Abstract
Background: Circadian rhythm is associated with the aging process and sex differences; however, how age and sex can change circadian regulation systems remains unclear. Thus, we aimed to evaluate age- and sex-related changes in gene expression and identify sex-specific target molecules that can regulate aging. Methods: Rat livers were categorized into four groups, namely, young male, old male, young female, and old female, and the expression of several genes involved in the regulation of the circadian rhythm was confirmed by in silico and in vitro studies. Results: Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses showed that the expression of genes related to circadian rhythms changed more in males than in females during liver aging. In addition, differentially expressed gene analysis and quantitative real-time polymerase chain reaction/western blotting analysis revealed that Nr1d1 and Nr1d2 expression was upregulated in males during liver aging. Furthermore, the expression of other circadian genes, such as Arntl, Clock, Cry1/2, Per1/2, and Rora/c, decreased in males during liver aging; however, these genes showed various gene expression patterns in females during liver aging. Conclusions: Age-related elevation of Nr1d1/2 downregulates the expression of other circadian genes in males, but not females, during liver aging. Consequently, age-related upregulation of Nr1d1/2 may play a more crucial role in the change in circadian rhythms in males than in females during liver aging.
1. Introduction
Aging is associated with a decline and molecular changes in circadian rhythms via age-associated pathways, which aggravate the aging process [,,]. In the literature, transcriptomic data confirmed changes in circadian gene expression in various organs of several aged species []. In addition, the disruption and dysfunction of circadian rhythms cause unbalanced homeostasis and age-associated diseases [,]. Therefore, malfunctions in circadian rhythms can adversely affect human health and lifespan, indicating that investigations of the circadian system in the aging process are necessary to promote healthy aging and longevity [,,]. These studies suggest that understanding the role of the circadian rhythm in the aging process is important.
Sex has been reported to be one of the factors on the modulation of the circadian rhythm [,], which indicates that sex differences are related to differences in sleep patterns, circadian timing, and physiology [,]. It is well known that light and food intake are the major cues that entrain circadian rhythm [,,,,]. Moreover, sex is associated with changes in the circadian system when exposed to light and food intake [,,,,]. In addition, sex differences in circadian rhythms contribute to the regulation of other biological pathways. For example, sex differences in circadian gene expression cause the infection response and inflammation in males and females to differ [,]. Furthermore, circadian genes were reported to be involved in metabolic regulation in a sex-different manner []. These studies indicate that an understanding of sex differences in circadian rhythms is essential to alleviate various age-related problems; however, the results of studies on sex differences associated with age-related changes in the circadian rhythms remain inconclusive.
The regulation of circadian rhythms has been demonstrated. In mammals, aryl hydrocarbon receptor nuclear translocators, such as (Arntl or Bmal1) and the clock circadian regulator (Clock), organize heterodimers to promote the expression of cryptochrome circadian regulators (Cry), period circadian regulators (Per), and other circadian genes [,]. Cry and Per also organize heterodimers and repress the Arntl–Clock complex, which serves as a negative feedback loop in circadian rhythms [,]. In addition, nuclear receptor subfamily 1 group D member 1 (Nr1d1), nuclear receptor subfamily 1 group D member 2 (Nr1d2), RAR-related orphan receptor A (Rora), and RAR-related orphan receptor C (Rorc) are transcription factors that bind retinoic acid-related orphan receptor binding elements (ROREs); however, Nr1d1 and Nr1d2 serve as repressors of ROREs and inhibit the expression of Arntl and Clock, thereby establishing another negative feedback loop in the circadian rhythm. Rora and Rorc form a positive feedback loop to induce the expression of Arntl and Clock [,,]. These studies have demonstrated that circadian genes such as Nr1d1/2, Arntl, Clock, Cry 1/2, Per1/2, and Rora/c construct regulatory systems with several feedback loops; however, how age and sex can change these circadian modulation systems remains unclear.
Therefore, in this study, we analyzed transcriptomic data of rat livers to examine age- and sex-differential expression of the genes and pathways using a systems-biological methodology. Next, we identified expression changes in the age- and sex-differential genes Nr1d1 and Nr1d2, which are involved in the circadian rhythm of male and female rats during liver aging. Finally, we validated the sex-specific expression of Nr1d1/2 and other circadian genes during liver aging.
2. Results
2.1. Age- and Sex-Related Changes in Gene Expression in Rat Liver
To determine the differences in gene expression according to age and sex in the liver, we separated RNA-Seq data from the liver tissues of rats into four groups: YM, OM, YF, and OF (young: aged 5 months, old: aged 20 months). Furthermore, DEGs were calculated after comparing the RNA-Seq data of the groups (OM vs. YM, OF vs. YF, OM vs. OF, and YM vs. YF) (Figure 1). We identified 285 upregulated genes and 161 downregulated genes in OM compared with YM, and 244 upregulated genes and 123 downregulated genes were identified in OF compared with YF. Furthermore, we identified 271 upregulated genes and 176 downregulated genes that were altered in OM compared with OF, and 221 upregulated genes and 186 downregulated genes were identified in YM compared with YF (Figure 2). Our data revealed age- and sex-related changes in gene expression during liver aging.
Figure 1.
Study designs for transcriptomic analysis of age and sex differences. RNA-Seq data of rat liver tissues were categorized into four groups, and DEGs were calculated by two groups. OM, old male; YM, young male; OF, old female; YF, young female.
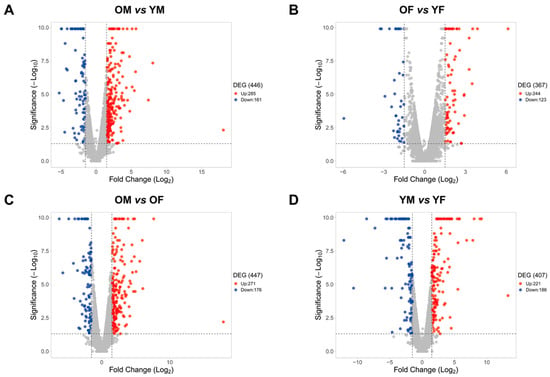
Figure 2.
Age- and sex-differentially expressed genes from RNA-Seq data of (A) OM vs. YM, (B) OF vs. YF, (C) OM vs. OF, and (D) YM vs. YF from SD rats (n = 3 in each group). Red dots represent upregulated genes between two groups. Blue dots represent downregulated genes between two groups. Grey dots represent genes that showed no change between two groups. The criteria for a DEG are |FC| > 1.5 and p-value < 0.05. OM, old male; YM, young male; OF, old female; YF, young female.
