Expression of MicroRNAs in Sepsis-Related Organ Dysfunction: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Search Criteria and Critical Appraisal
3. Results
3.1. Search Results and Included Studies
3.2. Risk of Bias
3.3. Expression of miRNAs in Sepsis-Related Organ Dysfunction
3.3.1. miRNA Expression in Sepsis-Related Brain Dysfunction
3.3.2. miRNA Expression in Sepsis-Related Heart Dysfunction
3.3.3. miRNA Expression in Blood during Sepsis
3.3.4. miRNA Expression in Sepsis-Related Lung Dysfunction
3.3.5. miRNA Expression in Sepsis-Related Liver Dysfunction
3.3.6. miRNA Expression in Sepsis-Related Kidney Dysfunction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AIFM1 | Apoptosis-inducing mitochondrial factor |
| AKAP1 | A-kinase anchor protein 1 |
| AKT1 | Serine/Threonine Kinase 1 |
| AKT2 | Serine/Threonine Kinase 2 |
| AKT3 | Serine/Threonine Kinase 3 |
| AQP1 | Aquaporin 1 |
| ARG1 | Arginase 1 |
| ARRB2 | Arrestin Beta 2 |
| ATG4B | Autophagy-Related 4B |
| BRD4 | Bromodomain-containing protein 4 |
| C/EBPβ | CCAAT-enhancer-binding protein beta |
| CASP3 | Caspase 3 |
| CAV2 | Caveolin-2; |
| CD64 | Cluster of Differentiation 64 |
| CDK6 | Cyclin-Dependent Kinase 6 |
| CUL3 | Cullin 3; |
| CXCL12 | C-X-C Motif Chemokine Ligand 12 |
| CYP2E1 | Cytochrome P450 2E1 |
| DUSP1 | Dual-Specificity Phosphatase 1 |
| DUSP7 | Dual-Specificity Phosphatase 7 |
| EGFL7 | EGF-Like Domain Multiple 7 |
| EIF4E | Eukaryotic Initiation Factor 4E |
| ELANE | Elastase, Neutrophil-Expressed |
| FBXW7 | F-Box And WD Repeat Domain Containing 7 |
| Fli-1 | Friend leukemia integration 1 |
| FOSL2 | FOS-Like 2 |
| FOXO1 | Forkhead Box O1 |
| FOXO3A | Forkhead Box O3A |
| HDAC4 | Histone Deacetylase 4 |
| HIF-1α | Hypoxia-inducible factor 1-alpha |
| HMGA2 | High-mobility group AT-hook 2 |
| HMGB1 | High mobility group box 1 |
| HO-1 | Heme oxygenase-1 |
| HSPA4 | Heat shock 70 kDa protein 4 |
| ICAM1 | Intercellular Adhesion Molecule 1 |
| IKKβ | Inhibitor of nuclear factor kappa-B kinase subunit beta |
| IL2RB | Interleukin 2 Receptor Subunit Beta |
| ILF3 | Interleukin Enhancer-Binding Factor 3 |
| IRAK1 | Interleukin 1 Receptor-Associated Kinase 1 |
| IRF2BP2 | Interferon regulatory factor 2-binding protein 2 |
| JAK | Janus Kinase |
| JAK2 | Janus Kinase 2 |
| JNK | c-Jun N-terminal kinase |
| LCN2 | Lipocalin-2; |
| LPCAT3 | Lysophosphatidylcholine acyltransferase 3 |
| LRP1 | Low-density lipoprotein receptor-related protein 1 |
| LRRFIP1 | LRR Binding FLII Interacting Protein 1 |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 |
| MAPK14 | Mitogen-Activated Protein Kinase 14 |
| MCPIP1 | Monocyte Chemotactic Protein-Induced Protein 1 |
| MD-2 | Myeloid Differentiation factor 2 |
| MFG-E8 | Milk fat globule-EGF factor 8 |
| MRP4 | Multidrug resistance protein 4 |
| mTOR | Mammalian Target Of Rapamycin |
| MyD88 | Myeloid differentiation primary response 88 |
| NAMPT | Nicotinamide Phosphoribosyltransferase |
| NCALD | Neurocalcin Delta |
| NFI-A | Nuclear Factor I A |
| NF-κB | Nuclear Factor kappa B |
| NLRP3 | NLR family pyrin domain containing 3 |
| Notch1 | Neurogenic locus notch homolog protein 1 |
| NOX4 | NADPH oxidase 4 |
| NOX5 | NADPH oxidase 5 |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| p38 MAPK | p38 mitogen-activated protein kinase |
| PDK4 | Pyruvate dehydrogenase lipoamide kinase isozyme 4 |
| Pea15a | Proliferation and apoptosis adaptor protein 15A |
| Peli2 | Pellino E3 Ubiquitin Protein Ligase Family Member 2 |
| PI3K | Phosphoinositide 3-kinases |
| PIAS1 | Protein Inhibitor Of Activated STAT 1 |
| PIK3C3 | Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 |
| PPAR γ | Peroxisome proliferator-activated receptor gamma |
| PPARGC1A | PPARG Coactivator 1 Alpha |
| PTEN | Phosphatase and tensin homolog |
| PTENP1 | Hosphatase and tensin homolog pseudogene 1 |
| RIP1 | Receptor-interacting serine/threonine-protein kinase 1 |
| ROCK1 | Rho-associated kinase 1 |
| ROCK2 | Rho-associated kinase 2 |
| RUNX2 | Runt-related transcription factor 2 |
| S1PR1 | Sphingosine-1-phosphate receptor 1 |
| SELP | Selectin P |
| SEMA3A | Semaphorin 3A |
| SHIP1 | Src homology 2 domain containing inositol polyphosphate 5-phosphatase 1 |
| SIRT1 | Sirtuin 1 |
| SMAD2 | Small Mother Against Decapentaplegic 2 |
| SMAD3 | Small Mother Against Decapentaplegic 3 |
| SOCS1 | Suppressor of cytokine signaling 1 |
| SOCS2 | Suppressor of cytokine signaling 2 |
| SOCS6 | Suppressor of cytokine signaling 6 |
| SORBS2 | Sorbin and SH3 Domain Containing 2 |
| SOX6 | SRY-Box Transcription Factor 6 |
| STAT3 | Signal transducer and activator of transcription 3 |
| STAT4 | Signal transducer and activator of transcription 4 |
| STX2 | Syntaxin 2 |
| TAB2 | TGF-beta activated kinase 1 (MAP3K7)-binding protein 2 |
| TAB3 | TGF-beta activated kinase 1 (MAP3K7)-binding protein 3 |
| TAK1 [MAP3K7] | Mitogen-activated protein kinase kinase kinase 7 |
| TGF-β1 | Transforming Growth Factor Beta Receptor 1 |
| TGF-β2 | Transforming Growth Factor Beta Receptor 2 |
| TGIF1 | TGFB-Induced Factor Homeobox 1 |
| THBS2 | Thrombospondin 2 |
| TLR4 | Toll-Like Receptor 4 |
| TLR6 | Toll-Like Receptor 6 |
| TNFAIP3 | TNF alpha-induced protein 3 |
| TOP2A | DNA Topoisomerase II Alpha |
| TRAF6 | TNF Receptor Associated Factor 6 |
| TREM1 | Triggering Receptor Expressed On Myeloid Cells 1 |
| TRIM37 | Tripartite Motif Containing 37 |
| TRIM8 | Tripartite Motif Containing 8 |
| TRPM7 | Transient Receptor Potential Cation Channel Subfamily M Member 7 |
| TUG1 | Taurine Upregulated 1 |
| USP13 | Ubiquitin Specific Peptidase 13 |
| VCAM1 | Vascular Cell Adhesion Molecule 1 |
| VOPP1 | VOPP1 WW Domain-Binding Protein |
| WNT1 | Wnt family member 1 |
| WNT5A | Wnt Family Member 5A |
| XIAP | X-Linked Inhibitor Of Apoptosis |
| XIST | X-inactive-specific transcript |
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Intensiv. Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef]
- Fleischmann-Struzek, C.; Mellhammar, L.; Rose, N.; Cassini, A.; Rudd, K.E.; Schlattmann, P.; Allegranzi, B.; Reinhart, K. Incidence and mortality of hospital- and ICU-treated sepsis: Results from an updated and expanded systematic review and meta-analysis. Intensiv. Care Med. 2020, 46, 1552–1562. [Google Scholar] [CrossRef]
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States—An Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and clinical management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef]
- Pool, R.; Gomez, H.; Kellum, J.A. Mechanisms of Organ Dysfunction in Sepsis. Crit. Care Clin. 2018, 34, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, M.; Evans, L.; Levy, M.; Rhodes, A. Sepsis and septic shock. Lancet 2018, 392, 75–87. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Healthcare Interventions: Explanation and Elaboration. BMJ (Clin. Res. Ed.) 2009, 339, b2700. [Google Scholar] [CrossRef]
- Hopkins, R.O. Acquired Brain Injury Following Sepsis. Crit. Care Med. 2019, 47, 1658–1659. [Google Scholar] [CrossRef]
- Boudreau, R.; Jiang, P.; Gilmore, B.L.; Spengler, R.; Tirabassi, R.; Nelson, J.A.; Ross, C.A.; Xing, Y.; Davidson, B.L. Transcriptome-wide Discovery of microRNA Binding Sites in Human Brain. Neuron 2014, 81, 294–305. [Google Scholar] [CrossRef]
- Wang, H.-F.; Li, Y.; Wang, Y.-Q.; Li, H.-J.; Dou, L. MicroRNA-494-3p alleviates inflammatory response in sepsis by targeting TLR6. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2971–2977. [Google Scholar]
- Tong, L.; Tang, C.; Cai, C.; Guan, X. Upregulation of the microRNA rno-miR-146b-5p may be involved in the development of intestinal injury through inhibition of Kruppel-like factor 4 in intestinal sepsis. Bioengineered 2020, 11, 1334–1349. [Google Scholar] [CrossRef]
- Dong, R.; Hu, D.; Li, Q.; Xu, T.; Sui, Y.; Qu, Y. Protective effects of microRNA-181b on aged rats with sepsis-induced hippocampus injury in vivo. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2019, 31, 857–861. [Google Scholar] [CrossRef]
- Chen, S.-L.; Cai, G.-X.; Ding, H.-G.; Liu, X.-Q.; Wang, Z.-H.; Jing, Y.-W.; Han, Y.-L.; Jiang, W.-Q.; Wen, M.-Y. JAK/STAT signaling pathway-mediated microRNA-181b promoted blood-brain barrier impairment by targeting sphingosine-1-phosphate receptor 1 in septic rats. Ann. Transl. Med. 2020, 8, 1458. [Google Scholar] [CrossRef]
- Yu, J.; Chen, J.; Yang, H.; Chen, S.; Wang, Z. Overexpression of miR-200a-3p promoted inflammation in sepsis-induced brain injury through ROS-induced NLRP3. Int. J. Mol. Med. 2019, 44, 1811–1823. [Google Scholar] [CrossRef]
- Visitchanakun, P.; Tangtanatakul, P.; Trithiphen, O.; Soonthornchai, W.; Wongphoom, J.; Tachaboon, S.; Srisawat, N.; Leelahavanichkul, A. Plasma miR-370-3P as a Biomarker of Sepsis-Associated Encephalopathy, the Transcriptomic Profiling Analysis of Microrna-Arrays from Mouse Brains. Shock 2020, 54, 347–357. [Google Scholar] [CrossRef]
- Kim, B.; Guaregua, V.; Chen, X.; Zhao, C.; Yeow, W.; Berg, N.K.; Eltzschig, H.K.; Yuan, X. Characterization of a Murine Model System to Study MicroRNA-147 During Inflammatory Organ Injury. Inflammation 2021, 44, 1426–1440. [Google Scholar] [CrossRef]
- Nong, A.; Li, Q.; Huang, Z.; Xu, Y.; He, K.; Jia, Y.; Cen, Z.; Liao, L.; Huang, Y. MicroRNA miR-126 attenuates brain injury in septic rats via NF-κB signaling pathway. Bioengineered 2021, 12, 2639–2648. [Google Scholar] [CrossRef]
- Rani, A.; Barter, J.; Kumar, A.; Stortz, J.A.; Hollen, M.; Nacionales, D.; Moldawer, L.L.; Efron, P.A.; Foster, T.C. Influence of age and sex on microRNA response and recovery in the hippocampus following sepsis. Aging 2022, 14, 728–746. [Google Scholar] [CrossRef]
- Zou, L.; He, J.; Gu, L.; Shahror, R.A.; Li, Y.; Cao, T.; Wang, S.; Zhu, J.; Huang, H.; Chen, F.; et al. Brain innate immune response via miRNA-TLR7 sensing in polymicrobial sepsis. Brain Behav. Immun. 2022, 100, 10–24. [Google Scholar] [CrossRef]
- Zhang, C.; Mo, M.; Ding, W.; Liu, W.; Yan, D.; Deng, J.; Luo, X.; Liu, J. High-mobility group box 1 (HMGB1) impaired cardiac excitation–contraction coupling by enhancing the sarcoplasmic reticulum (SR) Ca2+ leak through TLR4–ROS signaling in cardiomyocytes. J. Mol. Cell. Cardiol. 2014, 74, 260–273. [Google Scholar] [CrossRef]
- Pinto, B.B.; Dyson, A.; Umbrello, M.; Carre, J.; Ritter, C.; Clatworthy, I.; Duchen, M.R.; Singer, M. Improved Survival in a Long-Term Rat Model of Sepsis Is Associated with Reduced Mitochondrial Calcium Uptake Despite Increased Energetic Demand. Crit. Care Med. 2017, 45, e840–e848. [Google Scholar] [CrossRef]
