: A potential diagnostic pitfall but useful discriminatory marker. Int. J. Clin. Exp. Pathol. 2013, 6, 142–147. [Google Scholar]
- Chmelarova, M.; Krepinska, E.; Spacek, J.; Laco, J.; Nekvindova, J.; Palicka, V. Methylation analysis of tumour suppressor genes in ovarian cancer using ms-mlpa. Folia. Biol. 2012, 58, 246–250. [Google Scholar]
- Moelans, C.B.; Verschuur-Maes, A.H.; van Diest, P.J. Frequent promoter hypermethylation of brca2, cdh13, msh6, pax5, pax6 and wt1 in ductal carcinoma in situ and invasive breast cancer. J. Pathol. 2011, 225, 222–231. [Google Scholar]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef]
- Micalizzi, D.; Farabaugh, S.; Ford, H. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia 2010, 15, 117–134. [Google Scholar]
- Chao, Y.L.; Shepard, C.R.; Wells, A. Breast carcinoma cells re-express e-cadherin during mesenchymal to epithelial reverting transition. Mol. Cancer 2010, 9, 179. [Google Scholar]
- Hugo, H.; Ackland, M.L.; Blick, T.; Lawrence, M.G.; Clements, J.A.; Williams, E.D.; Thompson, E.W. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J. Cell Physiol. 2007, 213, 374–383. [Google Scholar]
- Chaffer, C.L.; Brennan, J.P.; Slavin, J.L.; Blick, T.; Thompson, E.W.; Williams, E.D. Mesenchymal-to-epithelial transition facilitates bladder cancer metastasis: Role of fibroblast growth factor receptor-2. Cancer Res. 2006, 66, 11271–11278. [Google Scholar] [PubMed]
- Benzina, S.; Beauregard, A.P.; Guerrette, R.; Jean, S.; Faye, M.D.; Laflamme, M.; Maïcas, E.; Crapoulet, N.; Ouellette, R.J.; Robichaud, G.A. Pax-5 is a potent transcriptional regulator of e-cadherin and breast cancer malignancy. Submitt. Mol. Cancer Res. MCR 2015, 8, 12052–12066. [Google Scholar]
- Cano, A.; Perez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing e-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar]
- Hajra, K.M.; Chen, D.Y.; Fearon, E.R. The slug zinc-finger protein represses e-cadherin in breast cancer. Cancer Res. 2002, 62, 1613–1618. [Google Scholar]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. Deltaef1 is a transcriptional repressor of e-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar]
- Baranwal, S.; Alahari, S.K. Molecular mechanisms controlling e-cadherin expression in breast cancer. Biochem. Biophys Res. Commun. 2009, 384, 6–11. [Google Scholar]
- Wells, A.; Yates, C.; Shepard, C.R. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin. Exp. Metastasis 2008, 25, 621–628. [Google Scholar]
- Conacci-Sorrell, M.; Simcha, I.; Ben-Yedidia, T.; Blechman, J.; Savagner, P.; Ben-Ze’ev, A. Autoregulation of e-cadherin expression by cadherin-cadherin interactions: The roles of beta-catenin signaling, slug, and mapk. J. Cell Biol. 2003, 163, 847–857. [Google Scholar] [PubMed]
- Ellsworth, R.E.; Seebach, J.; Field, L.A.; Heckman, C.; Kane, J.; Hooke, J.A.; Love, B.; Shriver, C.D. A gene expression signature that defines breast cancer metastases. Clin. Exp. Metastasis 2009, 26, 205–213. [Google Scholar] [PubMed]
- Heinaniemi, M.; Vuorenmaa, T.; Teppo, S.; Kaikkonen, M.U.; Bouvy-Liivrand, M.; Mehtonen, J.; Niskanen, H.; Zachariadis, V.; Laukkanen, S.; Liuksiala, T.; et al. Transcription-coupled genetic instability marks acute lymphoblastic leukemia structural variation hotspots. eLife 2016, 5, e13087. [Google Scholar]
- Kasprzyk, M.E.; Sura, W.; Dzikiewicz-Krawczyk, A. Enhancing b-cell malignancies-on repurposing enhancer activity towards cancer. Cancers 2021, 13, 3270. [Google Scholar] [PubMed]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding rnas as regulators in epigenetics (review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar]
- Dunn, B.K.; Verma, M.; Umar, A. Epigenetics in cancer prevention: Early detection and risk assessment: Introduction. Ann. N. Y. Acad. Sci. 2003, 983, 1–4. [Google Scholar]