2.2. Biological Functions of Genes Whose Expression Was Changed According to Age and Sex in Rat Livers, as Detected by GO Enrichment Analysis
To identify the biological functions of genes whose expression changed according to age and sex, we used The Database for Annotation, Visualization, and Integrated Discovery (DAVID) to perform a GO enrichment analysis of the DEGs for each dataset that was identified in the results of the DEG calculation. Furthermore, we listed the top 15 GOs for each dataset in Figure 3. As a result, upregulated genes in OM vs. YM were involved in 178 significantly enriched GO terms, including immune response and T cell proliferation. Downregulated genes in OM vs. YM were involved in 31 significantly enriched GO terms, including the circadian regulation of gene expression, circadian rhythms, and the oxidation-reduction process (Figure 3A). In addition, upregulated genes in OF vs. YF were involved in 77 significantly enriched GO terms, including innate immune response and leukocyte migration, and downregulated genes in OF vs. YF were involved in 37 significantly enriched GO terms, which included the circadian regulation of gene expression, circadian rhythms, and the oxidation-reduction process (Figure 3B). In addition, upregulated genes in OM vs. OF were involved in 134 significantly enriched GO terms, including immune response and T cell differentiation/proliferation/activation. Downregulated genes in OM vs. OF were involved in 37 significantly enriched GO terms, which included the circadian regulation of gene expression, circadian rhythms, and various metabolic processes (Figure 3C). In YM vs. YF, no significant sex differences were observed in age-related gene expression, indicating that the genes involved in metabolic processes were differentially expressed in YM and YF (Figure 3D). Notably, OM and OF showed an age-related downregulation of circadian rhythm and the circadian regulation of gene expression. Furthermore, OM showed a substantial reduction in circadian rhythm and the circadian regulation of gene expression compared with OF. These data collectively indicate that males and females showed an elevation in immune response as well as a reduction in metabolism and circadian rhythm during liver aging. We focused on circadian-related GOs, which showed more significant age-related downregulation in OM than in OF.
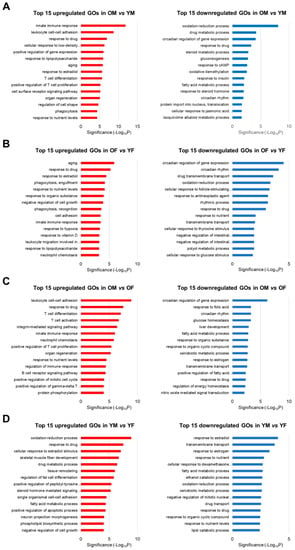
Figure 3.
Top 15 Gene Ontology (GO) enrichment analysis of RNA-Seq data of (A) OM vs. YM, (B) OF vs. YF, (C) OM vs. OF, and (D) YM vs. YF from SD rats (each n = 3). Upregulated GOs of immune response and downregulated GOs of metabolism and circadian rhythm were detected during aging in both sexes. OM, old male; YM, young male; OF, old female; YF, young female.
2.3. Biological Pathways of Genes Whose Expression Was Changed According to Age and Sex in Rat Liver, as Detected by KEGG Enrichment Analysis
To confirm the biological pathways of genes whose expressions changed according to age and sex, we performed KEGG enrichment analyses of the DEGs for each dataset that was identified in the results of the previous analysis and then listed pathways in which the DEGs were involved (Table 1). Inflammatory pathways, including the Jak-STAT signaling pathway and cytokine–cytokine receptor interactions, were upregulated in OM vs. YM, while metabolic pathways and circadian rhythms were downregulated in OM vs. YM. In addition, circadian rhythms were downregulated in OF vs. YF, although the metabolisms showed little age-related differences because the genes related to metabolic pathways were upregulated or downregulated, which indicated that metabolic genes were not changed in an age-specific manner. By contrast, immune pathways, including the T cell receptor signaling pathway and B cell receptor signaling pathway, were upregulated in OM vs. OF, while various metabolic pathways, drug metabolism, and circadian rhythms were downregulated in OM vs. OF. Finally, in YM vs. YF, both upregulated and downregulated pathways were involved in various metabolic processes, indicating that YM and YF showed no significant differences in immunity/inflammation and metabolic/circadian gene expression. These results showed that immune and inflammatory pathways were upregulated, while metabolic and circadian pathways were downregulated, in OM compared with OF. Furthermore, as in the GO analysis, KEGG enrichment analyses revealed that circadian rhythms were downregulated in both older males and females; however, these differences were more pronounced in older males than in older females. These results suggest that the circadian rhythm may play a crucial role in the liver aging of males but not females.

Table 1.
KEGG pathways of the DEGs involved in immunity, inflammation, metabolism, and circadian rhythm from RNA-Seq data in OM vs. YM, OF vs. YF, OM vs. OF, and YM vs. YF.
2.4. Gene Expression of Circadian Genes That Were Differentially Expressed According to Age and Sex in Rat Livers, as Detected by Transcriptomic Analysis
GO and KEGG analyses revealed that the expression of circadian rhythms showed sex differences during aging. Therefore, we selected genes related to the regulation of the circadian rhythm and confirmed their gene expression changes from each dataset. As shown in Table 2, Nr1d1 and Nr1d2 were upregulated in OM compared with YM, and their expression was downregulated in OF compared with YF. Conversely, the expression level of Arntl (or Bmal1) and Clock (or Npas2) decreased in males during aging, and the expression level of these genes was elevated in females during aging. In addition, Cry1, Rora, and Rorc showed age-related decreases only in males. By contrast, Per2 expression was downregulated in both sexes during aging. Furthermore, Cry2 and Per1 showed no age-related differences between men and women. These results reveal that several circadian genes, such as Nr1d1, Nr1d2, Arntl, and Clock, showed sex-specific changes in gene expression during liver aging, which resulted in the differential expression of circadian genes between OM and OF.

Table 2.
Fold change in genes related to circadian rhythm.
2.5. Gene Expression of Circadian Genes That Were Differentially Expressed According to Age and Sex in Rat Livers, as Determined via qRT-PCR and Western Blotting
Transcriptomic analysis revealed that several circadian genes, such as Nr1d1, Nr1d2, Arntl, and Clock, show sex differences in gene expression during the aging process. qRT-PCR was performed to validate the expression of the circadian genes (Figure 4). The expression of Nr1d1 and Nr1d2 was significantly increased in males and decreased in females during aging. Conversely, Arntl was significantly downregulated in males and upregulated in females during the aging process. Clock expression levels also significantly decreased in OM compared with YM, and there was no age-related change in females. In addition, Cry2, Per1, and Per2 were significantly downregulated in both sexes during aging. Cry1, Rora, and Rorc were significantly downregulated in old males, and the genes showed no gene expression changes in old females. Furthermore, western blotting was performed to detect the levels of Nr1d1, Nr1d2, Arntl, and Clock (Figure 5). The protein levels of Nr1d1 and Nr1d2 showed a tendency to be higher in males during aging. On the other hand, the protein level of Arntl was lower in old males than in young males, while its protein level was not changed in females during the aging process. In addition, the protein level of Clock was not changed in both sexes during aging. These results indicate that Nr1d1 and Nr1d2 were upregulated in old males, which downregulated the protein expression of their target gene, Arntl in aged males. Although Nr1d1/2 were not significantly overexpressed in protein levels, their protein expression and activity may be involved in the regulation of Arntl. Collectively, our study suggested that upregulation of Nr1d1 and Nr1d2 may regulate other circadian genes such as Arntl and Clock to a greater degree in males than in females during liver aging. These results highlight that sex differences can affect the expression of circadian genes during the aging of the liver.