- Ince, C.; Mayeux, P.R.; Nguyen, T.; Gomez, H.; Kellum, J.A.; Ospina-Tascón, G.A.; Hernandez, G.; Murray, P.; De Backer, D. The Endothelium in Sepsis. Shock 2016, 45, 259–270. [Google Scholar] [CrossRef]
- Vellinga, N.A.R.; Boerma, E.C.; Koopmans, M.; Donati, A.; Dubin, A.; Shapiro, N.I.; Pearse, R.; Machado, F.R.; Fries, M.; Akarsu-Ayazoglu, T.; et al. International Study on Microcirculatory Shock Occurrence in Acutely Ill Patients*. Crit. Care Med. 2015, 43, 48–56. [Google Scholar] [CrossRef]
- Hollenberg, S.M.; Piotrowski, M.J.; Parrillo, J.E. Nitric oxide synthase inhibition reverses arteriolar hyporesponsiveness to endothelin-1 in septic rats. Am. J. Physiol. Integr. Comp. Physiol. 1997, 272, R969–R974. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Nicholson, D.W. Apoptosis and caspases regulate death and inflammation in sepsis. Nat. Rev. Immunol. 2006, 6, 813–822. [Google Scholar] [CrossRef]
- Brealey, D.; Brand, M.; Hargreaves, I.; Heales, S.; Land, J.; Smolenski, R.; Davies, N.A.; Cooper, C.E.; Singer, M. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet 2002, 360, 219–223. [Google Scholar] [CrossRef]
- Crouser, E.D. Mitochondrial dysfunction in septic shock and multiple organ dysfunction syndrome. Mitochondrion 2004, 4, 729–741. [Google Scholar] [CrossRef]
- Stanzani, G.; Duchen, M.R.; Singer, M. The role of mitochondria in sepsis-induced cardiomyopathy. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2019, 1865, 759–773. [Google Scholar] [CrossRef]
- Hollenberg, S.M.; Singer, M. Pathophysiology of sepsis-induced cardiomyopathy. Nat. Rev. Cardiol. 2021, 18, 424–434. [Google Scholar] [CrossRef]
- Conway-Morris, A.; Wilson, J.; Shankar-Hari, M. Immune Activation in Sepsis. Crit. Care Clin. 2018, 34, 29–42. [Google Scholar] [CrossRef]
- Manetti, A.C.; Maiese, A.; Di Paolo, M.; De Matteis, A.; La Russa, R.; Turillazzi, E.; Frati, P.; Fineschi, V. MicroRNAs and Sepsis-Induced Cardiac Dysfunction: A Systematic Review. Int. J. Mol. Sci. 2020, 22, 321. [Google Scholar] [CrossRef]
- Kumar, A.; Thota, V.; Dee, L.; Olson, J.; Uretz, E.; Parrillo, J.E. Tumor necrosis factor alpha and interleukin 1beta are responsible for in vitro myocardial cell depression induced by human septic shock serum. J. Exp. Med. 1996, 183, 949–958. [Google Scholar] [CrossRef]
- Vasilescu, C.; Rossi, S.; Shimizu, M.; Tudor, S.; Veronese, A.; Ferracin, M.; Nicoloso, M.; Barbarotto, E.; Popa, M.; Stanciulea, O.; et al. MicroRNA Fingerprints Identify miR-150 as a Plasma Prognostic Marker in Patients with Sepsis. PLoS ONE 2009, 4, e7405. [Google Scholar] [CrossRef]
- Roderburg, C.; Luedde, M.; Cardenas, D.V.; Vucur, M.; Scholten, D.; Frey, N.; Koch, A.; Trautwein, C.; Tacke, F.; Luedde, T. Circulating MicroRNA-150 Serum Levels Predict Survival in Patients with Critical Illness and Sepsis. PLoS ONE 2013, 8, e54612. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-F.; Yu, M.-L.; Yu, G.; Bian, J.-J.; Deng, X.-M.; Wan, X.-J.; Zhu, K.-M. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, K.; Chen, W.J.; Feng, D.; Jia, Y.; Xie, L. Serum miR-574-5p. Shock 2012, 37, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Vilanova, D.; Atalar, K.; Delfour, O.; Edgeworth, J.; Ostermann, M.; Hernández-Fuentes, M.; Razafimahatratra, S.; Michot, B.; Persing, D.H.; et al. Genome-Wide Sequencing of Cellular microRNAs Identifies a Combinatorial Expression Signature Diagnostic of Sepsis. PLoS ONE 2013, 8, e75918. [Google Scholar] [CrossRef]
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Pinilla-Vera, M.; Blackwell, T.S.; Baron, R.M.; et al. MicroRNA-181b regulates NF-κB–mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990. [Google Scholar] [CrossRef]
- Li, Y.; Dalli, J.; Chiang, N.; Baron, R.M.; Quintana, C.; Serhan, C.N. Plasticity of Leukocytic Exudates in Resolving Acute Inflammation Is Regulated by MicroRNA and Proresolving Mediators. Immunity 2013, 39, 885–898. [Google Scholar] [CrossRef]
- Tacke, F.; Roderburg, C.; Benz, F.; Cardenas, D.V.; Luedde, M.; Hippe, H.-J.; Frey, N.; Vucur, M.; Gautheron, J.; Koch, A.; et al. Levels of Circulating miR-133a Are Elevated in Sepsis and Predict Mortality in Critically Ill Patients. Crit. Care Med. 2014, 42, 1096–1104. [Google Scholar] [CrossRef]
- Benz, F.; Roy, S.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating MicroRNAs as Biomarkers for Sepsis. Int. J. Mol. Sci. 2016, 17, 78. [Google Scholar] [CrossRef]
- Shankar-Hari, M.; Lord, G.M. How might a diagnostic microRNA signature be used to speed up the diagnosis of sepsis? Expert Rev. Mol. Diagn. 2014, 14, 249–251. [Google Scholar] [CrossRef]
- Ruparelia, N.; Chai, J.T.; Fisher, E.A.; Choudhury, R.P. Inflammatory processes in cardiovascular disease: A route to targeted therapies. Nat. Rev. Cardiol. 2017, 14, 133–144. [Google Scholar] [CrossRef]
- Yende, S.; Linde-Zwirble, W.; Mayr, F.; Weissfeld, L.A.; Reis, S.; Angus, D.C. Risk of Cardiovascular Events in Survivors of Severe Sepsis. Am. J. Respir. Crit. Care Med. 2014, 189, 1065–1074. [Google Scholar] [CrossRef]
- Hu, Y.-F.; Chen, Y.-J.; Lin, Y.-J.; Chen, S.-A. Inflammation and the pathogenesis of atrial fibrillation. Nat. Rev. Cardiol. 2015, 12, 230–243. [Google Scholar] [CrossRef]
- Beltrán-García, J.; Osca-Verdegal, R.; Nácher-Sendra, E.; Cardona-Monzonís, A.; Sanchis-Gomar, F.; Carbonell, N.; Pallardó, F.V.; Lavie, C.J.; García-Giménez, J.L. Role of non-coding RNAs as biomarkers of deleterious cardiovascular effects in sepsis. Prog. Cardiovasc. Dis. 2021, 68, 70–77. [Google Scholar] [CrossRef]
- Wang, X.; Huang, W.; Yang, Y.; Wang, Y.; Peng, T.; Chang, J.; Caldwell, C.C.; Zingarelli, B.; Fan, G.-C. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2014, 1842, 701–711. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, P.; Chen, W.; Feng, D.; Jia, Y.; Xie, L. Serum MicroRNA Signatures Identified by Solexa Sequencing Predict Sepsis Patients’ Mortality: A Prospective Observational Study. PLoS ONE 2012, 7, e38885. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Z.; Wang, H.; Ren, F.; Li, Y.; Zou, S.; Xu, J.; Xie, L. The protective role of miR-223 in sepsis-induced mortality. Sci. Rep. 2020, 10, 17691. [Google Scholar] [CrossRef]
- M’Baya-Moutoula, E.; Louvet, L.; Molinié, R.; Guerrera, I.C.; Cerutti, C.; Fourdinier, O.; Nourry, V.; Gutierrez, L.; Morlière, P.; Mesnard, F.; et al. A multi-omics analysis of the regulatory changes induced by miR-223 in a monocyte/macrophage cell line. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2018, 1864, 2664–2678. [Google Scholar] [CrossRef]
- Xue, W.-L.; Bai, X.; Zhang, L. rhTNFR:Fc increases Nrf2 expression via miR-27a mediation to protect myocardium against sepsis injury. Biochem. Biophys. Res. Commun. 2015, 464, 855–861. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell Survival Responses to Environmental Stresses Via the Keap1-Nrf2-ARE Pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Gao, M.; Wang, X.; Zhang, X.; Ha, T.; Ma, H.; Liu, L.; Kalbfleisch, J.H.; Gao, X.; Kao, R.L.; Williams, D.L.; et al. Attenuation of Cardiac Dysfunction in Polymicrobial Sepsis by MicroRNA-146a Is Mediated via Targeting of IRAK1 and TRAF6 Expression. J. Immunol. 2015, 195, 672–682. [Google Scholar] [CrossRef]
- An, R.; Feng, J.; Xi, C.; Xu, J.; Sun, L. miR-146a Attenuates Sepsis-Induced Myocardial Dysfunction by Suppressing IRAK1 and TRAF6 via Targeting ErbB4 Expression. Oxidative Med. Cell. Longev. 2018, 2018, 7163057. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, L.; Fan, X.; Dong, X.; Zhang, Z.; Fan, W. MicroRNA-146a improves sepsis-induced cardiomyopathy by regulating the TLR-4/NF-κB signaling pathway. Exp. Ther. Med. 2019, 18, 779–785. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Y. MiR-146b protect against sepsis induced mice myocardial injury through inhibition of Notch1. Histochem. J. 2018, 49, 411–417. [Google Scholar] [CrossRef]
- Ma, H.; Wang, X.; Ha, T.; Gao, M.; Liu, L.; Wang, R.; Yu, K.; Kalbfleisch, J.H.; Kao, R.L.; Williams, D.L.; et al. MicroRNA-125b Prevents Cardiac Dysfunction in Polymicrobial Sepsis by Targeting TRAF6-Mediated Nuclear Factor κB Activation and p53-Mediated Apoptotic Signaling. J. Infect. Dis. 2016, 214, 1773–1783. [Google Scholar] [CrossRef]
- Wei, S.; Liu, Q. Long noncoding RNA MALAT1 modulates sepsis-induced cardiac inflammation through the miR-150-5p/NF-κB axis. Int. J. Clin. Exp. Pathol. 2019, 12, 3311–3319. [Google Scholar]
- Zhu, X.G.; Zhang, T.N.; Wen, R.; Liu, C.F. Overexpression of miR-150-5p Alleviates Apoptosis in Sepsis-Induced Myocardial Depression. BioMed Res. Int. 2020, 2020, 3023186. [Google Scholar] [CrossRef]
- Wang, H.; Bei, Y.; Shen, S.; Huang, P.; Shi, J.; Zhang, J.; Sun, Q.; Chen, Y.; Yang, Y.; Xu, T.; et al. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J. Mol. Cell. Cardiol. 2016, 94, 43–53. [Google Scholar] [CrossRef]
- Wang, H.; Bei, Y.; Huang, P.; Zhou, Q.; Shi, J.; Sun, Q.; Zhong, J.; Li, X.; Kong, X.; Xiao, J. Inhibition of miR-155 Protects Against LPS-induced Cardiac Dysfunction and Apoptosis in Mice. Mol. Ther. Nucleic Acids 2016, 5, e374. [Google Scholar] [CrossRef]
- Diao, X.; Sun, S. PMicroRNA-124a regulates LPS-induced septic cardiac dysfunction by targeting STX2. Biotechnol. Lett. 2017, 39, 1335–1342. [Google Scholar] [CrossRef]
- Zheng, G.; Pan, M.; Jin, W.; Jin, G.; Huang, Y. MicroRNA-135a is up-regulated and aggravates myocardial depression in sepsis via regulating p38 MAPK/NF-κB pathway. Int. Immunopharmacol. 2017, 45, 6–12. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, Y.; Shaikh, Z.; Li, H.; Zhang, H.; Caudle, Y.; Zheng, S.; Yan, H.; Hu, D.; Stuart, C.; et al. MicroRNA-155 attenuates late sepsis-induced cardiac dysfunction through JNK and β-arrestin 2. Oncotarget 2017, 8, 47317–47329. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Liu, J.; Dong, S. miRNA-214 Protects Sepsis-Induced Myocardial Injury. Shock 2018, 50, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Sang, Z.; Zhang, P.; Wei, Y.; Dong, S. miR-214-3p Attenuates Sepsis-Induced Myocardial Dysfunction in Mice by Inhibiting Autophagy through PTEN/AKT/mTOR Pathway. BioMed Res. Int. 2020, 2020, 1409038. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Hu, J.; Wang, Z.; Zong, H.; Zhang, L.; Zhang, R.; Sun, L. LncRNA H19 functions as an Aquaporin 1 competitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. Biomed. Pharmacother. 2018, 105, 1183–1191. [Google Scholar] [CrossRef]