- Bao, Y.; Cao, X. Epigenetic control of b cell development and b-cell-related immune disorders. Clin. Rev. Allergy Immunol. 2016, 50, 301–312. [Google Scholar]
- Kurogi, T.; Inoue, H.; Guo, Y.; Nobukiyo, A.; Nohara, K.; Kanno, M. A methyl-deficient diet modifies early b cell development. Pathobiology 2012, 79, 209–218. [Google Scholar]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and function of mammalian DNA methyltransferases. Chembiochem 2011, 12, 206–222. [Google Scholar] [PubMed]
- Mohr, F.; Dohner, K.; Buske, C.; Rawat, V.P. Tet genes: New players in DNA demethylation and important determinants for stemness. Exp. Hematol. 2011, 39, 272–281. [Google Scholar] [PubMed]
- Li, G.; Zan, H.; Xu, Z.; Casali, P. Epigenetics of the antibody response. Trends Immunol. 2013, 34, 460–470. [Google Scholar] [PubMed]
- Choukrallah, M.A.; Matthias, P. The interplay between chromatin and transcription factor networks during b cell development: Who pulls the trigger first? Front. Immunol. 2014, 5, 156. [Google Scholar]
- Cherry, S.R.; Beard, C.; Jaenisch, R.; Baltimore, D. V(d)j recombination is not activated by demethylation of the kappa locus. Proc. Natl. Acad. Sci. USA 2000, 97, 8467–8472. [Google Scholar]
- Lio, C.W.; Zhang, J.; Gonzalez-Avalos, E.; Hogan, P.G.; Chang, X.; Rao, A. Tet2 and tet3 cooperate with b-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 2016, 5, e18290. [Google Scholar]
- Maier, H. Early b cell factor cooperates with runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immunol. 2004, 5, 1069–1077. [Google Scholar]
- Gao, H.; Lukin, K.; Ramírez, J.; Fields, S.; Lopez, D.; Hagman, J. Opposing effects of swi/snf and mi-2/nurd chromatin remodeling complexes on epigenetic reprogramming by ebf and pax5. Proc. Natl. Acad. Sci. USA 2009, 106, 11258–11263. [Google Scholar]
- McManus, S.; Ebert, A.; Salvagiotto, G.; Medvedovic, J.; Sun, Q.; Tamir, I.; Jaritz, M.; Tagoh, H.; Busslinger, M. The transcription factor pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed b cells. EMBO J. 2011, 30, 2388–2404. [Google Scholar]
- Danbara, M.; Kameyama, K.; Higashihara, M.; Takagaki, Y. DNA methylation dominates transcriptional silencing of pax5 in terminally differentiated b cell lines. Mol. Immunol. 2002, 38, 1161–1166. [Google Scholar]
- Dominguez, P.M.; Shaknovich, R. Epigenetic function of activation-induced cytidine deaminase and its link to lymphomagenesis. Front. Immunol. 2014, 5, 642. [Google Scholar] [PubMed]
- Muramatsu, M.; Kinoshita, K.; Fagarasan, S.; Yamada, S.; Shinkai, Y.; Honjo, T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (aid), a potential rna editing enzyme. Cell 2000, 102, 553–563. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Neumeister, P.; Goossens, T.; Nanjangud, G.; Chaganti, R.S.; Kuppers, R.; Dalla-Favera, R. Hypermutation of multiple proto-oncogenes in b-cell diffuse large-cell lymphomas. Nature 2001, 412, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Deng, Y.; Feng, Y.; Long, D.; Ma, K.; Wang, X.; Zhao, M.; Lu, L.; Lu, Q. Epigenetic regulation in b-cell maturation and its dysregulation in autoimmunity. Cell. Mol. Immunol. 2018, 15, 676–684. [Google Scholar] [PubMed]
- Hütter, G.; Kaiser, M.; Neumann, M.; Mossner, M.; Nowak, D.; Baldus, C.D.; Gökbuget, N.; Hoelzer, D.; Thiel, E.; Hofmann, W.-K. Epigenetic regulation of pax5 expression in acute t-cell lymphoblastic leukemia. Leuk. Res. 2011, 35, 614–619. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Downing, J.R. Global genomic characterization of acute lymphoblastic leukemia. Semin. Hematol. 2009, 46, 3–15. [Google Scholar] [PubMed]
- Nordlund, J.; Bäcklin, C.L.; Zachariadis, V.; Cavelier, L.; Dahlberg, J.; Öfverholm, I.; Barbany, G.; Nordgren, A.; Övernäs, E.; Abrahamsson, J.; et al. DNA methylation-based subtype prediction for pediatric acute lymphoblastic leukemia. Clin. Epigenet. 2015, 7, 11. [Google Scholar] [CrossRef]