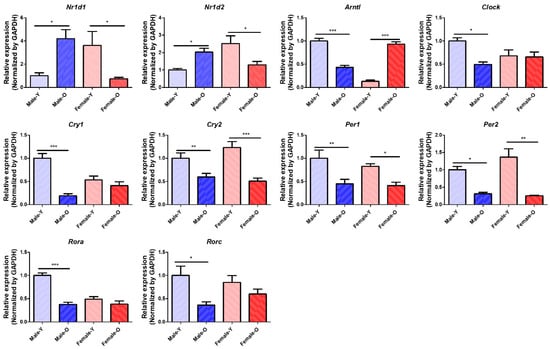
Figure 4.
Relative mRNA expression of circadian genes differentially expressed across sex and age in SD rats (n = 6 per group). Nr1d1, Nr1d2, Arntl, Clock, Cry1, Cry2, Per1, Per2, Rora, and Rorc showed sex differences in their gene expression during liver aging. Nr1d1, nuclear receptor subfamily 1 group D member 1; Nr1d2, nuclear receptor subfamily 1 group D member 2; Arntl, aryl hydrocarbon receptor nuclear translocator like; Clock, clock circadian regulator; Cry1, cryptochrome circadian regulator 1; Cry2, cryptochrome circadian regulator 2; Per1, period circadian regulator 1; Per2, period circadian regulator 1; Rora, RAR-related orphan receptor A; Rorc, RAR-related orphan receptor C; Y, young; O, old; * p < 0.05, ** p < 0.01, and *** p < 0.001 between the two groups.
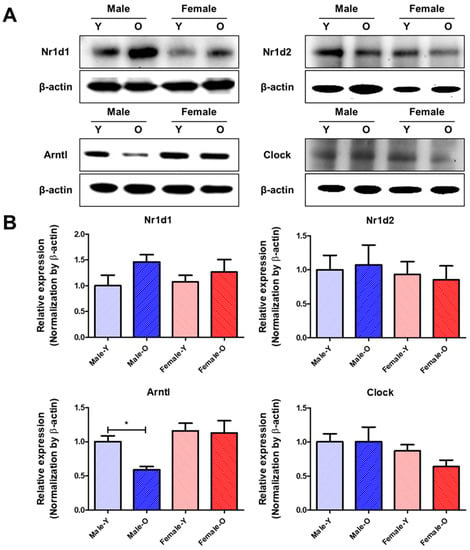
Figure 5.
Protein expressions of Nr1d1/2, Arntl, and Clock which were differentially expressed according to sex and age in SD rats (n = 4 or 6 per group). (A) Representative band images of Nr1d1/2, Arntl, and Clock. (B) Relative expression of Nr1d1/2, Arntl, and Clock. These proteins showed sex differences in their expression during liver aging. Nr1d1, nuclear receptor subfamily 1 group D member 1; Nr1d2, nuclear receptor subfamily 1 group D member 2; Arntl, aryl hydrocarbon receptor nuclear translocator like; Clock, clock circadian regulator; Y, young; O, old. * p < 0.05 between the two groups.
3. Discussion
In this study, we conducted a transcriptomic analysis of RNA-Seq data from rat livers, which revealed that the DEGs, GOs, and KEGG pathways related to inflammation were upregulated and that those related to metabolism were downregulated during aging. Notably, DEGs, GOs, and KEGG pathways related to circadian rhythms were altered in both sexes during the aging process. In addition, transcriptomic and experimental analyses also showed that the expressions of circadian genes Arntl, Clock, Cry1/2, Per1/2, and Rora/c were downregulated in males and that the expression level of Nr1d1/2 was higher in males than in females during liver aging.
Circadian rhythms interact with inflammatory pathways, and disruption induces inflammation and inflammatory diseases [,]. In addition, inflammation disturbs the circadian rhythm and contributes to a vicious cycle of circadian rhythm disruption and inflammation []. Furthermore, circadian genes were demonstrated to be related to the immune system and further regulate the expression of various cytokines, chemokines, and immune-related genes [,]. Our study also agrees with these studies and suggests that downregulation of circadian genes in males can cause male-specific disruptions of the circadian rhythm and worsen inflammatory responses in males. Therefore, further research is necessary to discover inflammatory transcription factors that affect the expression of circadian genes and confirm the sex-specific changes in circadian genes under inflammatory conditions, such as LPS stimulation.
The literature has reported that the circadian rhythm interacts with various metabolic proteins and systemically regulates various metabolic pathways [,,]. In addition, disruption of the circadian rhythm can cause several metabolic problems such as obesity, insulin resistance, chronic inflammation, and hepatic diseases [,,]. These results suggest the importance of circadian rhythms, which are involved in the regulation of the metabolism. Our study also showed that genes related to several metabolic pathways were downregulated, especially in males, during aging. Therefore, we suggest that the male-specific disruption of circadian genes may be related to the male-specific downregulation of metabolic genes.
Nr1d1 regulated the expression of cytokines under LPS stimulation []. Nr1d1 has been reported to alleviate inflammation during inflammatory diseases such as rheumatoid arthritis and hepatitis [,]. Furthermore, Nr1d1 alleviated inflammation under LPS conditions by regulating inflammatory pathways such as the NF-κB signaling pathway [,]. Nr1d1 also affects immunity via interactions with molecules and pathways associated with immune response [,]. In addition, Nr1d1/2 regulated metabolic functions including steroid metabolism [,,,]. Other studies have suggested that elevated Nr1d1 expression may improve metabolism [,]. Furthermore, activating Nr1d1/2 with agonists has been demonstrated to result in anti-inflammatory effects, the regulation of metabolism, and improvements in inflammatory and metabolic diseases [,,,]. Our study revealed that Nr1d1/2 were overexpressed in aged male livers. This finding indicates that these genes may regulate inflammatory and metabolic changes to maintain liver homeostasis. However, this compensation response could disrupt circadian rhythms because chronic or irregularly high expressions of Nr1d1/2 may suppress the expression of other circadian genes, which is consistent with our results. Therefore, molecular mechanisms regulating Nr1d1/2 are necessary to study the male-specific increase in inflammation and deterioration of the metabolism during the aging process.