- Wang, W.; Li, F.; Sun, Y.; Lei, L.; Zhou, H.; Lei, T.; Xia, Y.; Verkman, A.S.; Yang, B. Aquaporin-1 retards renal cyst development in polycystic kidney disease by inhibition of Wnt signaling. FASEB J. 2015, 29, 1551–1563. [Google Scholar] [CrossRef]
- Wang, C.; Yan, M.; Jiang, H.; Wang, Q.; Guan, X.; Chen, J.; Wang, C. Protective effects of puerarin on acute lung and cerebrum injury induced by hypobaric hypoxia via the regulation of aquaporin (AQP) via NF-κB signaling pathway. Int. Immunopharmacol. 2016, 40, 300–309. [Google Scholar] [CrossRef]
- Tomita, Y.; Dorward, H.; Yool, A.J.; Smith, E.; Townsend, A.R.; Price, T.J.; Hardingham, J.E. Role of Aquaporin 1 Signalling in Cancer Development and Progression. Int. J. Mol. Sci. 2017, 18, 299. [Google Scholar] [CrossRef]
- Tang, B.; Xuan, L.; Tang, M.; Wang, H.; Zhou, J.; Liu, J.; Wu, S.; Li, M.; Wang, X.; Zhang, H. miR-93-3p alleviates lipopolysaccharide-induced inflammation and apoptosis in H9c2 cardiomyocytes by inhibiting toll-like receptor 4. Pathol. Res. Pract. 2018, 214, 1686–1693. [Google Scholar] [CrossRef]
- Yao, Y.; Sun, F.; Lei, M. miR-25 inhibits sepsis-induced cardiomyocyte apoptosis by targetting PTEN. Biosci. Rep. 2018, 38, BSR20171511. [Google Scholar] [CrossRef]
- Wu, P.; Kong, L.; Li, J. MicroRNA-494-3p protects rat cardiomyocytes against septic shock via PTEN. Exp. Ther. Med. 2018, 17, 1706–1716. [Google Scholar] [CrossRef]
- Zhang, H.; Caudle, Y.; Shaikh, A.; Yao, B.; Yin, D. Inhibition of microRNA-23b prevents polymicrobial sepsis-induced cardiac dysfunction by modulating TGIF1 and PTEN. Biomed. Pharmacother. 2018, 103, 869–878. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Q.; Xiao, J.; Lei, J.; Liu, Y.; Xu, H.; Hong, Z. Molecular validation of the precision-cut kidney slice (PCKS) model of renal fibrosis through assessment of TGF-β1-induced Smad and p38/ERK signaling. Int. Immunopharmacol. 2016, 34, 32–36. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Quaggin, S.E.; Vaughan, D.E. Molecular basis of organ fibrosis: Potential therapeutic approaches. Exp. Biol. Med. 2013, 238, 461–481. [Google Scholar] [CrossRef]
- Cao, C.; Zhang, Y.; Chai, Y.; Wang, L.; Yin, C.; Shou, S.; Jin, H. Attenuation of Sepsis-Induced Cardiomyopathy by Regulation of MicroRNA-23b Is Mediated Through Targeting of MyD88-Mediated NF-κB Activation. Inflammation 2019, 42, 973–986. [Google Scholar] [CrossRef]
- Guo, H.; Tang, L.; Xu, J.; Lin, C.; Ling, X.; Lu, C.; Liu, Z. MicroRNA-495 serves as a diagnostic biomarker in patients with sepsis and regulates sepsis-induced inflammation and cardiac dysfunction. Eur. J. Med. Res. 2019, 24, 37. [Google Scholar] [CrossRef]
- Zhu, J.; Lin, X.; Yan, C.; Yang, S.; Zhu, Z. microRNA-98 protects sepsis mice from cardiac dysfunction, liver and lung injury by negatively regulating HMGA2 through inhibiting NF-κB signaling pathway. Cell Cycle 2019, 18, 1948–1964. [Google Scholar] [CrossRef]
- Ouyang, H.; Tan, Y.; Li, Q.; Xia, F.; Xiao, X.; Zheng, S.; Lu, J.; Zhong, J.; Hu, Y. MicroRNA-208-5p regulates myocardial injury of sepsis mice via targeting SOCS2-mediated NF-κB/HIF-1α pathway. Int. Immunopharmacol. 2020, 81, 106204. [Google Scholar] [CrossRef]
- Sun, B.; Luan, C.; Guo, L.; Zhang, B.; Liu, Y. Low expression of microRNA-328 can predict sepsis and alleviate sepsis-induced cardiac dysfunction and inflammatory response. Braz. J. Med. Biol. Res. 2020, 53, e9501. [Google Scholar] [CrossRef]
- Zhang, T.-N.; Yang, N.; Goodwin, J.E.; Mahrer, K.; Li, D.; Xia, J.; Wen, R.; Zhou, H.; Zhang, T.; Song, W.-L.; et al. Characterization of Circular RNA and microRNA Profiles in Septic Myocardial Depression: A Lipopolysaccharide-Induced Rat Septic Shock Model. Inflammation 2019, 42, 1990–2002. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Zhu, Y.; Sun, A.; Meng, T.; Li, H. RETRACTED: Protective role of long noncoding RNA CRNDE in myocardial tissues from injury caused by sepsis through the microRNA-29a/SIRT1 axis. Life Sci. 2020, 255, 117849. [Google Scholar] [CrossRef]
- Song, Y.-X.; Ou, Y.-M.; Zhou, J.-Y. Gracillin inhibits apoptosis and inflammation induced by lipopolysaccharide (LPS) to alleviate cardiac injury in mice via improving miR-29a. Biochem. Biophys. Res. Commun. 2020, 523, 580–587. [Google Scholar] [CrossRef]
- Chen, T.; Zhu, C.; Ye, C. LncRNA CYTOR attenuates sepsis-induced myocardial injury via regulating miR-24/XIAP. Cell Biochem. Funct. 2020, 38, 976–985. [Google Scholar] [CrossRef]
- Sun, F.; Yuan, W.; Wu, H.; Chen, G.; Sun, Y.; Yuan, L.; Zhang, W.; Lei, M. LncRNA KCNQ1OT1 attenuates sepsis-induced myocardial injury via regulating miR-192-5p/XIAP axis. Exp. Biol. Med. 2020, 245, 620–630. [Google Scholar] [CrossRef]
- Wei, J.-L.; Wu, C.-J.; Chen, J.-J.; Shang, F.-T.; Guo, S.-G.; Zhang, X.-C.; Liu, H. LncRNA NEAT1 promotes the progression of sepsis-induced myocardial cell injury by sponging miR-144-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 851–861. [Google Scholar]
- Xing, P.-C.; An, P.; Hu, G.-Y.; Wang, D.-L.; Zhou, M.-J. LncRNA MIAT Promotes Inflammation and Oxidative Stress in Sepsis-Induced Cardiac Injury by Targeting miR-330-5p/TRAF6/NF-κB Axis. Biochem. Genet. 2020, 58, 783–800. [Google Scholar] [CrossRef]
- Han, Y.; Cai, Y.; Lai, X.; Wang, Z.; Wei, S.; Tan, K.; Xu, M.; Xie, H. lncRNA RMRP Prevents Mitochondrial Dysfunction and Cardiomyocyte Apoptosis via the miR-1-5p/hsp70 Axis in LPS-Induced Sepsis Mice. Inflammation 2020, 43, 605–618. [Google Scholar] [CrossRef]
- Li, Z.; Yi, N.; Chen, R.; Meng, Y.; Wang, Y.; Liu, H.; Cao, W.; Hu, Y.; Gu, Y.; Tong, C.; et al. miR-29b-3p protects cardiomyocytes against endotoxin-induced apoptosis and inflammatory response through targeting FOXO3A. Cell. Signal. 2020, 74, 109716. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Tang, L.; Chen, J.; Chen, D.; Wen, W.; Han, F. Inhibition of miR-101-3p protects against sepsis-induced myocardial injury by inhibiting MAPK and NF-κB pathway activation via the upregulation of DUSP1. Int. J. Mol. Med. 2021, 47, 20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, L.; Zhang, J. Protective role of matrine in sepsis-associated cardiac dysfunction through regulating the lncRNA PTENP1/miR-106b-5p axis. Biomed. Pharmacother. 2021, 134, 111112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yu, L.; Sheng, Y. Clinical value and role of microRNA-29c-3p in sepsis-induced inflammation and cardiac dysfunction. Eur. J. Med. Res. 2021, 26, 90. [Google Scholar] [CrossRef]
- Yang, C.; Wen, K. Predictive value and regulatory mechanism of serum miR-499a-5p on myocardial dysfunction in sepsis. J. Cardiothorac. Surg. 2021, 16, 301. [Google Scholar] [CrossRef]
- Henriquez-Camacho, C.; Losa, J. Biomarkers for Sepsis. BioMed Res. Int. 2014, 2014, 547818. [Google Scholar] [CrossRef]
- Limper, M.; de Kruif, M.; Duits, A.; Brandjes, D.; van Gorp, E. The diagnostic role of Procalcitonin and other biomarkers in discriminating infectious from non-infectious fever. J. Infect. 2010, 60, 409–416. [Google Scholar] [CrossRef]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review. Biosens. Bioelectron. 2018, 124–125, 205–215. [Google Scholar] [CrossRef]
- Reddy, B.; Hassan, U.; Seymour, C.; Angus, D.C.; Isbell, T.S.; White, K.; Weir, W.; Yeh, L.; Vincent, A.; Bashir, R. Point-of-care sensors for the management of sepsis. Nat. Biomed. Eng. 2018, 2, 640–648. [Google Scholar] [CrossRef]
- Li, J.-M.; Zhang, H.; Zuo, Y.-J. MicroRNA-218 alleviates sepsis inflammation by negatively regulating VOPP1 via JAK/STAT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5620–5626. [Google Scholar] [CrossRef]
- El-Khier, N.A.; Zaki, M.E.; Alkasaby, N.M. Study of MicroRNA-122 as a Diagnostic Biomarker of Sepsis. Egypt. J. Immunol. 2019, 26, 105–116. [Google Scholar]
- Xue, Z.; Xi, Q.; Liu, H.; Guo, X.; Zhang, J.; Zhang, Z.; Li, Y.; Yang, G.; Zhou, D.; Yang, H.; et al. miR-21 promotes NLRP3 inflammasome activation to mediate pyroptosis and endotoxic shock. Cell Death Dis. 2019, 10, 461. [Google Scholar] [CrossRef]
- Jia, P.; Wu, X.; Dai, Y.; Teng, J.; Fang, Y.; Hu, J.; Zou, J.; Liang, M.; Ding, X. MicroRNA-21 Is Required for Local and Remote Ischemic Preconditioning in Multiple Organ Protection Against Sepsis*. Crit. Care Med. 2017, 45, e703–e710. [Google Scholar] [CrossRef]
- Yao, M.; Cui, B.; Zhang, W.; Ma, W.; Zhao, G.; Xing, L. Exosomal miR-21 secreted by IL-1β-primed-mesenchymal stem cells induces macrophage M2 polarization and ameliorates sepsis. Life Sci. 2021, 264, 118658. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, D.; Gao, J.; Xiang, X.; Hu, X.; Li, S.; Wu, W.; Cai, J.; Tang, C.; Zhang, D.; et al. Discovery and validation of miR-452 as an effective biomarker for acute kidney injury in sepsis. Theranostics 2020, 10, 11963–11975. [Google Scholar] [CrossRef]
- Rocchi, A.; Chiti, E.; Maiese, A.; Turillazzi, E.; Spinetti, I. MicroRNAs: An Update of Applications in Forensic Science. Diagnostics 2020, 11, 32. [Google Scholar] [CrossRef]
- Maiese, A.; Manetti, A.C.; Iacoponi, N.; Mezzetti, E.; Turillazzi, E.; Di Paolo, M.; La Russa, R.; Frati, P.; Fineschi, V. State-of-the-Art on Wound Vitality Evaluation: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 6881. [Google Scholar] [CrossRef]
- Zhou, M.-H.; Zhang, L.; Song, M.-J.; Sun, W.-J. MicroRNA-218 prevents lung injury in sepsis by inhibiting RUNX2. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8438–8446. [Google Scholar] [PubMed]
- Rahmel, T.; Schäfer, S.T.; Frey, U.; Adamzik, M.; Peters, J. Increased circulating microRNA-122 is a biomarker for discrimination and risk stratification in patients defined by sepsis-3 criteria. PLoS ONE 2018, 13, e0197637. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, X.; Li, C.; Sun, G.; Ding, P.; Li, Y.; Yang, P.; Zhang, M.; Wang, L. Identification and Validation of Immune-Related Biomarker Gene and Construction of ceRNA Networks in Septic Cardiomyopathy. Front. Cell. Infect. Microbiol. 2022, 12, 912492. [Google Scholar] [CrossRef]
- Na, L.; Ding, H.; Xing, E.; Zhang, Y.; Gao, J.; Liu, B.; Yu, J.; Zhao, Y. The predictive value of microRNA-21 for sepsis risk and its correlation with disease severity, systemic inflammation, and 28-day mortality in sepsis patients. J. Clin. Lab. Anal. 2020, 34, e23103. [Google Scholar] [CrossRef]
- Lin, R.; Hu, H.; Li, L.; Chen, G.; Luo, L.; Rao, P. The potential of microRNA-126 in predicting disease risk, mortality of sepsis, and its correlation with inflammation and sepsis severity. J. Clin. Lab. Anal. 2020, 34, e23408. [Google Scholar] [CrossRef]