- Palmisano, W.A.; Crume, K.P.; Grimes, M.J.; Winters, S.A.; Toyota, M.; Esteller, M.; Joste, N.; Baylin, S.B.; Belinsky, S.A. Aberrant promoter methylation of the transcription factor genes pax5 α and β in human cancers. Cancer Res. 2003, 63, 4620–4625. [Google Scholar] [PubMed]
- Zhang, W.; Yan, W.; Qian, N.; Han, Q.; Zhang, W.; Dai, G. Paired box 5 increases the chemosensitivity of esophageal squamous cell cancer cells by promoting p53 signaling activity. Chin. Med. J. 2022, 135, 606–618. [Google Scholar] [CrossRef]
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys Acta 2014, 1839, 627–643. [Google Scholar] [PubMed]
- Liu, G.J.; Cimmino, L.; Jude, J.G.; Hu, Y.; Witkowski, M.T.; McKenzie, M.D.; Kartal-Kaess, M.; Best, S.A.; Tuohey, L.; Liao, Y.; et al. Pax5 loss imposes a reversible differentiation block in b-progenitor acute lymphoblastic leukemia. Genes Dev. 2014, 28, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Bullerwell, C.E.; Robichaud, P.P.; Deprez, P.M.L.; Joy, A.P.; Wajnberg, G.; D’Souza, D.; Chacko, S.; Fournier, S.; Crapoulet, N.; Barnett, D.A.; et al. Ebf1 drives hallmark b cell gene expression by enabling the interaction of pax5 with the mll h3k4 methyltransferase complex. Sci. Rep. 2021, 11, 1537. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Angelin-Duclos, C.; Greenwood, J.; Liao, J.; Calame, K. Transcriptional repression by blimp-1 (prdi-bf1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 2000, 20, 2592–2603. [Google Scholar] [CrossRef] [PubMed]
- Gyory, I.; Wu, J.; Fejer, G.; Seto, E.; Wright, K.L. Prdi-bf1 recruits the histone h3 methyltransferase g9a in transcriptional silencing. Nat. Immunol. 2004, 5, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Kung, J.; Holmes, A.B.; Wells, V.A.; Mo, T.; Basso, K.; Dalla-Favera, R. The foxo1 transcription factor instructs the germinal center dark zone program. Immunity 2015, 43, 1064–1074. [Google Scholar] [CrossRef]
- Lin, Y.C.; Jhunjhunwala, S.; Benner, C.; Heinz, S.; Welinder, E.; Mansson, R.; Sigvardsson, M.; Hagman, J.; Espinoza, C.A.; Dutkowski, J.; et al. A global network of transcription factors, involving e2a, ebf1 and foxo1, that orchestrates b cell fate. Nat. Immunol. 2010, 11, 635–643. [Google Scholar] [CrossRef]
- Jin, L.; Ma, X.; Lei, X.; Tong, J.a.; Wang, R. Cyclophosphamide inhibits pax5 methylation to regulate the growth of retinoblastoma via the notch1 pathway. Hum. Exp. Toxicol. 2021, 40, S497–S508. [Google Scholar] [CrossRef]
- Gangaraju, V.K.; Bartholomew, B. Mechanisms of atp dependent chromatin remodeling. Mutat. Res. 2007, 618, 3–17. [Google Scholar] [CrossRef]
- Balasenthil, S.; Gururaj, A.; Talukder, A.; Bagheri-Yarmand, R.; Arrington, T.; Haas, B.; Braisted, J.; Kim, I.; Lee, N.; Kumar, R. Identification of pax5 as a target of mta1 in b-cell lymphomas. Cancer Res. 2007, 67, 7132–7138. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. Rna-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Butler, A.A.; Webb, W.M.; Lubin, F.D. Regulatory rnas and control of epigenetic mechanisms: Expectations for cognition and cognitive dysfunction. Epigenomics 2016, 8, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian micrornas in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular roles and function of circular rnas in eukaryotic cells. Cell. Mol. Life Sci. 2018, 75, 1071–1098. [Google Scholar] [CrossRef] [PubMed]
- Harquail, J.; LeBlanc, N.; Ouellette, R.J.; Robichaud, G.A. Mirnas 484 and 210 regulate pax-5 expression and function in breast cancer cells. Carcinogenesis 2019, 40, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Rothe, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microrna expression profiling identifies mir-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef]