Except for Nr1d1/2, our data demonstrated that the expression of circadian genes decreased in males during the aging process. Aging induces dysfunction in Arntl and Clock, which worsens the aging process and metabolic diseases [,,]. In addition, the suppression of cellular senescence and inflammatory diseases that are aggravated by Cry1/2 has been demonstrated [,]. Furthermore, the expression level of Per1/2 decreases during aging, and the downregulation of Per1/2 contributes to cytokine production and inflammatory disease pathogenesis [,,]. Studies have indicated that the repression of Rora enhances inflammation and cytokine production and contributes to the deterioration of the metabolism [,,]. Although the role of Rorc, as to whether the protein has proinflammatory or anti-inflammatory effects [,,], remains unclear. It is clear that molecules targeting Rora/c can be effective agents to regulate inflammation- and metabolism-related diseases [,,,,]. Furthermore, Nr1d1/2 and Rora/c compete to bind to ROREs; therefore, modulation of Rora/c may play a key role in regulating Nr1d1/2 activation [].
Our study revealed that the circadian rhythm is affected by sex differences. The effects of hormones related to circadian rhythm occur in a sex-dependent manner []. In addition, sex hormones have been reported to regulate circadian rhythms and behavior []. Furthermore, Nr1d1 is affected by sex hormones. For example, testosterone inhibits Nr1d1 expression, and Nr1d1 is involved in the inhibition of testosterone synthesis [,]. By contrast, progesterone and estradiol induce the expression of Nr1d1, and Nr1d1 inhibits estradiol synthesis [,]. These studies indicate that a decrease in testosterone during male aging induces Nr1d1 expression in OM, and a decrease in female sex hormones during female aging alleviates Nr1d1 expression in old females, which corresponds with our results. Furthermore, upregulated Nr1d1 may deepen aging in males because it can inhibit testosterone synthesis. In addition, calorie restriction, an anti-aging strategy, affects the expression of circadian genes, including Nr1d1 and Rorc, in a sex-dependent manner, which proves the presence of sex differences in circadian rhythms during aging []. Additionally, the disruption of Rora expression has been reported to be more critical in males than females because Rora was highly correlated with its target genes in males []. These studies suggest that males may exhibit greater changes in circadian rhythm during liver aging, which supports our data that showed a more significant downregulation in OM than in OF.
4. Materials and Methods
4.1. Animals
Six young male rats (YM; 5 months old), six old male rats (OM; 20 months old), six young female rats (YF; 5 months old), and six old female rats (OF; 20 months old) were purchased from Samtako [,,,]. All Sprague Dawley (SD) rats were maintained at 23 ± 2 °C with a relative humidity of 60 ± 5% and a 12 h light and dark cycle. The rats were fed ad libitum with a water vehicle and a normal chow diet purchased from Biopia (Gunpo, Gyeonggi-do, Korea). Their tissues were immediately frozen in liquid nitrogen for isolation and analysis. All animal experiments were approved by the Pusan National University Institutional Animal Care and Use Committee. This study complied with all animal testing guidelines (approval number PNU-2019-2282) issued by the Pusan National University.
4.2. RNA Sequencing (RNA-Seq)
The total RNA was isolated from the liver samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and samples from each group were pooled in equal quantities for RNA-Seq (n = 3 in each group). RNA integrity was assessed using an Agilent 2100 BioAnalyzer and an Infinite F200 (concentration value greater than 65 ng/µL, quantity value greater than 1 µg, and RNA integrity number value greater than 6). Libraries were prepared for 150-bp paired-end sequencing using the TruSeq stranded mRNA Sample Preparation Kit (Illumina, CA, USA). mRNA molecules were purified and fragmented from 1 μg of the total RNA using oligo (dT) magnetic beads. Fragmented mRNAs were synthesized as single-stranded cDNAs through random hexamer priming. Double-stranded cDNA was prepared by applying it as a template for second strand synthesis. After the sequential process of end repair, A-tailing, and adapter ligation, cDNA libraries were amplified with a polymerase chain reaction (PCR). The quality of the cDNA libraries was evaluated using an Agilent 2100 BioAnalyzer (Agilent, Santa Clara, CA, USA). They were measured using the KAPA library quantification kit (Kapa Biosystems, Wilmington, MA, USA) according to the manufacturer’s library quantification protocol. After the cluster amplification of denatured templates was performed, paired-end sequencing (2 × 150 bp) was performed using Illumina NovaSeq6000 (Illumina, San Diego, CA, USA).
4.3. Differential Expressed Gene (DEG) Analysis
The adapter sequences and any ends of the reads with a Phred quality score less than 20 were trimmed and reads shorter than 50 bp were removed simultaneously using cutadapt v.2.8 []. Filtered reads were mapped to the reference genome related to the species using the aligner STAR v.2.7.1a, by applying ENCODE standard options (refer to “Alignment” in the “Help” section in the html report) with the “-quant Mode Transcriptome SAM” option for estimating the transcriptome expression level []. Gene expression estimation was performed using RSEM v.1.3.1, considering the direction of the reads that corresponded to the library protocol using the “--strandedness” option. To improve the accuracy of the measurement, we applied the “--estimate-rspd” option []. All other options were set to default values. FPKM and TPM values were calculated to normalize the sequencing depth among the samples. Based on the estimated read counts in the previous step, DEGs were identified using R package TCC v.1.26.0 []. The TCC package applies robust normalization strategies to compare tag count data. The normalization factors were calculated using the iterative edgeR method []. After calculation, we selected DEGs whose expression was changed by 1.5-fold or more and whose p value was less than 0.05. DEGs are presented in Table S1. A volcano plot of the DEGs for each dataset was constructed using the criteria, which are based on the VolcaNoseR website [].
4.4. Enrichment Analysis of the DEGs
Gene Ontology (GO) enrichment analysis was used to analyze the biological functions of the genes, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed to investigate the signaling pathways related to the unique DEGs. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used to perform GO and KEGG enrichment analyses (filtering options: p < 0.05). The results of the GO and KEGG enrichment analyses are listed in Tables S2 and S3. For the overlapping KEGG pathways, we overlapped the enriched KEGG pathways from the datasets and identified common KEGG pathways that were consistently altered in the datasets.
4.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Primers for qRT-PCR were synthesized by Bioneer, Inc. (Daejeon, Korea). Total RNA was isolated from rat liver tissues (20 mg) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) (n = 6 per group) and was reverse-transcribed using the cDNA synthesis kit from GenDEPOT. qRT-PCR analysis was performed to quantify mRNA levels using SYBR Green (Bioneer, Daejeon, Korea) and the CFX Connect System (Bio-Rad Laboratories Inc., Hercules, CA, USA). Primer sequences are listed in Table S4.
4.6. Cytosolic Protein Extraction from Liver Tissues
Frozen liver tissues (150–200 mg) were ground using liquid nitrogen in a mortar and pestle. Ground liver tissues were homogenized in 1 mL hypotonic lysis buffer. Buffer A was composed of KCl (10 mM), MgCl2 (2 mM), dithiothreitol (DTT) (1 mM), EDTA (0.1 mM), PMSF (0.1 mM), pepstatin (1 μM), leupeptin (2 μM), β-glycerophosphate (20 mM), NaF (20 mM), Na3VO4 (2 mM), and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4 (10 mM). The tissue homogenizer was used for 20 s. After homogenates were incubated on ice for 15 min, 125 μL of 10% Nonidet P-40 (NP-40) was added to the homogenates and mixed for 15 s, and the mixture was centrifuged at 14,000× g for 2 min at 4 °C. The supernatants were deemed cytosolic fractions.