- Zhao, D.; Li, S.; Cui, J.; Wang, L.; Ma, X.; Li, Y. Plasma miR-125a and miR-125b in sepsis: Correlation with disease risk, inflammation, severity, and prognosis. J. Clin. Lab. Anal. 2020, 34, e23036. [Google Scholar] [CrossRef]
- Song, Y.; Dou, H.; Li, X.; Zhao, X.; Li, Y.; Liu, D.; Ji, J.; Liu, F.; Ding, L.; Ni, Y.; et al. Exosomal miR-146a Contributes to the Enhanced Therapeutic Efficacy of Interleukin-1β-Primed Mesenchymal Stem Cells Against Sepsis. Stem Cells 2017, 35, 1208–1221. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yu, L.; Zhang, R.; Zhu, L.; Shen, W. Correlation of microRNA-146a/b with disease risk, biochemical indices, inflammatory cytokines, overall disease severity, and prognosis of sepsis. Medicine 2020, 99, e19754. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, J.; Cao, M.; Han, S.; Liu, Y.; Wang, K.; Han, F.; Li, X.; Jia, Y.; Wang, X.; et al. MicroRNA-146a protects against LPS-induced organ damage by inhibiting Notch1 in macrophage. Int. Immunopharmacol. 2018, 63, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Brudecki, L.; Ferguson, D.A.; McCall, C.E.; El Gazzar, M. MicroRNA-146a and RBM4 form a negative feed-forward loop that disrupts cytokine mRNA translation following TLR4 responses in human THP-1 monocytes. Immunol. Cell Biol. 2013, 91, 532–540. [Google Scholar] [CrossRef]
- Banerjee, S.; Meng, J.; Das, S.; Krishnan, A.; Haworth, J.; Charboneau, R.; Zeng, Y.; Ramakrishnan, S.; Roy, S. Morphine induced exacerbation of sepsis is mediated by tempering endotoxin tolerance through modulation of miR-146a. Sci. Rep. 2013, 3, 1977. [Google Scholar] [CrossRef]
- Möhnle, P.; Schütz, S.V.; van der Heide, V.; Hübner, M.; Luchting, B.; Sedlbauer, J.; Limbeck, E.; Hinske, L.C.; Briegel, J.; Kreth, S. MicroRNA-146a controls Th1-cell differentiation of human CD4+ T lymphocytes by targeting PRKCε. Eur. J. Immunol. 2015, 45, 260–272. [Google Scholar] [CrossRef]
- Paik, S.; Choe, J.H.; Choi, G.-E.; Kim, J.-E.; Kim, J.-M.; Song, G.Y.; Jo, E.-K. Rg6, a rare ginsenoside, inhibits systemic inflammation through the induction of interleukin-10 and microRNA-146a. Sci. Rep. 2019, 9, 4342. [Google Scholar] [CrossRef]
- Zhang, N.; Gao, Y.; Yu, S.; Sun, X.; Shen, K. Berberine attenuates Aβ42-induced neuronal damage through regulating circHDAC9/miR-142-5p axis in human neuronal cells. Life Sci. 2020, 252, 117637. [Google Scholar] [CrossRef]
- Gao, N.G.N.; Dong, L.D.L. MicroRNA-146 regulates the inflammatory cytokines expression in vascular endothelial cells during sepsis. Die Pharm. Int. J. Pharm. Sci. 2017, 72, 700–704. [Google Scholar] [CrossRef]
- Zou, Q.; Zhao, S.; Wu, Q.; Wang, H.; He, X.; Liu, C. Correlation analysis of microRNA-126 expression in peripheral blood lymphocytes with apoptosis and prognosis in patients with sepsis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2020, 32, 938–942. [Google Scholar] [CrossRef]
- Dang, C.P.; Leelahavanichkul, A. Over-expression of miR-223 induces M2 macrophage through glycolysis alteration and attenuates LPS-induced sepsis mouse model, the cell-based therapy in sepsis. PLoS ONE 2020, 15, e0236038. [Google Scholar] [CrossRef]
- Yao, L.; Liu, Z.; Zhu, J.; Li, B.; Chai, C.; Tian, Y. Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. Int. J. Clin. Exp. Pathol. 2015, 8, 7675–7684. [Google Scholar]
- Manetti, A.C.; Maiese, A.; Baronti, A.; Mezzetti, E.; Frati, P.; Fineschi, V.; Turillazzi, E. MiRNAs as New Tools in Lesion Vitality Evaluation: A Systematic Review and Their Forensic Applications. Biomedicines 2021, 9, 1731. [Google Scholar] [CrossRef]
- Benz, F.; Tacke, F.; Luedde, M.; Trautwein, C.; Luedde, T.; Koch, A.; Roderburg, C. Circulating MicroRNA-223 Serum Levels Do Not Predict Sepsis or Survival in Patients with Critical Illness. Dis. Markers 2015, 2015, 384208. [Google Scholar] [CrossRef]
- Wu, X.; Yang, J.; Yu, L.; Long, D. Plasma miRNA-223 correlates with risk, inflammatory markers as well as prognosis in sepsis patients. Medicine 2018, 97, e11352. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, J.; Li, Z.; Li, J.; Sang, M. MicroRNA-205-5b inhibits HMGB1 expression in LPS-induced sepsis. Int. J. Mol. Med. 2016, 38, 312–318. [Google Scholar] [CrossRef]
- Wu, Y.; Li, P.; Goodwin, A.J.; Cook, J.A.; Halushka, P.V.; Zingarelli, B.; Fan, H. miR-145a Regulation of Pericyte Dysfunction in a Murine Model of Sepsis. J. Infect. Dis. 2020, 222, 1037–1045. [Google Scholar] [CrossRef]
- Ma, F.; Li, Z.; Cao, J.; Kong, X.; Gong, G. A TGFBR2/SMAD2/DNMT1/miR-145 negative regulatory loop is responsible for LPS-induced sepsis. Biomed. Pharmacother. 2019, 112, 108626. [Google Scholar] [CrossRef]
- Cao, X.; Zhang, C.; Zhang, X.; Chen, Y.; Zhang, H. MiR-145 negatively regulates TGFBR2 signaling responsible for sepsis-induced acute lung injury. Biomed. Pharmacother. 2019, 111, 852–858. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, J.; Xue, Y.; Zhao, J.; Li, D.; Zhang, S.; Li, K.; Hou, Y.; Fan, H. GSKJ4 Protects Mice Against Early Sepsis via Reducing Proinflammatory Factors and Up-Regulating MiR-146a. Front. Immunol. 2018, 9, 2272. [Google Scholar] [CrossRef]
- Sisti, F.; Wang, S.; Brandt, S.L.; Glosson-Byers, N.; Mayo, L.D.; Son, Y.M.; Sturgeon, S.; Filgueiras, L.; Jancar, S.; Wong, H.; et al. Nuclear PTEN enhances the maturation of a microRNA regulon to limit MyD88-dependent susceptibility to sepsis. Sci. Signal. 2018, 11, eaai9085. [Google Scholar] [CrossRef]
- Zhu, X. MiR-125b but not miR-125a is upregulated and exhibits a trend to correlate with enhanced disease severity, inflammation, and increased mortality in sepsis patients. J. Clin. Lab. Anal. 2020, 34, e23094. [Google Scholar] [CrossRef]
- Sun, J.; Sun, X.; Chen, J.; Liao, X.; He, Y.; Wang, J.; Chen, R.; Hu, S.; Qiu, C. microRNA-27b shuttled by mesenchymal stem cell-derived exosomes prevents sepsis by targeting JMJD3 and downregulating NF-κB signaling pathway. Stem Cell Res. Ther. 2021, 12, 14. [Google Scholar] [CrossRef]
- Zheng, G.; Qiu, G.; Ge, M.; Meng, J.; Zhang, G.; Wang, J.; Huang, R.; Shu, Q.; Xu, J. miR-10a in Peripheral Blood Mononuclear Cells Is a Biomarker for Sepsis and Has Anti-Inflammatory Function. Mediat. Inflamm. 2020, 2020, 4370983. [Google Scholar] [CrossRef]
- Liu, J.; Shi, K.; Chen, M.; Xu, L.; Hong, J.; Hu, B.; Yang, X.; Sun, R. Elevated miR-155 expression induces immunosuppression via CD39+ regulatory T-cells in sepsis patient. Int. J. Infect. Dis. 2015, 40, 135–141. [Google Scholar] [CrossRef]
- Zhang, W.; Jia, J.; Liu, Z.; Si, D.; Ma, L.; Zhang, G. Circulating microRNAs as biomarkers for Sepsis secondary to pneumonia diagnosed via Sepsis 3.0. BMC Pulm. Med. 2019, 19, 93. [Google Scholar] [CrossRef]
- Cai, X.; Jiang, Y.; Zhou, H.; Ma, D.; Chen, Z.K. MicroRNA-19a and CD22 Comprise a Feedback Loop for B Cell Response in Sepsis. Med. Sci. Monit. 2015, 21, 1548–1555. [Google Scholar] [CrossRef]
- Xu, H.; Liu, X.; Ni, H. Clinical significance of miR-19b-3p in patients with sepsis and its regulatory role in the LPS-induced inflammatory response. Eur. J. Med. Res. 2020, 25, 9. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Y.; Zheng, Q.; Dong, X.; Xie, Z.; Panayi, A.; Bai, X.; Li, Z. MicroRNA-150 inhibits myeloid-derived suppressor cells proliferation and function through negative regulation of ARG-1 in sepsis. Life Sci. 2021, 278, 119626. [Google Scholar] [CrossRef]
- Sheng, B.; Zhao, L.; Zang, X.; Zhen, J.; Chen, W. miR-375 ameliorates sepsis by downregulating miR-21 level via inhibiting JAK2-STAT3 signaling. Biomed. Pharmacother. 2017, 86, 254–261. [Google Scholar] [CrossRef]
- McClure, C.; Brudecki, L.; Ferguson, D.A.; Yao, Z.Q.; Moorman, J.P.; McCall, C.E.; El Gazzar, M. MicroRNA 21 (miR-21) and miR-181b Couple with NFI-A to Generate Myeloid-Derived Suppressor Cells and Promote Immunosuppression in Late Sepsis. Infect. Immun. 2014, 82, 3816–3825. [Google Scholar] [CrossRef] [PubMed]
- McClure, C.; McPeak, M.B.; Youssef, D.; Yao, Z.Q.; McCall, C.E.; El Gazzar, M. Stat3 and C/EBPβ synergize to induce miR-21 and miR-181b expression during sepsis. Immunol. Cell Biol. 2017, 95, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Dai, Q.-C.; Shen, H.-L.; Zhang, X.-W. Diagnostic value of elevated serum miRNA-143 levels in sepsis. J. Int. Med. Res. 2016, 44, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Zhang, L.; Chen, Y.; Du, H.; Wen, Z.; Wang, T.; Chen, D. Down-regulation of circDMNT3B is conducive to intestinal mucosal permeability dysfunction of rats with sepsis via sponging miR-20b-5p. J. Cell. Mol. Med. 2020, 24, 6731–6740. [Google Scholar] [CrossRef]
- Ji, Z.-R.; Xue, W.-L.; Zhang, L. Schisandrin B Attenuates Inflammation in LPS-Induced Sepsis Through miR-17-5p Downregulating TLR4. Inflammation 2019, 42, 731–739. [Google Scholar] [CrossRef]
- Szilágyi, B.; Fejes, Z.; Póliska, S.; Pócsi, M.; Czimmerer, Z.; Patsalos, A.; Fenyvesi, F.; Rusznyák, Á.; Nagy, G.; Kerekes, G.; et al. Reduced miR-26b Expression in Megakaryocytes and Platelets Contributes to Elevated Level of Platelet Activation Status in Sepsis. Int. J. Mol. Sci. 2020, 21, 866. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y.; Dai, L.; Wang, N. MiR-96-5p alleviates inflammatory responses by targeting NAMPT and regulating the NF-κB pathway in neonatal sepsis. Biosci. Rep. 2020, 40, BSR20201267. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, Z.; Mao, Y.; Dong, W.; Zhang, Y.; Yin, N.; Jiang, L. miR-27a is up regulated and promotes inflammatory response in sepsis. Cell. Immunol. 2014, 290, 190–195. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, D.; Li, Y.; Li, Y.; Li, Y. Paclitaxel alleviated liver injury of septic mice by alleviating inflammatory response via microRNA-27a/TAB3/NF-κB signaling pathway. Biomed. Pharmacother. 2018, 97, 1424–1433. [Google Scholar] [CrossRef]
- Yang, Q.; Cao, K.; Jin, G.; Zhang, J. Hsa-miR-346 plays a role in the development of sepsis by downregulating SMAD3 expression and is negatively regulated by lncRNA MALAT1. Mol. Cell. Probes 2019, 47, 101444. [Google Scholar] [CrossRef]
- Bai, X.; Li, J.; Li, L.; Liu, M.; Liu, Y.; Cao, M.; Tao, K.; Xie, S.; Hu, D. Extracellular Vesicles from Adipose Tissue-Derived Stem Cells Affect Notch-miR148a-3p Axis to Regulate Polarization of Macrophages and Alleviate Sepsis in Mice. Front. Immunol. 2020, 11, 1391. [Google Scholar] [CrossRef]
- Zhang, T.; Xiang, L. Honokiol alleviates sepsis-induced acute kidney injury in mice by targeting the miR-218-5p/heme oxygenase-1 signaling pathway. Cell. Mol. Biol. Lett. 2019, 24, 15. [Google Scholar] [CrossRef]