- Huang, X.; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.T.; Giaccia, A.J. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell 2009, 35, 856–867. [Google Scholar]
- Qin, Q.; Furong, W.; Baosheng, L. Multiple functions of hypoxia-regulated mir-210 in cancer. J. Exp. Clin. Cancer Res. 2014, 33, 50. [Google Scholar]
- Devlin, C.; Greco, S.; Martelli, F.; Ivan, M. Mir-210: More than a silent player in hypoxia. IUBMB Life 2011, 63, 94–100. [Google Scholar] [CrossRef]
- Huang, X.; Le, Q.T.; Giaccia, A.J. Mir-210--micromanager of the hypoxia pathway. Trends Mol. Med. 2010, 16, 230–237. [Google Scholar] [CrossRef]
- Harquail, J.; LeBlanc, N.; Landry, C.; Crapoulet, N.; Robichaud, G.A. Pax-5 inhibits nf-kappab activity in breast cancer cells through ikkepsilon and mirna-155 effectors. J. Mammary Gland. Biol. Neoplasia 2018, 23, 177–187. [Google Scholar] [CrossRef]
- Lu, D.; Nakagawa, R.; Lazzaro, S.; Staudacher, P.; Abreu-Goodger, C.; Henley, T.; Boiani, S.; Leyland, R.; Galloway, A.; Andrews, S.; et al. The mir-155–pu.1 axis acts on pax5 to enable efficient terminal b cell differentiation. J. Exp. Med. 2014, 211, 2183–2198. [Google Scholar] [PubMed]
- Calame, K. Microrna-155 function in b cells. Immunity 2007, 27, 825–827. [Google Scholar] [CrossRef] [PubMed]
- MacMurray, E.; Barr, M.; Bruce, A.; Epp, L.; Zwollo, P. Alternative splicing of the trout pax5 gene and identification of novel b cell populations using pax5 signatures. Dev. Comp. Immunol. 2013, 41, 270–281. [Google Scholar]
- Anspach, J.; Poulsen, G.; Kaattari, I.; Pollock, R.; Zwollo, P. Reduction in DNA binding activity of the transcription factor pax-5a in b lymphocytes of aged mice. J. Immunol. 2001, 166, 2617–2626. [Google Scholar] [PubMed]
- Arseneau, J.R.; Laflamme, M.; Lewis, S.M.; Maicas, E.; Ouellette, R.J. Multiple isoforms of pax5 are expressed in both lymphomas and normal b-cells. Br. J. Haematol. 2009, 147, 328–338. [Google Scholar] [PubMed]
- Sadakane, Y.; Zaitsu, M.; Nishi, M.; Sugita, K.; Mizutani, S.; Matsuzaki, A.; Sueoka, E.; Hamasaki, Y.; Ishii, E. Expression and production of aberrant pax5 with deletion of exon 8 in b-lineage acute lymphoblastic leukaemia of children. Br. J. Haematol. 2007, 136, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Andreassi, C.; Riccio, A. To localize or not to localize: Mrna fate is in 3’utr ends. Trends Cell Biol. 2009, 19, 465–474. [Google Scholar] [PubMed]
- Sachs, A. The role of poly(a) in the translation and stability of mrna. Curr. Opin. Cell Biol. 1990, 2, 1092–1098. [Google Scholar]
- Tian, B.; Hu, J.; Zhang, H.; Lutz, C. A large-scale analysis of mrna polyadenylation of human and mouse genes. Nucleic Acids Res. 2005, 33, 201–212. [Google Scholar] [CrossRef]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating cells express mrnas with shortened 3’ untranslated regions and fewer microrna target sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef]
- Mayr, C.; Bartel, D. Widespread shortening of 3′utrs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009, 138, 673–684. [Google Scholar] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular rnas are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Lu, M. Circular rna: Functions, applications and prospects. ExRNA 2020, 2, 1. [Google Scholar]
- Santer, L.; Bär, C.; Thum, T. Circular rnas: A novel class of functional rna molecules with a therapeutic perspective. Mol. Ther. 2019, 27, 1350–1363. [Google Scholar] [PubMed]
- Panda, A.C. Circular rnas act as mirna sponges. Circ. RNAs 2018, 1087, 67–79. [Google Scholar]
- Prats, A.-C.; David, F.; Diallo, L.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular rna, the key for translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y. Extensive translation of circular rnas driven by n6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar]