4.7. Western Blotting
Total protein extraction samples of the rat liver tissues were boiled for 5 min in a loading buffer containing 0.2% bromophenol blue, 125 mM Tris-HCl, 10% 2-mercaptoethanol, pH 6.8, and 4% sodium dodecyl sulfate. Equal amounts of protein (8–10 μg) were loaded and separated via SDS-PAGE using 10% gels and then transferred to PVDF membranes at 25 V for 10 min using a semidry transfer method. Protein-transferred membranes were soaked in 5% nonfat milk buffer containing 100 mM NaCl, 10 mM Tris (pH 7.5), and 0.1% Tween-20 for 2 h. The membranes were washed four times in TBS-Tween buffer for 8 min and were immunoblotted with specific primary antibodies (1:1000) dilution at 4 °C overnight. This was followed by HRP-conjugated secondary incubation (1:10,000 dilution) according to the manufacturer’s instructions of each primary antibody for 1 h at RT. Antibody labeling was detected using enhanced chemiluminescence in accordance with the manufacturer’s instructions. Molecular weights were determined using wide-range protein markers. The resulting immunoblots were visualized using Chemiluminescent HRP Substrate (Advansta, San Jose, CA, USA), Davinchchemi CAS-400 (Davinch-K, Seoul, Korea), and ImageJ software (NIH, Bethesda, MD, USA), according to the manufacturers’ instructions. Antibodies against Nr1d1 (sc-100910), Nr1d2 (sc-398252), Arntl (sc-365645), and β-Actin (sc-47778) were purchased from Santa Cruz (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Clock antibody (#5157) was obtained from Cell Signaling Technology (Danvers, MA, USA), and secondary antibodies (GTX213110-01, GTX213111-01) were purchased from Genetax (Irvine, CA, USA).
4.8. Statistical Analysis
The Student’s t-test was used to analyze the differences between the two groups. Statistical significance was set at p < 0.05. Statistical analyses were performed using GraphPad Prism 5 (La Jolla, CA, USA).
5. Conclusions
In conclusion, via transcriptomic analysis, this study revealed that the genes involved in most circadian rhythms were downregulated in males, but not in females, during liver aging. In detail, Nr1d1/2 expressions were upregulated in males and downregulated in females, leading to sex-different disruptions of the negative feedback loop in the circadian rhythm via regulation of their target genes, such as Arntl and Clock, during liver aging. Therefore, upregulation of Nr1d1/2 expression may play an important role in disrupting the negative feedback loop in the circadian rhythm of sex-specific male rats during the liver aging process (Figure 6). Our analysis highlighted that male-specific upregulated genes Nr1d1 and Nr1d2 play an important role in the circadian rhythm of male rats, but not in female rats, in the liver aging process.
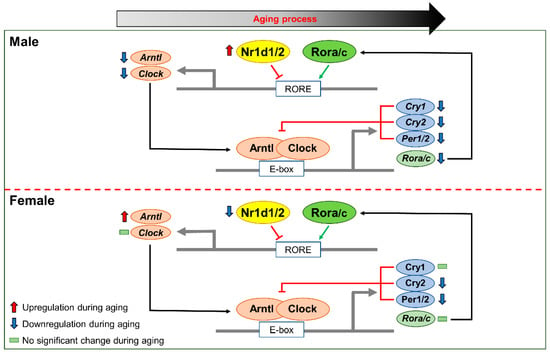
Figure 6.
Changes in the expression of circadian genes due to sex-different expression of Nr1d1/2. During the liver aging process, Nr1d1/2 upregulation suppressed expression of Arntl and Clock and resulted in the downregulation of other circadian genes, such as Cry1/2, Per1/2, and Rora/c. Male-specific upregulation of Nr1d1/2 might be involved in the change in circadian rhythm during male liver aging.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231710032/s1.
Author Contributions
Conceptualization, S.G.N. and H.Y.C.; Data curation, S.G.N.; Formal analysis, S.G.N. and H.J.J.; Methodology, S.G.N., D.P. and H.Y.C.; Software, S.G.N.; Supervision, H.Y.C.; Visualization, S.G.N. and H.J.J.; Writing—original draft, S.G.N.; Writing—review & editing, S.G.N., H.J.J., S.K., R.A., D.H.K., D.P. and H.Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a National Research Foundation (NRF) grant funded by the Korean Government (NRF-2018R1A2A3075425).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kondratova, A.A.; Kondratov, R.V. The circadian clock and pathology of the ageing brain. Nat. Rev. Neurosci. 2012, 13, 325–335. [Google Scholar] [CrossRef]
- Welz, P.S.; Benitah, S.A. Molecular Connections Between Circadian Clocks and Aging. J. Mol. Biol. 2020, 432, 3661–3679. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Rodriguez, V.A.; Rijo-Ferreira, F.; Green, C.B.; Takahashi, J.S. Importance of circadian timing for aging and longevity. Nat. Commun. 2021, 12, 2862. [Google Scholar] [CrossRef] [PubMed]
- Barth, E.; Srivastava, A.; Wengerodt, D.; Stojiljkovic, M.; Axer, H.; Witte, O.W.; Kretz, A.; Marz, M. Age-dependent expression changes of circadian system-related genes reveal a potentially conserved link to aging. Aging 2021, 13, 25694–25716. [Google Scholar] [CrossRef] [PubMed]
- Yanar, K.; Simsek, B.; Cakatay, U. Integration of Melatonin Related Redox Homeostasis, Aging, and Circadian Rhythm. Rejuvenation Res. 2019, 22, 409–419. [Google Scholar] [CrossRef]
- Sharma, A.; Sethi, G.; Tambuwala, M.M.; Aljabali, A.A.A.; Chellappan, D.K.; Dua, K.; Goyal, R. Circadian Rhythm Disruption and Alzheimer’s Disease: The Dynamics of a Vicious Cycle. Curr. Neuropharmacol. 2021, 19, 248–264. [Google Scholar] [CrossRef]
- Hood, S.; Amir, S. The aging clock: Circadian rhythms and later life. J. Clin. Investig. 2017, 127, 437–446. [Google Scholar] [CrossRef]
- De Nobrega, A.K.; Lyons, L.C. Aging and the clock: Perspective from flies to humans. Eur. J. Neurosci. 2020, 51, 454–481. [Google Scholar] [CrossRef]
- Banks, G.; Nolan, P.M.; Peirson, S.N. Reciprocal interactions between circadian clocks and aging. Mamm. Genome 2016, 27, 332–340. [Google Scholar] [CrossRef]
- Nicolaides, N.C.; Chrousos, G.P. Sex differences in circadian endocrine rhythms: Clinical implications. Eur. J. Neurosci. 2020, 52, 2575–2585. [Google Scholar] [CrossRef]