- Ma, J.; Xu, L.-Y.; Sun, Q.-H.; Wan, X.-Y.; Li, B. Inhibition of miR-1298-5p attenuates sepsis lung injury by targeting SOCS6. Mol. Cell. Biochem. 2021, 476, 3745–3756. [Google Scholar] [CrossRef]
- Gu, W.; Wen, D.; Lu, H.; Zhang, A.; Wang, H.; Du, J.; Zeng, L.; Jiang, J. MiR-608 Exerts Anti-inflammatory Effects by Targeting ELANE in Monocytes. J. Clin. Immunol. 2020, 40, 147–157. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.-J.; Tang, X.-M.; Wei, Y.-K. Urinary miR-26b as a potential biomarker for patients with sepsis-associated acute kidney injury: A Chinese population-based study. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4604–4610. [Google Scholar]
- Huo, R.; Dai, M.; Fan, Y.; Zhou, J.-Z.; Li, L.; Zu, J. Predictive value of miRNA-29a and miRNA-10a-5p for 28-day mortality in patients with sepsis-induced acute kidney injury. Nan Fang Yi Ke Da Xue Xue Bao 2017, 37, 646–651. [Google Scholar] [CrossRef]
- Cao, Y.-Y.; Wang, Z.; Wang, Z.-H.; Jiang, X.-G.; Lu, W.-H. Inhibition of miR-155 alleviates sepsis-induced inflammation and intestinal barrier dysfunction by inactivating NF-κB signaling. Int. Immunopharmacol. 2021, 90, 107218. [Google Scholar] [CrossRef]
- Ma, F.; Liu, F.; Ding, L.; You, M.; Yue, H.; Zhou, Y.; Hou, Y. Anti-inflammatory effects of curcumin are associated with down regulating microRNA-155 in LPS-treated macrophages and mice. Pharm. Biol. 2017, 55, 1263–1273. [Google Scholar] [CrossRef]
- Wang, Y.; Li, T.; Wu, B.; Liu, H.; Luo, J.; Feng, D.; Shi, Y. STAT1 Regulates MD-2 Expression in Monocytes of Sepsis via miR-30a. Inflammation 2014, 37, 1903–1911. [Google Scholar] [CrossRef]
- Mei, L.; He, M.; Zhang, C.; Miao, J.; Wen, Q.; Liu, X.; Xu, Q.; Ye, S.; Ye, P.; Huang, H.; et al. Paeonol attenuates inflammation by targeting HMGB1 through upregulating miR-339-5p. Sci. Rep. 2019, 9, 19370. [Google Scholar] [CrossRef]
- Wang, X.; Hao, L.; Bu, H.-F.; Scott, A.W.; Tian, K.; Liu, F.; De Plaen, I.G.; Liu, Y.; Mirkin, C.A.; Tan, X.-D. Spherical nucleic acid targeting microRNA-99b enhances intestinal MFG-E8 gene expression and restores enterocyte migration in lipopolysaccharide-induced septic mice. Sci. Rep. 2016, 6, 31687. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, K.; Sun, Y.; Tian, T.; Shen, W.; Sun, F.; Yuan, W.; Wu, H.; Chen, G.; Yuan, L.; et al. MiR-215-5p inhibits the inflammation injury in septic H9c2 by regulating ILF3 and LRRFIP1. Int. Immunopharmacol. 2020, 78, 106000. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, P.; Chen, W.; Feng, D.; Jia, Y.; Xie, L.-X. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin. Chem. Lab. Med. (CCLM) 2012, 50, 1423–1428. [Google Scholar] [CrossRef]
- Liang, G.; Wu, Y.; Guan, Y.; Dong, Y.; Jiang, L.; Mao, G.; Wu, R.; Huang, Z.; Jiang, H.; Qi, L.; et al. The correlations between the serum expression of miR-206 and the severity and prognosis of sepsis. Ann. Palliat. Med. 2020, 9, 3222–3234. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zhang, C.; Zhang, C.; Yang, H.; Gao, R.; Tong, Z. Plasma Hsa-miR-92a-3p in correlation with lipocalin-2 is associated with sepsis-induced coagulopathy. BMC Infect. Dis. 2020, 20, 155. [Google Scholar] [CrossRef]
- Matthay, M.A.; Zemans, R.L.; Zimmerman, G.A.; Arabi, Y.; Beitler, J.R.; Mercat, A.; Herridge, M.; Randolph, A.G.; Calfee, C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019, 5, 18. [Google Scholar] [CrossRef]
- Thompson, B.T.; Chambers, R.C.; Liu, K.D. Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2017, 377, 562–572. [Google Scholar] [CrossRef]
- Fan, E.; Brodie, D.; Slutsky, A.S. Acute Respiratory Distress Syndrome. JAMA 2018, 319, 698–710. [Google Scholar] [CrossRef]
- Heyland, D.; Muscedere, J.; Wischmeyer, P.E.; Cook, D.; Jones, G.; Albert, M.; Elke, G.; Berger, M.M.; Day, A.G. A Randomized Trial of Glutamine and Antioxidants in Critically Ill Patients. N. Engl. J. Med. 2013, 368, 1489–1497. [Google Scholar] [CrossRef]
- Bernard, G.R.; Wheeler, A.P.; Arons, M.M.; Morris, P.E.; Paz, H.L.; Russell, J.A.; Wright, P.E. A Trial of Antioxidants N-acetylcysteine and Procysteine in ARDS. Chest 1997, 112, 164–172. [Google Scholar] [CrossRef]
- Jepsen, S.; Herlevsen, P.; Knudsen, P.; Bud, M.I.; Klausen, N.-O. Antioxidant treatment with N-acetylcysteine during adult respiratory distress syndrome. Crit. Care Med. 1992, 20, 918–923. [Google Scholar] [CrossRef] [PubMed]
- ARDS Clinical Trials Network; National Institutes of Health. Randomized, placebo-controlled trial of lisofylline for early treatment of acute lung injury and acute respiratory distress syndrome. Crit. Care Med. 2002, 30, 1–6. [Google Scholar] [CrossRef]
- Abraham, E.; Baughman, R.; Fletcher, E.; Heard, S.; Lamberti, J.; Levy, H.; Nelson, L.; Rumbak, M.; Steingrub, J.; Taylor, J.; et al. Liposomal prostaglandin E1 (TLC C-53) in acute respiratory distress syndrome. Crit. Care Med. 1999, 27, 1478–1485. [Google Scholar] [CrossRef] [PubMed]
- Bone, R.C.; Slotman, G.; Maunder, R.; Silverman, H.; Hyers, T.M.; Kerstein, M.D.; Ursprung, J.J. Randomized Double-Blind, Multicenter Study of Prostaglandin E1 in Patients with the Adult Respiratory Distress Syndrome. Chest 1989, 96, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Zeiher, B.G.; Artigas, A.; Vincent, J.-L.; Dmitrienko, A.; Jackson, K.; Thompson, B.T.; Bernard, G. Neutrophil elastase inhibition in acute lung injury: Results of the STRIVE study. Crit. Care Med. 2004, 32, 1695–1702. [Google Scholar] [CrossRef]
- Liu, K.D.; Levitt, J.; Zhuo, H.; Kallet, R.H.; Brady, S.; Steingrub, J.; Tidswell, M.; Siegel, M.D.; Soto, G.; Peterson, M.W.; et al. Randomized Clinical Trial of Activated Protein C for the Treatment of Acute Lung Injury. Am. J. Respir. Crit. Care Med. 2008, 178, 618–623. [Google Scholar] [CrossRef]
- The National Heart, Lung, and Blood Institute ARDS Clinical Trials Network; Truwit, J.D.; Bernard, G.R.; Steingrub, J.; Matthay, M.A.; Liu, K.D.; Albertson, T.E.; Brower, R.G.; Shanholtz, C.; Rock, P.; et al. Rosuvastatin for Sepsis-Associated Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2014, 370, 2191–2200. [Google Scholar] [CrossRef]
- McAuley, D.F.; Laffey, J.G.; O’Kane, C.M.; Perkins, G.D.; Mullan, B.; Trinder, T.J.; Johnston, P.; Hopkins, P.A.; Johnston, A.J.; McDowell, C.; et al. Simvastatin in the Acute Respiratory Distress Syndrome. N. Engl. J. Med. 2014, 371, 1695–1703. [Google Scholar] [CrossRef]
- McAuley, D.F.; Cross, L.M.; Hamid, U.; Gardner, E.; Elborn, J.S.; Cullen, K.M.; Dushianthan, A.; Grocott, M.; Matthay, M.A.; O’Kane, C. Keratinocyte growth factor for the treatment of the acute respiratory distress syndrome (KARE): A randomised, double-blind, placebo-controlled phase 2 trial. Lancet Respir. Med. 2017, 5, 484–491. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.P.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Liu, F.; Nie, C.; Zhao, N.; Wang, Y.; Liu, Y.; Li, Y.; Zeng, Z.; Ding, C.; Shao, Q.; Qing, C.; et al. MiR-155 Alleviates Septic Lung Injury by Inducing Autophagy Via Inhibition of Transforming Growth Factor-β-Activated Binding Protein 2. Shock 2017, 48, 61–68. [Google Scholar] [CrossRef]
- Jiang, K.; Yang, J.; Guo, S.; Zhao, G.; Wu, H.; Deng, G. Peripheral Circulating Exosome-Mediated Delivery of miR-155 as a Novel Mechanism for Acute Lung Inflammation. Mol. Ther. 2019, 27, 1758–1771. [Google Scholar] [CrossRef]
- Yuan, Z.; Syed, M.A.; Panchal, D.; Joo, M.; Bedi, C.; Lim, S.; Önyüksel, H.; Rubinstein, I.; Colonna, M.; Sadikot, R.T. TREM-1-accentuated lung injury via miR-155 is inhibited by LP17 nanomedicine. Am. J. Physiol. Cell. Mol. Physiol. 2016, 310, L426–L438. [Google Scholar] [CrossRef]
- Liau, N.P.D.; Laktyushin, A.; Lucet, I.S.; Murphy, J.M.; Yao, S.; Whitlock, E.; Callaghan, K.; Nicola, N.A.; Kershaw, N.J.; Babon, J.J. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 2018, 9, 1558. [Google Scholar] [CrossRef]
- Li, H.-F.; Wu, Y.-L.; Tseng, T.-L.; Chao, S.-W.; Lin, H.; Chen, H.-H. Inhibition of miR-155 potentially protects against lipopolysaccharide-induced acute lung injury through the IRF2BP2-NFAT1 pathway. Am. J. Physiol. Cell Physiol. 2020, 319, C1070–C1081. [Google Scholar] [CrossRef]
- Han, Y.; Li, Y.; Jiang, Y. The Prognostic Value of Plasma MicroRNA-155 and MicroRNA-146a Level in Severe Sepsis and Sepsis-Induced Acute Lung Injury Patients. Clin. Lab. 2016, 62, 2355–2360. [Google Scholar] [CrossRef]
- Vergadi, E.; Vaporidi, K.; Theodorakis, E.E.; Doxaki, C.; Lagoudaki, E.; Ieronymaki, E.; Alexaki, V.I.; Helms, M.; Kondili, E.; Soennichsen, B.; et al. Akt2 Deficiency Protects from Acute Lung Injury via Alternative Macrophage Activation and miR-146a Induction in Mice. J. Immunol. 2014, 192, 394–406. [Google Scholar] [CrossRef]
- Zeng, Z.; Gong, H.; Li, Y.; Jie, K.; Ding, C.; Shao, Q.; Liu, F.; Zhan, Y.; Nie, C.; Zhu, W.; et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp. Lung Res. 2013, 39, 275–282. [Google Scholar] [CrossRef]
- Xu, R.; Shao, Z.; Cao, Q. MicroRNA-144-3p enhances LPS induced septic acute lung injury in mice through downregulating Caveolin-2. Immunol. Lett. 2021, 231, 18–25. [Google Scholar] [CrossRef]
- Ren, H.; Mu, W.; Xu, Q. miR-19a-3p inhibition alleviates sepsis-induced lung injury via enhancing USP13 expression. Acta Biochim. Pol. 2021, 68, 201–206. [Google Scholar] [CrossRef]
- Liu, Y.; Guan, H.; Zhang, J.-L.; Zheng, Z.; Wang, H.-T.; Tao, K.; Han, S.-C.; Su, L.-L.; Hu, D. Acute downregulation of miR-199a attenuates sepsis-induced acute lung injury by targeting SIRT1. Am. J. Physiol. Physiol. 2018, 314, C449–C455. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ding, R.; Hu, Z.; Yin, X.; Xiao, F.; Zhang, W.; Yan, S.; Lv, C. MicroRNA-34a Inhibition Alleviates Lung Injury in Cecal Ligation and Puncture Induced Septic Mice. Front. Immunol. 2020, 11, 1829. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.-C.; Song, W.-W.; Zhao, M.-L.; Zhang, R.-M.; Tian, X. Effect of miR-132 on lung injury in sepsis rats via regulating Sirt1 expression. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1042–1049. [Google Scholar] [PubMed]
- Jiao, Y.; Zhang, T.; Zhang, C.; Ji, H.; Tong, X.; Xia, R.; Wang, W.; Ma, Z.; Shi, X. Exosomal miR-30d-5p of neutrophils induces M1 macrophage polarization and primes macrophage pyroptosis in sepsis-related acute lung injury. Crit. Care 2021, 25, 356. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.-H.; Yang, Z.-T.; Cheng, N.-T.; Tang, H.-X.; Xu, M. The function and mechanism of microRNA-92a-3p in lipopolysaccharide-induced acute lung injury. Immunopharmacol. Immunotoxicol. 2022, 44, 47–57. [Google Scholar] [CrossRef]