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights into the role of circrnas: Biogenesis, characterization, functional, and clinical impact in human malignancies. Front. Cell Dev. Biol. 2021, 9, 617281. [Google Scholar]
- Fontemaggi, G.; Turco, C.; Esposito, G.; Di Agostino, S. New molecular mechanisms and clinical impact of circrnas in human cancer. Cancers 2021, 13, 3154. [Google Scholar]
- King, J.K.; Ung, N.M.; Paing, M.H.; Contreras, J.R.; Alberti, M.O.; Fernando, T.R.; Zhang, K.; Pellegrini, M.; Rao, D.S. Regulation of marginal zone b-cell differentiation by microrna-146a. Front. Immunol. 2016, 7, 670. [Google Scholar]
- Lai, M.; Gonzalez-Martin, A.; Cooper, A.B.; Oda, H.; Jin, H.Y.; Shepherd, J.; He, L.; Zhu, J.; Nemazee, D.; Xiao, C. Regulation of b-cell development and tolerance by different members of the mir-17 approximately 92 family micrornas. Nat. Commun. 2016, 7, 12207. [Google Scholar]
- Psathas, J.N.; Doonan, P.J.; Raman, P.; Freedman, B.D.; Minn, A.J.; Thomas-Tikhonenko, A. The myc-mir-17-92 axis amplifies b-cell receptor signaling via inhibition of itim proteins: A novel lymphomagenic feed-forward loop. Blood 2013, 122, 4220–4229. [Google Scholar] [PubMed]
- Dal Bo, M.; Bomben, R.; Hernández, L.; Gattei, V. The myc/mir-17-92 axis in lymphoproliferative disorders: A common pathway with therapeutic potential. Oncotarget 2015, 6, 19381–19392. [Google Scholar] [PubMed]
- Kovac, C.R.; Emelyanov, A.; Singh, M.; Ashouian, N.; Birshtein, B.K. Bsap (pax5)-importin alpha 1 (rch1) interaction identifies a nuclear localization sequence. J. Biol Chem. 2000, 275, 16752–16757. [Google Scholar] [PubMed]
- Tiacci, E.; Pileri, S.; Orleth, A.; Pacini, R.; Tabarrini, A.; Frenguelli, F.; Liso, A.; Diverio, D.; Lo-Coco, F.; Falini, B. Pax5 expression in acute leukemias: Higher b-lineage specificity than cd79a and selective association with t(8;21)-acute myelogenous leukemia. Cancer Res. 2004, 64, 7399–7404. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Choi, J.E.; She, C.J.; Hwang, S.M.; Shin, H.Y.; Ahn, H.S.; Yoon, S.S.; Kim, B.K.; Park, M.H.; Lee, D.S. Pax5 deletion is common and concurrently occurs with cdkn2a deletion in b-lineage acute lymphoblastic leukemia. Blood Cells Mol. Dis. 2011, 47, 62–66. [Google Scholar] [PubMed]
- Erickson, P.; Gao, J.; Chang, K.S.; Look, T.; Whisenant, E.; Raimondi, S.; Lasher, R.; Trujillo, J.; Rowley, J.; Drabkin, H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, aml1/eto, with similarity to drosophila segmentation gene, runt. Blood 1992, 80, 1825–1831. [Google Scholar]
- Russell, L.J.; Akasaka, T.; Majid, A.; Sugimoto, K.J.; Loraine Karran, E.; Nagel, I.; Harder, L.; Claviez, A.; Gesk, S.; Moorman, A.V.; et al. T(6;14)(p22;q32): A new recurrent igh@ translocation involving id4 in b-cell precursor acute lymphoblastic leukemia (bcp-all). Blood 2008, 111, 387–391. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasri Nasrabadi, P.; Martin, D.; Gharib, E.; Robichaud, G.A. The Pleiotropy of PAX5 Gene Products and Function. Int. J. Mol. Sci. 2022, 23, 10095. https://doi.org/10.3390/ijms231710095
Nasri Nasrabadi P, Martin D, Gharib E, Robichaud GA. The Pleiotropy of PAX5 Gene Products and Function. International Journal of Molecular Sciences. 2022; 23(17):10095. https://doi.org/10.3390/ijms231710095
Chicago/Turabian StyleNasri Nasrabadi, Parinaz, Danick Martin, Ehsan Gharib, and Gilles A. Robichaud. 2022. "The Pleiotropy of PAX5 Gene Products and Function" International Journal of Molecular Sciences 23, no. 17: 10095. https://doi.org/10.3390/ijms231710095
APA StyleNasri Nasrabadi, P., Martin, D., Gharib, E., & Robichaud, G. A. (2022). The Pleiotropy of PAX5 Gene Products and Function. International Journal of Molecular Sciences, 23(17), 10095. https://doi.org/10.3390/ijms231710095