- Hirao, J.; Nishimura, M.; Arakawa, S.; Niino, N.; Mori, K.; Furukawa, T.; Sanbuissho, A.; Manabe, S.; Nishihara, M.; Mori, Y. Sex and circadian modulatory effects on rat liver as assessed by transcriptome analyses. J. Toxicol. Sci. 2011, 36, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.; Silver, R. Sex differences in circadian timing systems: Implications for disease. Front. Neuroendocr. 2014, 35, 111–139. [Google Scholar] [CrossRef] [PubMed]
- Spitschan, M.; Santhi, N.; Ahluwalia, A.; Fischer, D.; Hunt, L.; Karp, N.A.; Levi, F.; Pineda-Torra, I.; Vidafar, P.; White, R. Sex differences and sex bias in human circadian and sleep physiology research. Elife 2022, 11, e65419. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Julius, A.A.; Wen, J.T. Optimization of light exposure and sleep schedule for circadian rhythm entrainment. PLoS ONE 2021, 16, e0251478. [Google Scholar] [CrossRef]
- Emens, J.S.; Burgess, H.J. Effect of Light and Melatonin and Other Melatonin Receptor Agonists on Human Circadian Physiology. Sleep Med. Clin. 2015, 10, 435–453. [Google Scholar] [CrossRef] [PubMed]
- Adafer, R.; Messaadi, W.; Meddahi, M.; Patey, A.; Haderbache, A.; Bayen, S.; Messaadi, N. Food Timing, Circadian Rhythm and Chrononutrition: A Systematic Review of Time-Restricted Eating’s Effects on Human Health. Nutrients 2020, 12, 3770. [Google Scholar] [CrossRef]
- Page, A.J. Gastrointestinal Vagal Afferents and Food Intake: Relevance of Circadian Rhythms. Nutrients 2021, 13, 844. [Google Scholar] [CrossRef]
- Yamamuro, D.; Takahashi, M.; Nagashima, S.; Wakabayashi, T.; Yamazaki, H.; Takei, A.; Takei, S.; Sakai, K.; Ebihara, K.; Iwasaki, Y.; et al. Peripheral circadian rhythms in the liver and white adipose tissue of mice are attenuated by constant light and restored by time-restricted feeding. PLoS ONE 2020, 15, e0234439. [Google Scholar] [CrossRef]
- Roenneberg, T.; Merrow, M. Entrainment of the human circadian clock. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 293–299. [Google Scholar] [CrossRef]
- Kuljis, D.A.; Loh, D.H.; Truong, D.; Vosko, A.M.; Ong, M.L.; McClusky, R.; Arnold, A.P.; Colwell, C.S. Gonadal- and sex-chromosome-dependent sex differences in the circadian system. Endocrinology 2013, 154, 1501–1512. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Loh, D.H.; Colwell, C.S.; Tache, Y.; Reue, K.; Arnold, A.P. Sex differences in diurnal rhythms of food intake in mice caused by gonadal hormones and complement of sex chromosomes. Horm. Behav. 2015, 75, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Okyar, A.; Kumar, S.A.; Filipski, E.; Piccolo, E.; Ozturk, N.; Monje, H.X.; Pala, Z.; Abraham, K.; Gomes, A.; Orman, M.N.; et al. Sex-, feeding-, and circadian time-dependency of P-glycoprotein expression and activity-implications for mechanistic pharmacokinetics modeling. Sci. Rep. 2019, 9, 10505. [Google Scholar] [CrossRef] [PubMed]
- Textoris, J.; Ban, L.H.; Capo, C.; Raoult, D.; Leone, M.; Mege, J.L. Sex-Related Differences in Gene Expression Following Coxiella burnetii Infection in Mice: Potential Role of Circadian Rhythm. PLoS ONE 2010, 5, e12190. [Google Scholar] [CrossRef] [PubMed]
- Eftekhar Vaghefi, S.S.; Mousavi, F.; Khaksari, M.; Asadikaram, G.; Soltani, Z. Sex-related Changes in Circadian Rhythm of Inflammatory and Oxidative Stress Markers in CKD. Iran. J. Kidney Dis. 2021, 15, 351–363. [Google Scholar]
- Bur, I.M.; Cohen-Solal, A.M.; Carmignac, D.; Abecassis, P.Y.; Chauvet, N.; Martin, A.O.; van der Horst, G.T.; Robinson, I.C.; Maurel, P.; Mollard, P.; et al. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J. Biol. Chem. 2009, 284, 9066–9073. [Google Scholar] [CrossRef]
- Cox, K.H.; Takahashi, J.S. Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 2019, 63, R93–R102. [Google Scholar] [CrossRef]
- Trott, A.J.; Menet, J.S. Regulation of circadian clock transcriptional output by CLOCK: BMAL1. PLoS Genet. 2018, 14, e1007156. [Google Scholar] [CrossRef]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef]
- Cao, X.M.; Yang, Y.Y.; Selby, C.P.; Liu, Z.X.; Sancar, A. Molecular mechanism of the repressive phase of the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2020, 118, e2021174118. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solt, L.A.; Kojetin, D.J.; Burris, T.P. The REV-ERBs and RORs: Molecular links between circadian rhythms and lipid homeostasis. Future Med. Chem. 2011, 3, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Li, L. Circadian Clock Regulates Inflammation and the Development of Neurodegeneration. Front. Cell. Infect. Microbiol. 2021, 11, 696554. [Google Scholar] [CrossRef] [PubMed]
- Gombert, M.; Carrasco-Luna, J.; Pin-Arboledas, G.; Codoner-Franch, P. The connection of circadian rhythm to inflammatory bowel disease. Transl. Res. 2019, 206, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Huang, L.; Zhao, J.; Chen, S.; Liu, J.; Li, G. The circadian clock and inflammation: A new insight. Clin. Chim. Acta 2021, 512, 12–17. [Google Scholar] [CrossRef]
- Hergenhan, S.; Holtkamp, S.; Scheiermann, C. Molecular Interactions between Components of the Circadian Clock and the Immune System. J. Mol. Biol. 2020, 432, 3700–3713. [Google Scholar] [CrossRef]
- Curtis, A.M.; Bellet, M.M.; Sassone-Corsi, P.; O’Neill, L.A. Circadian clock proteins and immunity. Immunity 2014, 40, 178–186. [Google Scholar] [CrossRef]
- Huang, W.; Ramsey, K.M.; Marcheva, B.; Bass, J. Circadian rhythms, sleep, and metabolism. J. Clin. Investig. 2011, 121, 2133–2141. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J. Bile acid metabolism and circadian rhythms. Am. J. Physiol. Liver Physiol. 2020, 319, G549–G563. [Google Scholar] [CrossRef]