- Yang, J.; Mao, M.; Zhen, Y.-Y. miRNA-23a has effects to improve lung injury induced by sepsis in vitro and vivo study. Biomed. Pharmacother. 2018, 107, 81–89. [Google Scholar] [CrossRef]
- Meng, L.; Cao, H.; Wan, C.; Jiang, L. MiR-539-5p alleviates sepsis-induced acute lung injury by targeting ROCK1. Folia Histochem. Cytobiol. 2020, 57, 168–178. [Google Scholar] [CrossRef]
- Pan, W.; Wei, N.; Xu, W.; Wang, G.; Gong, F.; Li, N. MicroRNA-124 alleviates the lung injury in mice with septic shock through inhibiting the activation of the MAPK signaling pathway by downregulating MAPK14. Int. Immunopharmacol. 2019, 76, 105835. [Google Scholar] [CrossRef]
- Leng, C.; Sun, J.; Xin, K.; Ge, J.; Liu, P.; Feng, X. High expression of miR-483-5p aggravates sepsis-induced acute lung injury. J. Toxicol. Sci. 2020, 45, 77–86. [Google Scholar] [CrossRef]
- Lou, W.; Yan, J.; Wang, W. Downregulation of miR-497-5p Improves Sepsis-Induced Acute Lung Injury by Targeting IL2RB. BioMed Res. Int. 2021, 2021, 6624702. [Google Scholar] [CrossRef]
- You, Q.; Wang, J.; Jia, D.; Jiang, L.; Chang, Y.; Li, W. MiR-802 alleviates lipopolysaccharide-induced acute lung injury by targeting Peli2. Agents Actions 2020, 69, 75–85. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, J.; Yang, F.; Wang, D.; Lu, Y.; Liu, L. MicroRNA-326 prevents sepsis-induced acute lung injury via targeting TLR4. Free Radic. Res. 2020, 54, 408–418. [Google Scholar] [CrossRef]
- Yang, P.; Xiong, W.; Chen, X.; Liu, J.; Ye, Z. Overexpression of miR-129-5p Mitigates Sepsis-Induced Acute Lung Injury by Targeting High Mobility Group Box 1. J. Surg. Res. 2020, 256, 23–30. [Google Scholar] [CrossRef]
- Lin, J.; Lin, Z.; Lin, L. MiR-490 alleviates sepsis-induced acute lung injury by targeting MRP4 in new-born mice. Acta Biochim. Pol. 2021, 68, 151–158. [Google Scholar] [CrossRef]
- Yao, W.; Xu, L.; Jia, X.; Li, S.; Wei, L. MicroRNA-129 plays a protective role in sepsis-induced acute lung injury through the suppression of pulmonary inflammation via the modulation of the TAK1/NF-κB pathway. Int. J. Mol. Med. 2021, 48, 139. [Google Scholar] [CrossRef]
- Xie, T.; Liang, J.; Liu, N.; Wang, Q.; Li, Y.; Noble, P.W.; Jiang, D. MicroRNA-127 Inhibits Lung Inflammation by Targeting IgG Fcγ Receptor I. J. Immunol. 2012, 188, 2437–2444. [Google Scholar] [CrossRef]
- Jiang, L.; Ni, J.; Shen, G.; Xia, Z.; Zhang, L.; Xia, S.; Pan, S.; Qu, H.; Li, X. Upregulation of endothelial cell-derived exosomal microRNA-125b-5p protects from sepsis-induced acute lung injury by inhibiting topoisomerase II alpha. Agents Actions 2021, 70, 205–216. [Google Scholar] [CrossRef]
- Zhang, W.; Li, J.; Yao, H.; Li, T. Restoring microRNA-499-5p Protects Sepsis-Induced Lung Injury Mice Via Targeting Sox6. Nanoscale Res. Lett. 2021, 16, 89. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, D.; Liu, H.; Xu, H. miR-942-5p prevents sepsis-induced acute lung injury via targeting TRIM37. Int. J. Exp. Pathol. 2021, 102, 192–199. [Google Scholar] [CrossRef]
- Yin, J.; Han, B.; Shen, Y. LncRNA NEAT1 inhibition upregulates miR-16-5p to restrain the progression of sepsis-induced lung injury via suppressing BRD4 in a mouse model. Int. Immunopharmacol. 2021, 97, 107691. [Google Scholar] [CrossRef]
- Qiu, N.; Xu, X.; He, Y. LncRNA TUG1 alleviates sepsis-induced acute lung injury by targeting miR-34b-5p/GAB1. BMC Pulm. Med. 2020, 20, 49. [Google Scholar] [CrossRef]
- Wang, H.-R.; Guo, X.-Y.; Liu, X.-Y.; Song, X. Down-regulation of lncRNA CASC9 aggravates sepsis-induced acute lung injury by regulating miR-195-5p/PDK4 axis. Agents Actions 2020, 69, 559–568. [Google Scholar] [CrossRef]
- Zhu, L.; Shi, D.; Cao, J.; Song, L. LncRNA CASC2 Alleviates Sepsis-induced Acute Lung Injury by Regulating the miR-152-3p/PDK4 Axis. Immunol. Investig. 2022, 51, 1257–1271. [Google Scholar] [CrossRef]
- Xie, H.; Chai, H.; Du, X.; Cui, R.; Dong, Y. Overexpressing long non-coding RNA OIP5-AS1 ameliorates sepsis-induced lung injury in a rat model via regulating the miR-128-3p/Sirtuin-1 pathway. Bioengineered 2021, 12, 9723–9738. [Google Scholar] [CrossRef]
- Chen, H.; Hu, X.; Li, R.; Liu, B.; Zheng, X.; Fang, Z.; Chen, L.; Chen, W.; Min, L.; Hu, S. LncRNA THRIL aggravates sepsis-induced acute lung injury by regulating miR-424/ROCK2 axis. Mol. Immunol. 2020, 126, 111–119. [Google Scholar] [CrossRef]
- Liu, B.; Chen, F.; Cheng, N.T.; Tang, Z.; Wang, X.G.; Xu, M. MicroRNA-1224-5p Aggravates Sepsis-Related Acute Lung Injury in Mice. Oxid Med Cell Longev. 2022, 2022, 9493710. [Google Scholar] [CrossRef]
- He, Q.; Yin, J.; Zou, B.; Guo, H. WIN55212-2 alleviates acute lung injury by inhibiting macrophage glycolysis through the miR-29b-3p/FOXO3/PFKFB3 axis. Mol Immunol. 2022, 149, 119–128. [Google Scholar] [CrossRef]
- Li, A.; Li, J.; Bao, Y.; Yuan, D.; Huang, Z. Xuebijing injection alleviates cytokine-induced inflammatory liver injury in CLP-induced septic rats through induction of suppressor of cytokine signaling 1. Exp. Ther. Med. 2016, 12, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-B.; Chen, W.-W.; Chen, H.-P.; Cai, S.-X.; Lin, J.-D.; Qiu, L.-Z. MiR-155 aggravated septic liver injury by oxidative stress-mediated ER stress and mitochondrial dysfunction via targeting Nrf-2. Exp. Mol. Pathol. 2018, 105, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Zhang, S.-H.; Zhi, L.-D.; Li, H.; Wen, Q.-K.; Li, G.; Zhang, W.-J. MicroRNA-30e promotes hepatocyte proliferation and inhibits apoptosis in cecal ligation and puncture-induced sepsis through the JAK/STAT signaling pathway by binding to FOSL2. Biomed. Pharmacother. 2018, 104, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Chen, Y.; Zhao, Y.; Liu, Z.; Nan, C.; Zheng, B.; Liu, X.; Chen, X. microRNA-30a inhibits the liver cell proliferation and promotes cell apoptosis through the JAK/STAT signaling pathway by targeting SOCS-1 in rats with sepsis. J. Cell. Physiol. 2019, 234, 17839–17853. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, Q. Inhibition of miR-103a-3p suppresses lipopolysaccharide-induced sepsis and liver injury by regulating FBXW7 expression. Cell Biol. Int. 2020, 44, 1798–1810. [Google Scholar] [CrossRef]
- Chen, L.; Lu, Q.; Deng, F.; Peng, S.; Yuan, J.; Liu, C.; Du, X. miR-103a-3p Could Attenuate Sepsis-Induced Liver Injury by Targeting HMGB1. Inflammation 2020, 43, 2075–2086. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, H.; Pan, L.; Zhang, B.; Che, H. microRNA-103a-3p confers protection against lipopolysaccharide-induced sepsis and consequent multiple organ dysfunction syndrome by targeting HMGB1. Infect. Genet. Evol. 2021, 89, 104681. [Google Scholar] [CrossRef]
- Gu, C.; Hou, C.; Zhang, S. miR-425-5p improves inflammation and septic liver damage through negatively regulating the RIP1-mediated necroptosis. Agents Actions 2020, 69, 299–308. [Google Scholar] [CrossRef]
- Zhou, Y.-X.; Han, W.-W.; Song, D.-D.; Li, Z.-P.; Ding, H.-J.; Zhou, T.; Jiang, L.; Hu, E.-C. Effect of miR-10a on sepsis-induced liver injury in rats through TGF-β1/Smad signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 862–869. [Google Scholar] [CrossRef]
- Xu, N.; Chen, Y.-S.; Zhu, H.; Li, W.-S.; Sun, Z.-W.; Zhang, H.-B.; Li, X.-Y.; Zhang, B.; Zhang, C.; Wen, Z.-Q.; et al. Upregulation of miR-142-5p induced by lipopolysaccharide contributes to apoptosis of hepatocytes and hepatic failure. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5293–5303. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, H.; Li, C.; Liang, Z. Evidence for the circulating microRNA hsa-let-7d-3p as a potential new biomarker for sepsis in human subjects. Eur J Med Res. 2022, 27, 137. [Google Scholar] [CrossRef]
- Li, L.; He, Y.; He, X.-J.; Bi, M.-R.; Qi, Y.-H.; Zhu, W.-W. Down-regulation of long noncoding RNA LINC00472 alleviates sepsis-induced acute hepatic injury by regulating miR-373-3p/TRIM8 axis. Exp. Mol. Pathol. 2020, 117, 104562. [Google Scholar] [CrossRef]
- Li, Y.; Song, J.; Xie, Z.; Liu, M.; Sun, K. Long noncoding RNA colorectal neoplasia differentially expressed alleviates sepsis-induced liver injury via regulating miR-126-5p. IUBMB Life 2020, 72, 440–451. [Google Scholar] [CrossRef]
- Chen, X.; Song, D. LPS promotes the progression of sepsis by activation of lncRNA HULC/miR-204-5p/TRPM7 network in HUVECs. Biosci. Rep. 2020, 40, BSR20200740. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Li, Z.; Ji, P.; Jia, Y.; Bai, X.; Cai, W.; Li, X.; Yang, C.; Yang, Y.; Yang, K.; et al. MCPIP1 alleviated lipopolysaccharide-induced liver injury by regulating SIRT1 via modulation of microRNA-9. J. Cell. Physiol. 2019, 234, 22450–22462. [Google Scholar] [CrossRef] [PubMed]
- Maiese, A.; Bolino, G.; Mastracchio, A.; Frati, P.; Fineschi, V. An immunohistochemical study of the diagnostic value of TREM-1 as marker for fatal sepsis cases. Biotech Histochem. 2019, 94, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-X.; Pan, J.-Y.; Wang, Y.-J.; Huang, T.-C.; Li, X.-F. MiR-640 inhibition alleviates acute liver injury via regulating WNT signaling pathway and LRP1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8988–8996. [Google Scholar] [CrossRef]
- Zarbock, A.; Gomez, H.; Kellum, J.A. Sepsis-induced acute kidney injury revisited: Pathophysiology, prevention and future therapies. Curr. Opin. Crit. Care 2014, 20, 588–595. [Google Scholar] [CrossRef]
- Chawla, L.S.; Bellomo, R.; Bihorac, A.; Goldstein, S.L.; Siew, E.D.; Bagshaw, S.M.; Bittleman, D.; Cruz, D.; Endre, Z.; Fitzgerald, R.L.; et al. Acute kidney disease and renal recovery: Consensus report of the Acute Disease Quality Initiative (ADQI) 16 Workgroup. Nat. Rev. Nephrol. 2017, 13, 241–257. [Google Scholar] [CrossRef]
- Negi, S.; Koreeda, D.; Kobayashi, S.; Yano, T.; Tatsuta, K.; Mima, T.; Shigematsu, T.; Ohya, M. Acute kidney injury: Epidemiology, outcomes, complications, and therapeutic strategies. Semin. Dial. 2018, 31, 519–527. [Google Scholar] [CrossRef]
- Fleischmann, M.C.; Scherag, A.; Adhikari, N.K.J.; Hartog, C.S.; Tsaganos, T.; Schlattmann, P.; Angus, D.C.; Reinhart, K.; on behalf of International Forum of Acute Care Trialists. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am. J. Respir. Crit. Care Med. 2016, 193, 259–272. [Google Scholar] [CrossRef]
- Gómez, H.; Kellum, J.A. Sepsis-induced acute kidney injury. Curr. Opin. Crit. Care 2016, 22, 546–553. [Google Scholar] [CrossRef]