- Froy, O. The circadian clock and metabolism. Clin. Sci. 2011, 120, 65–72. [Google Scholar] [CrossRef]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, X.; Yin, L. Circadian rhythms in liver physiology and liver diseases. Compr. Physiol. 2013, 3, 917–940. [Google Scholar] [CrossRef] [PubMed]
- Serin, Y.; Acar Tek, N. Effect of Circadian Rhythm on Metabolic Processes and the Regulation of Energy Balance. Ann. Nutr. Metab. 2019, 74, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Huang, Y.; Chu, F.; Ji, S.; Liao, K.; Cui, Z.; Chen, J.; Tang, S. Clock Gene Nr1d1 Alleviates Retinal Inflammation Through Repression of Hmga2 in Microglia. J. Inflamm. Res. 2021, 14, 5901–5918. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Y.; Gao, Y.; Qi, D.; Zhao, L.; Zhao, L.; Liu, C.; Tao, T.; Zhou, C.; Sun, X.; et al. NR1D1 modulates synovial inflammation and bone destruction in rheumatoid arthritis. Cell Death Dis. 2020, 11, 129. [Google Scholar] [CrossRef]
- Pourcet, B.; Zecchin, M.; Ferri, L.; Beauchamp, J.; Sitaula, S.; Billon, C.; Delhaye, S.; Vanhoutte, J.; Mayeuf-Louchart, A.; Thorel, Q.; et al. Nuclear Receptor Subfamily 1 Group D Member 1 Regulates Circadian Activity of NLRP3 Inflammasome to Reduce the Severity of Fulminant Hepatitis in Mice. Gastroenterology 2018, 154, 1449–1464.e20. [Google Scholar] [CrossRef]
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198. [Google Scholar] [CrossRef]
- Griffin, P.; Dimitry, J.M.; Sheehan, P.W.; Lananna, B.V.; Guo, C.; Robinette, M.L.; Hayes, M.E.; Cedeno, M.R.; Nadarajah, C.J.; Ezerskiy, L.A.; et al. Circadian clock protein Rev-erbalpha regulates neuroinflammation. Proc. Natl. Acad. Sci. USA 2019, 116, 5102–5107. [Google Scholar] [CrossRef]
- Sato, S.; Sakurai, T.; Ogasawara, J.; Takahashi, M.; Izawa, T.; Imaizumi, K.; Taniguchi, N.; Ohno, H.; Kizaki, T. A circadian clock gene, Rev-erbalpha, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J. Immunol. 2014, 192, 407–417. [Google Scholar] [CrossRef]
- Hunter, A.L.; Pelekanou, C.E.; Barron, N.J.; Northeast, R.C.; Grudzien, M.; Adamson, A.D.; Downton, P.; Cornfield, T.; Cunningham, P.S.; Billaud, J.N.; et al. Adipocyte NR1D1 dictates adipose tissue expansion during obesity. Elife 2021, 10, e63324. [Google Scholar] [CrossRef]
- Dai, L.; Zhang, Q.; Shi, J.; Bai, X.; An, X.; Zhang, B.; Zhang, Y.; Zhao, X. The Distribution, Expression Patterns and Functional Analysis of NR1D1 and NR4A2 in the Reproductive Axis Tissues of the Male Tianzhu White Yak. Animals 2021, 11, 3117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, M.; Lu, D.; Wang, S.; Yu, F.; Guo, L.; Wen, S.; Wu, B. REV-ERBalpha Regulates CYP7A1 Through Repression of Liver Receptor Homolog-1. Drug Metab. Dispos. 2018, 46, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Woldt, E.; Sebti, Y.; Solt, L.A.; Duhem, C.; Lancel, S.; Eeckhoute, J.; Hesselink, M.K.; Paquet, C.; Delhaye, S.; Shin, Y.; et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 2013, 19, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Yin, L.; Collins, J.L.; Parks, D.J.; Orband-Miller, L.A.; Wisely, G.B.; Joshi, S.; Lazar, M.A.; Willson, T.M.; Zuercher, W.J. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbalpha. ACS Chem. Biol. 2010, 5, 925–932. [Google Scholar] [CrossRef]
- Solt, L.A.; Wang, Y.; Banerjee, S.; Hughes, T.; Kojetin, D.J.; Lundasen, T.; Shin, Y.; Liu, J.; Cameron, M.D.; Noel, R.; et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012, 485, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Bunger, M.K.; Walisser, J.A.; Sullivan, R.; Manley, P.A.; Moran, S.M.; Kalscheur, V.L.; Colman, R.J.; Bradfield, C.A. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis 2005, 41, 122–132. [Google Scholar] [CrossRef]
- Wyse, C.A.; Coogan, A.N. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res. 2010, 1337, 21–31. [Google Scholar] [CrossRef]
- Marcheva, B.; Ramsey, K.M.; Buhr, E.D.; Kobayashi, Y.; Su, H.; Ko, C.H.; Ivanova, G.; Omura, C.; Mo, S.; Vitaterna, M.H.; et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010, 466, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Su, B.; Mo, L.; Qiu, W.; Ying, J.; Lin, P.; Yang, B.; Li, D.; Wang, D.; Xu, L.; et al. Circadian clock protein CRY1 prevents paclitaxelinduced senescence of bladder cancer cells by promoting p53 degradation. Oncol. Rep. 2021, 45, 1033–1043. [Google Scholar] [CrossRef]
- Bekki, H.; Duffy, T.; Okubo, N.; Olmer, M.; Alvarez-Garcia, O.; Lamia, K.; Kay, S.; Lotz, M. Suppression of circadian clock protein cryptochrome 2 promotes osteoarthritis. Osteoarthr. Cartil. 2020, 28, 966–976. [Google Scholar] [CrossRef]
- Baburski, A.Z.; Sokanovic, S.J.; Bjelic, M.M.; Radovic, S.M.; Andric, S.A.; Kostic, T.S. Circadian rhythm of the Leydig cells endocrine function is attenuated during aging. Exp. Gerontol. 2016, 73, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Morioka, N.; Saeki, M.; Sugimoto, T.; Higuchi, T.; Zhang, F.F.; Nakamura, Y.; Hisaoka-Nakashima, K.; Nakata, Y. Downregulation of the spinal dorsal horn clock gene Per1 expression leads to mechanical hypersensitivity via c-jun N-terminal kinase and CCL2 production in mice. Mol. Cell. Neurosci. 2016, 72, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Nah, S.S.; Chang, S.H.; Kim, H.K.; Kwon, J.T.; Lee, S.; Cho, I.H.; Lee, S.W.; Kim, Y.O.; Hong, S.J.; et al. PER2 is downregulated by the LPS-induced inflammatory response in synoviocytes in rheumatoid arthritis and is implicated in disease susceptibility. Mol. Med. Rep. 2017, 16, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yuan, X.; Li, Y.; Chen, J.; Wu, S.; Jiang, A.; Miao, X.; Shu, Q. Circulating exosomal microRNA-18a-5p accentuates intestinal inflammation in Hirschsprung-associated enterocolitis by targeting RORA. Am. J. Transl. Res. 2021, 13, 4182–4196. [Google Scholar]