- Anders, H.-J. Signaling Danger: Toll-Like Receptors and their Potential Roles in Kidney Disease. J. Am. Soc. Nephrol. 2004, 15, 854–867. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Petejova, N.; Martinek, A.; Zadrazil, J.; Kanova, M.; Klementa, V.; Sigutova, R.; Kacirova, I.; Hrabovsky, V.; Svagera, Z.; Stejskal, D. Acute Kidney Injury in Septic Patients Treated by Selected Nephrotoxic Antibiotic Agents—Pathophysiology and Biomarkers—A Review. Int. J. Mol. Sci. 2020, 21, 7115. [Google Scholar] [CrossRef]
- Wei, Q.; Bhatt, K.; He, H.-Z.; Mi, Q.-S.; Haase, V.H.; Dong, Z. Targeted Deletion of Dicer from Proximal Tubules Protects against Renal Ischemia-Reperfusion Injury. J. Am. Soc. Nephrol. 2010, 21, 756–761. [Google Scholar] [CrossRef]
- Xu, G.; Mo, L.; Wu, C.; Shen, X.; Dong, H.; Yu, L.; Pan, P.; Pan, K. The miR-15a-5p-XIST-CUL3 regulatory axis is important for sepsis-induced acute kidney injury. Ren. Fail. 2019, 41, 955–966. [Google Scholar] [CrossRef]
- Fu, D.; Dong, J.; Li, P.; Tang, C.; Cheng, W.; Xu, Z.; Zhou, W.; Ge, J.; Xia, C.; Zhang, Z. MiRNA-21 has effects to protect kidney injury induced by sepsis. Biomed. Pharmacother. 2017, 94, 1138–1144. [Google Scholar] [CrossRef]
- Wei, W.; Yao, Y.-Y.; Bi, H.-Y.; Zhai, Z.; Gao, Y. miR-21 protects against lipopolysaccharide-stimulated acute kidney injury and apoptosis by targeting CDK6. Ann. Transl. Med. 2020, 8, 303. [Google Scholar] [CrossRef]
- Lin, Z.; Liu, Z.; Wang, X.; Qiu, C.; Zheng, S. MiR-21-3p Plays a Crucial Role in Metabolism Alteration of Renal Tubular Epithelial Cells during Sepsis Associated Acute Kidney Injury via AKT/CDK2-FOXO1 Pathway. BioMed Res. Int. 2019, 2019, 2821731. [Google Scholar] [CrossRef]
- Wei, W.; Yao, Y.; Bi, H.; Xu, W.; Gao, Y. Circular RNA circ_0068,888 protects against lipopolysaccharide-induced HK-2 cell injury via sponging microRNA-21–5p. Biochem. Biophys. Res. Commun. 2021, 540, 1–7. [Google Scholar] [CrossRef]
- Zou, Z.; Lin, Q.; Yang, H.; Liu, Z.; Zheng, S. Nrp-1 Mediated Plasmatic Ago2 Binding miR-21a-3p Internalization: A Novel Mechanism for miR-21a-3p Accumulation in Renal Tubular Epithelial Cells during Sepsis. BioMed Res. Int. 2020, 2020, 2370253. [Google Scholar] [CrossRef]
- Lv, K.-T.; Liu, Z.; Feng, J.; Zhao, W.; Hao, T.; Ding, W.-Y.; Chu, J.-P.; Gao, L.-J. MiR-22-3p Regulates Cell Proliferation and Inhibits Cell Apoptosis through Targeting the eIF4EBP3 Gene in Human Cervical Squamous Carcinoma Cells. Int. J. Med. Sci. 2018, 15, 142–152. [Google Scholar] [CrossRef]
- Zhang, P.; Yi, L.; Qu, S.; Dai, J.; Li, X.; Liu, B.; Li, H.; Ai, K.; Zheng, P.; Qiu, S.; et al. The Biomarker TCONS_00016233 Drives Septic AKI by Targeting the miR-22-3p/AIFM1 Signaling Axis. Mol. Ther. Nucleic Acids 2020, 19, 1027–1042. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Kong, M.; Yang, J. MiR-22-3p suppresses sepsis-induced acute kidney injury by targeting PTEN. Biosci. Rep. 2020, 40, BSR20200527. [Google Scholar] [CrossRef]
- Ye, J.; Feng, H.; Peng, Z. miR-23a-3p inhibits sepsis-induced kidney epithelial cell injury by suppressing Wnt/β-catenin signaling by targeting wnt5a. Braz. J. Med. Biol. Res. 2022, 55, e11571. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, X.; Wu, Y. The miR-26a-5p/IL-6 axis alleviates sepsis-induced acute kidney injury by inhibiting renal inflammation. Ren. Fail. 2022, 44, 551–561. [Google Scholar] [CrossRef]
- Ha, Z.; Yu, Z. Downregulation of miR -29b-3p aggravates podocyte injury by targeting HDAC4 in LPS-induced acute kidney injury. Kaohsiung J. Med. Sci. 2021, 37, 1069–1076. [Google Scholar] [CrossRef]
- Ding, G.; An, J.; Li, L. MicroRNA-103a-3p enhances sepsis-induced acute kidney injury via targeting CXCL12. Bioengineered 2022, 13, 10288–10298. [Google Scholar] [CrossRef]
- Shen, Y.; Yu, J.; Jing, Y.; Zhang, J. MiR-106a aggravates sepsis-induced acute kidney injury by targeting THBS2 in mice model. Acta Cir. Bras. 2019, 34, e201900602. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Wang, J.; Miao, H. MiR-107 induces TNF-α secretion in endothelial cells causing tubular cell injury in patients with septic acute kidney injury. Biochem. Biophys. Res. Commun. 2017, 483, 45–51. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, H.; Qian, J.; Sun, L.; Sang, L.; Wang, P.; Yuan, B.; Zhang, J. The regulation of LPCAT3 by miR-124-3p.1 in acute kidney injury suppresses cell proliferation by disrupting phospholipid metabolism. Biochem. Biophys. Res. Commun. 2022, 604, 37–42. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, Y.; Xia, Y.; Li, C.; Song, Z.; Zhu, J. MiR-150-5p protects against septic acute kidney injury via repressing the MEKK3/JNK pathway. Cell. Signal. 2021, 86, 110101. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Luo, F.; Liu, X.; Lu, L.; Xu, H.; Yang, Q.; Xue, J.; Shi, L.; Li, J.; Zhang, A.; et al. NF-kB-regulated exosomal miR-155 promotes the inflammation associated with arsenite carcinogenesis. Cancer Lett. 2017, 388, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Cui, Y.; Xiong, X.; Wang, C.; Zhang, Y. Inhibition of microRNA-155 alleviates lipopolysaccharide-induced kidney injury in mice. Int. J. Clin. Exp. Pathol. 2017, 10, 9362–9371. [Google Scholar] [PubMed]
- Lin, Y.; Ding, Y.; Song, S.; Li, M.; Wang, T.; Guo, F. Expression patterns and prognostic value of miR-210, miR-494, and miR-205 in middle-aged and old patients with sepsis-induced acute kidney injury. Bosn. J. Basic Med. Sci. 2019, 19, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Sang, Z.; Dong, S.; Zhang, P.; Wei, Y. miR-214 ameliorates sepsis-induced acute kidney injury via PTEN/AKT/mTOR-regulated autophagy. Mol. Med. Rep. 2021, 24, 683. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ye, F.; Jian, Y.; Liu, C.; Tu, Z.; Yang, D. MicroRNA -214-5p aggravates sepsis-related acute kidney injury in mice. Drug Dev. Res. 2022, 83, 339–350. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Q.; Dai, Z.; Pan, H. miR-22 alleviates sepsis-induced acute kidney injury via targeting the HMGB1/TLR4/NF-κB signaling pathway. Int Urol Nephrol. 2022. [Google Scholar] [CrossRef]
- Qi, Y.; Han, Y.; Chen, C.; Xue, Y.; Jiao, N.; Li, X. Inhibition of microRNA-665 Alleviates Septic Acute Kidney Injury by Targeting Bcl-2. J. Healthc. Eng. 2022, 2022, 2961187. [Google Scholar] [CrossRef]
- Ge, Q.-M.; Huang, C.-M.; Zhu, X.-Y.; Bian, F.; Pan, S.-M. Differentially expressed miRNAs in sepsis-induced acute kidney injury target oxidative stress and mitochondrial dysfunction pathways. PLoS ONE 2017, 12, e0173292. [Google Scholar] [CrossRef]
- Caserta, S.; Kern, F.; Cohen, J.; Drage, S.; Newbury, S.F.; Llewelyn, M. Circulating Plasma microRNAs can differentiate Human Sepsis and Systemic Inflammatory Response Syndrome (SIRS). Sci. Rep. 2016, 6, 28006. [Google Scholar] [CrossRef]
- Zou, Y.-F.; Wen, D.; Zhao, Q.; Shen, P.-Y.; Shi, H.; Chen, Y.-X.; Zhang, W. Urinary MicroRNA-30c-5p and MicroRNA-192-5p as potential biomarkers of ischemia–reperfusion-induced kidney injury. Exp. Biol. Med. 2017, 242, 657–667. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Agrawal, S.; Tapmeier, T.T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.T.; Becker, C.M. The miRNA Mirage: How Close Are We to Finding a Non-Invasive Diagnostic Biomarker in Endometriosis? A Systematic Review. Int. J. Mol. Sci. 2018, 19, 599. [Google Scholar] [CrossRef]
- Jadhav, K.B.; Nagraj, S.K.; Arora, S. miRNA for the assessment of lymph node metastasis in patients with oral squamous cell carcinoma: Systematic review and metanalysis. J. Oral Pathol. Med. 2021, 50, 345–352. [Google Scholar] [CrossRef]
- Leo, C.G.; Mincarone, P.; Tumolo, M.R.; Panico, A.; Guido, M.; Zizza, A.; Guarino, R.; De Santis, G.; Sedile, R.; Sabina, S. MiRNA expression profiling in HIV pathogenesis, disease progression and response to treatment: A systematic review. Epigenomics 2021, 13, 1653–1671. [Google Scholar] [CrossRef]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef]
- Zhong, S.; Golpon, H.; Zardo, P.; Borlak, J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. 2021, 230, 164–196. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas-García, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015, 29, 3595–3611. [Google Scholar] [CrossRef]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Chen, C.; Chu, X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol. Cancer 2018, 17, 22. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Crippa, M.P.; Manfredi, A.A.; Mezzapelle, R.; Querini, P.R.; Venereau, E. High-mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol. Rev. 2017, 280, 74–82. [Google Scholar] [CrossRef]
- Tak, P.P.; Firestein, G.S. NF-κB: A key role in inflammatory diseases. J. Clin. Investig. 2001, 107, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Suzuki, M.; Sakaguchi, R.; Hanada, T.; Yasukawa, H. SOCS, Inflammation, and Autoimmunity. Front. Immunol. 2012, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.B.; Coon, T.A.; Glasser, J.R.; McVerry, B.J.; Zhao, J.; Zhao, Y.; Zou, C.; Ellis, B.; Sciurba, F.C.; Zhang, Y.; et al. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat. Immunol. 2013, 14, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Patel, L.; Pass, I.; Coxon, P.; Downes, C.; Smith, S.A.; Macphee, C.H. Tumor suppressor and anti-inflammatory actions of PPARγ agonists are mediated via upregulation of PTEN. Curr. Biol. 2001, 11, 764–768. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 51. [Google Scholar] [CrossRef]
- Seminerio, I.; Descamps, G.; Dupont, S.; de Marrez, L.; Laigle, J.-A.; Lechien, J.R.; Kindt, N.; Journe, F.; Saussez, S. Infiltration of FoxP3+ Regulatory T Cells is a Strong and Independent Prognostic Factor in Head and Neck Squamous Cell Carcinoma. Cancers 2019, 11, 227. [Google Scholar] [CrossRef]
- de Moraes, G.N.; Carneiro, L.D.T.; Maia, R.C.; Lam, E.W.-F.; Sharrocks, A.D. FOXK2 Transcription Factor and Its Emerging Roles in Cancer. Cancers 2019, 11, 393. [Google Scholar] [CrossRef]
- Gurnari, C.; Falconi, G.; De Bellis, E.; Voso, M.T.; Fabiani, E. The Role of Forkhead Box Proteins in Acute Myeloid Leukemia. Cancers 2019, 11, 865. [Google Scholar] [CrossRef]
- Sinclair, D.; Guarente, L. Unlocking the Secrets of Longevity Genes. Sci. Am. 2006, 294, 48–51, 54–57. [Google Scholar] [CrossRef]
- Levy, D.E.; Lee, C.-K. What does Stat3 do? J. Clin. Investig. 2002, 109, 1143–1148. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Giordano, M.; Trotta, M.C.; Ciarambino, T.; D’Amico, M.; Schettini, F.; Di Sisto, A.; D’Auria, V.; Voza, A.; Malatino, L.S.; Biolo, G.; et al. Circulating miRNA-195-5p and -451a in Patients with Acute Hemorrhagic Stroke in Emergency Department. Life 2022, 12, 763. [Google Scholar] [CrossRef]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef]