- Han, S.; Li, Z.; Han, F.; Jia, Y.; Qi, L.; Wu, G.; Cai, W.; Xu, Y.; Li, C.; Zhang, W.; et al. ROR alpha protects against LPS-induced inflammation by down-regulating SIRT1/NF-kappa B pathway. Arch. Biochem. Biophys. 2019, 668, 1–8. [Google Scholar] [CrossRef]
- Chai, C.; Cox, B.; Yaish, D.; Gross, D.; Rosenberg, N.; Amblard, F.; Shemuelian, Z.; Gefen, M.; Korach, A.; Tirosh, O.; et al. Agonist of RORA Attenuates Nonalcoholic Fatty Liver Progression in Mice via Up-regulation of MicroRNA 122. Gastroenterology 2020, 159, 999–1014.e9. [Google Scholar] [CrossRef]
- El-Karaksy, S.M.; Raafat, H.A.; Abadir, M.N.; Hanna, M.O. Down-regulation of expression of retinoid acid-related orphan receptor C (RORC) in systemic lupus erythematosus. J. Recept. Signal Transduct. 2016, 36, 207–212. [Google Scholar] [CrossRef]
- Billon, C.; Murray, M.H.; Avdagic, A.; Burris, T.P. RORgamma regulates the NLRP3 inflammasome. J. Biol. Chem. 2019, 294, 10–19. [Google Scholar] [CrossRef]
- Nadeem, A.; Al-Harbi, N.O.; Al-Harbi, M.M.; El-Sherbeeny, A.M.; Ahmad, S.F.; Siddiqui, N.; Ansari, M.A.; Zoheir, K.M.A.; Attia, S.M.; Al-Hosaini, K.A.; et al. Imiquimod-induced psoriasis-like skin inflammation is suppressed by BET bromodomain inhibitor in mice through RORC/IL-17A pathway modulation. Pharmacol. Res. 2015, 99, 248–257. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, Z.; Luo, X.; Yang, H.; Li, F.; Wang, N. Retinoid acid receptor-related orphan receptor alpha (RORalpha) regulates macrophage M2 polarization via activation of AMPKalpha. Mol. Immunol. 2016, 80, 17–23. [Google Scholar] [CrossRef]
- Auclair, M.; Roblot, N.; Capel, E.; Feve, B.; Antoine, B. Pharmacological modulation of RORalpha controls fat browning, adaptive thermogenesis, and body weight in mice. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E219–E233. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Lyda, B.; Chang, M.R.; Lauer, J.L.; Solt, L.A.; Burris, T.P.; Kamenecka, T.M.; Griffin, P.R. Identification of SR2211: A potent synthetic RORgamma-selective modulator. ACS Chem. Biol. 2012, 7, 672–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauber, B.P.; Gobbi, A.; Robarge, K.; Zhou, A.H.; Barnard, A.; Cao, J.H.; Deng, Y.Z.; Eidenschenk, C.; Everett, C.; Ganguli, A.; et al. Discovery of imidazo [1,5-a]pyridines and -pyrimidines as potent and selective RORc inverse agonists. Bioorg. Med. Chem. Lett. 2015, 25, 2907–2912. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Su, M.; Wu, X.Z.; Wang, D.Z.; Kang, Y.Y.; Wang, C.G.; Assani, I.; Wang, M.X.; Zhao, S.F.; Lv, S.M.; et al. Discovery of 2H-chromone-4-one based sulfonamide derivatives as potent retinoic acid receptor-related orphan receptor gammat inverse agonists. Eur. J. Med. Chem. 2022, 229, 114065. [Google Scholar] [CrossRef]
- Guillaumond, F.; Dardente, H.; Giguere, V.; Cermakian, N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005, 20, 391–403. [Google Scholar] [CrossRef]
- Rohr, K.E.; Telega, A.; Savaglio, A.; Evans, J.A. Vasopressin regulates daily rhythms and circadian clock circuits in a manner influenced by sex. Horm. Behav. 2021, 127, 104888. [Google Scholar] [CrossRef]
- Iwahana, E.; Karatsoreos, I.; Shibata, S.; Silver, R. Gonadectomy reveals sex differences in circadian rhythms and suprachiasmatic nucleus androgen receptors in mice. Horm. Behav. 2008, 53, 422–430. [Google Scholar] [CrossRef]
- Kawamura, M.; Tasaki, H.; Misawa, I.; Chu, G.; Yamauchi, N.; Hattori, M.A. Contribution of testosterone to the clock system in rat prostate mesenchyme cells. Andrology 2014, 2, 225–233. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, J.; Yang, L.; Zhang, H.; Zhang, Y.; Gao, D.; Jiang, H.; Li, Y.; Dong, H.; Ma, T.; et al. Glyphosate exposure attenuates testosterone synthesis via NR1D1 inhibition of StAR expression in mouse Leydig cells. Sci. Total Environ. 2021, 785, 147323. [Google Scholar] [CrossRef]
- Isayama, K.; Chen, H.; Yamauchi, N.; Hattori, M.A. REV-ERBalpha inhibits the PTGS2 expression in bovine uterus endometrium stromal and epithelial cells exposed to ovarian steroids. J. Reprod. Dev. 2014, 60, 362–370. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Zhang, L.; Shi, S.; Zhou, X.; Hu, Y.; Gao, L.; Yang, G.; Pang, W.; Chen, H.; et al. NR1D1 targeting CYP19A1 inhibits estrogen synthesis in ovarian granulosa cells. Theriogenology 2022, 180, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Astafev, A.A.; Patel, S.A.; Kondratov, R.V. Calorie restriction effects on circadian rhythms in gene expression are sex dependent. Sci. Rep. 2017, 7, 9716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, V.W.; Sarachana, T.; Sherrard, R.M.; Kocher, K.M. Investigation of sex differences in the expression of RORA and its transcriptional targets in the brain as a potential contributor to the sex bias in autism. Mol. Autism 2015, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Gardner, R.S.; Newman, L.A.; Mohler, E.G.; Tunur, T.; Gold, P.E.; Korol, D.L. Aging is not equal across memory systems. Neurobiol. Learn. Mem. 2020, 172, 107232. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yu, S.H.; Kim, S.-O. Decreased urothelial expression of caveolin 1 and 2 in aging rats showing detrusor overactivity: Potential association with aging bladder. Investig. Clin. Urol. 2021, 62, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.; Pearl, C.A. Effects of Estrogen Treatment on Aging in the Rat Epididymis. Anat. Rec. 2019, 302, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Andreollo, N.A.; Santos, E.F.; Araujo, M.R.; Lopes, L.R. Rat’s age versus human’s age: What is the relationship? Arq. Bras. Cir. Dig. 2012, 25, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Sun, J.Q.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).