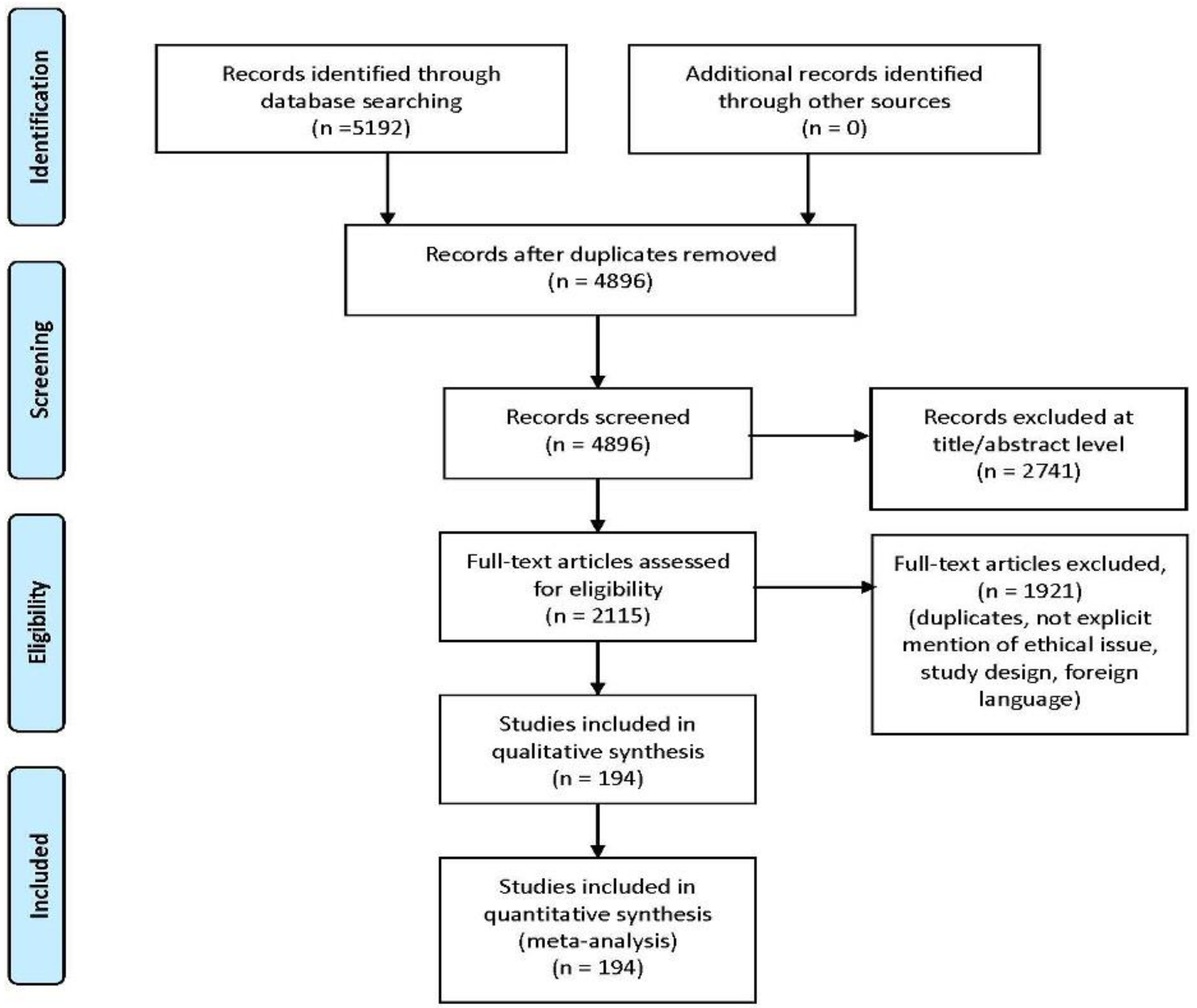
| Author and Reference | Year of Publication | Sepsis Model | Brief Description of miRNAs in Sepsis-Induced Brain Injury |
|---|---|---|---|
| Dong et al. [19] | 2019 | In vivo | miR-181b is downregulated in hippocampus |
| Yu et al. [21] | 2019 | In vitro | miR-200a-3p upregulation induced inflammatory response in sepsis-induced brain injury |
| Chen et al. [20] | 2020 | In vivo | miR-181b is overexpressed in septic cerebral cortex |
| Visitchanakun et al. [22] | 2020 | In vivo | miR-370-3p overexpression in SAE (in brain tissue and blood) miR-370-3p associated with TNF-α and increased brain apoptosis in SAE mice |
| Kim et al. [23] | 2021 | In vivo | miR-147 is downregulated in brain compared to other organ and also compared to controls |
| Nong et al. [24] | 2021 | In vivo | miR-126 downregulation is related to a protective role against sepsis-related blood–brain barrier damage |
| Rani et al. [25] | 2022 | In vivo | miR-190a-3p, let-7a-1-3p, miR-3085-3p were overexpressed in young and older females on day 1; miR-383-5p was instead downregulated in young males and females on day 4 |
| Zou et al. [26] | 2022 | In vivo In vitro | miR-146a induces CXCL2 in microglia and reduces CXCL” in astrocytes miR-146a knock-out is related to low levels of neutrophils and monocytes in brain tissue |
| Author and Reference | Year of Publication | Sepsis Model | Brief Description of miRNAs in Cardiac Tissue during Sepsis |
|---|---|---|---|
| Wang et al. [54] | 2014 | In vivo | mir-223 was downregulated; downregulation increased inflammation and myocardial injury |
| Liu et al. [56] | 2020 | In vivo | miR-223 was upregulated; upregulation reduced inflammation and apoptosis |
| M’baya-Moutoula et al. [57] | 2018 | In vitro | miR-223 expression regulates NF-κB pathway |
| Xue et al. [58] | 2015 | In vivo | miR-27a was upregulated; upregulation increased inflammation |
| Gao et al. [60] | 2015 | In vivo | miR-146a was upregulated; upregulation reduced inflammation and myocardial injury |
| An et al. [61] | 2018 | In vitro | miR-146a was upregulated; upregulation reduced inflammation and myocardial injury |
| Xie et al. [62] | 2019 | In vivo | miR-146a upregulation reduced inflammation and myocardial injury |
| Wang et al. [63] | 2018 | In vivo | miR-146b was upregulated; upregulation reduced inflammation and myocardial injury |
| Ma et al. [64] | 2016 | In vivo In vitro | miR-125b was downregulated and its upregulation reduced inflammation and myocardial injury |
| Wei et al. [65] | 2019 | In vitro | miR-150-5p was downregulated and its upregulation reduced inflammation and myocardial injury |
| Zhu et al. [66] | 2020 | In vivo In vitro | miR-150-5p was downregulated and its upregulation reduced inflammation and myocardial injury |
| Wang et al. [67] | 2016 | In vivo | miR-21-3p was upregulated and its downregulation reduced inflammation and cardiac disfunction |
| Wang et al. [68] | 2016 | In vivo | miR-155 was upregulated and its downregulation reduced inflammation and myocardial injury |
| Zhou et al. [71] | 2017 | In vivo | miR-155 was upregulated and overexpression reduced inflammation and myocardial injury |
| Diao et al.. [69] | 2017 | In vivo | miR-124a was downregulated and its upregulation reduced inflammation and myocardial injury |
| Zheng et al. [70] | 2017 | In vivo | miR-135a was upregulated; upregulation increased myocardial inflammation |
| Ge et al. [72] | 2018 | In vivo | miR-214 was upregulated; upregulation reduced inflammation and myocardial injury |
| Sang et al. [73] | 2020 | In vivo | miR-214-3p was upregulated; upregulation reduced inflammation and myocardial injury |
| Fang et al. [74] | 2018 | In vivo In vitro | miR-874 was upregulated; upregulation increased inflammation |
| Tang et al. [78] | 2018 | In vitro | miR-93-3p was downregulated and its upregulation reduced inflammation and myocardial injury |
| Yao et al. [79] | 2018 | In vivo In vitro | miR-25 was downregulated and its upregulation reduced inflammation and myocardial injury |
| Wu et al. [80] | 2018 | In vivo In vitro | miR-494-3p was downregulated and its upregulation reduced inflammation and myocardial injury |
| Zhang et al. [81] | 2018 | In vivo In vitro | miR-23b was upregulated; upregulation reduced inflammation and myocardial injury |
| Cao et al. [84] | 2019 | In vivo In vitro | miR-23b was upregulated; upregulation reduced inflammation and myocardial injury |
| Guo et al. [85] | 2019 | In vivo | miR-495 was downregulated and its upregulation reduced inflammation and myocardial injury |
| Zhu et al. [86] | 2019 | In vivo | miR-98 was downregulated and its upregulation reduced inflammation and myocardial injury |
| Ouyang et al. [87] | 2020 | In vivo | miR-208-5p was upregulated; upregulation increased inflammation and determined myocardial injury |
| Sun et al. [88] | 2020 | In vivo | miR-328 was upregulated; upregulation increased inflammation |
| Zhu et al. [91] | 2020 | In vivo | miR-29a was upregulated; upregulation increased inflammation and determined myocardial injury |
| Song et al. [92] | 2020 | In vivo In vitro | miR-29a was upregulated; upregulation reduced inflammation and apoptosis |
| Chen et al. [93] | 2020 | In vitro | miR-24 was downregulated; upregulation increased inflammation and determined myocardial injury |
| Sun et al. [94] | 2020 | In vivo In vitro | miR-192-5p was downregulated; downregulation reduced inflammation and cardiac disfunction |
| Wei et al. [95] | 2020 | In vitro | miR-144-3p was downregulated; upregulation reduced inflammation and myocardial injury |
| Xing et al. [96] | 2020 | In vivo In vitro | miR-330-5p was downregulated; upregulation reduced inflammation and myocardial injury |
| Han et al. [97] | 2020 | In vivo In vitro | miR-1-5p was downregulated; downregulation reduced inflammation and cardiac disfunction |
| Li et al. [98] | 2020 | In vivo | miR-29b-3p was downregulated; upregulation reduced inflammation and myocardial injury |
| Xin et al. [99] | 2021 | In vivo In vitro | miR-101-3p was upregulated; downregulation reduced inflammation and cardiac disfunction |
| Liu et al. [100] | 2021 | In vivo In vitro | miR-106b-5p was upregulated; upregulation reduced inflammation |
| Zhang et al. [101] | 2021 | In vivo | miR-29c-3p was upregulated; upregulation increased inflammation and determined myocardial injury |
| Yang et al. [102] | 2021 | In vivo In vitro | miR-499a-5p was downregulated; upregulation reduced inflammation and myocardial injury |
| Author and Reference | Year of Publication | Sepsis Model | Brief Description of miRNAs in Blood during Sepsis |
|---|---|---|---|
| Wang et al. [113] | 2019 | In vivo | miR-494-3p was downregulated; upregulation reduced inflammation through reduction in TNF-α levels and reduction in NF-κB p65 nuclear translocation |
| Li et al. [114] | 2018 | In vivo In vitro | miR-218 was downregulated; upregulation reduced inflammation |
| Zhou et al. [115] | 2018 | In vivo | miR-218 was downregulated; upregulation reduced inflammation |
| Abou El-Khier NT et al. [116] | 2018 | In vivo In vitro | miR-122 was upregulated, and can be used as a prognostic biomarker for sepsis |
| Sun et al. [117] | 2020 | In vivo | miR-328 was upregulated; downregulation reduced inflammation. Its expression was a good diagnostic marker for sepsis |
| Na et al. [118] | 2020 | In vivo In vitro | miR-21 was downregulated, representing a good predictor for sepsis risk |
| Lin et al. [119] | 2020 | In vivo | miR-126 was upregulated, representing a good marker for sepsis |
| Li et al. [120] | 2020 | In vivo | miR-125a was upregulated, representing a good marker for sepsis |
| Song et al. [121] | 2017 | In vivo In vitro | miR-146a was upregulated, upregulation improved survival in septic mice. It represents a good biomarker and predictor for high sepsis risk |
| Chen et al. [122] | 2020 | In vivo | miR-146a was upregulated; upregulation was associated with increased inflammation. It represents a good biomarker and predictor for high sepsis risk |
| Bai et al. [123] | 2018 | In vivo | miR-146a was upregulated; upregulation improved survival in septic mice. It represents a good biomarker and predictor for high sepsis risk |
| Brudecki et al. [124] | 2013 | In vivo In vitro | miR-146a was upregulated; upregulation improved survival in septic mice. It represents a good biomarker and predictor for high sepsis risk |
| Banarjee et al. [125] | 2013 | In vivo In vitro | miR-146a was upregulated; upregulation improved survival in septic mice. It represents a good biomarker and predictor for high sepsis risk |
| Mohnle et al. [126] | 2015 | In vivo In vitro | miR-146a was downregulated; downregulation induced inflammation |
| Paik et al. [127] | 2019 | In vivo | miR-146a was downregulated; downregulation induced inflammation |
| Chen et al. [122] | 2020 | In vivo | miR-146b was upregulated; upregulation was associated with increased inflammation. It represents a good biomarker and predictor for high sepsis risk |
| Zhang et al. [128] | 2020 | In vivo | miR-146b was upregulated; upregulation reduced inflammation |
| Gao et al. [129] | 2017 | In vivo | miR-146b was upregulated; upregulation reduced inflammation |
| Zou et al. [130] | 2020 | In vivo | miR-126 was upregulated, representing a good marker for sepsis |
| Dang et al. [131] | 2020 | In vivo In vitro | miR-223 was expressed in sepsis; upregulation reduced severity of sepsis |
| Yao et al. [132] | 2015 | In vivo | miR-25 upregulation reduced oxidative stress |
| Xue et al. [133] | 2019 | In vivo In vitro | miR-21 was upregulated; downregulation reduced inflammation |
| Liu et al. [56] | 2020 | In vivo | miR-223 was downregulated; represents a good biomarker for sepsis |
| Benz et al. [134] | 2015 | In vivo | miR-223 serum levels not correlated with sepsis, either in mouse models or in human patients. |
| Wu et al. [135] | 2018 | In vivo | miR-223 was upregulated; upregulation was associated with disease severity and inflammation |
| Zhou et al. [136] | 2016 | In vivo In vitro | miR-205-5b was downregulated with increased HMGB1 expression and inflammation |
| Wu et al. [137] | 2020 | In vivo In vitro | miR-145a was downregulated; upregulation reduced inflammation |
| Ma et al. [138] | 2019 | In vivo In vitro | miR-145a was downregulated; upregulation reduced inflammation |
| Cao et al. [139] | 2019 | In vivo In vitro | miR-145a was downregulated; upregulation reduced inflammation |
| Pan et al. [140] | 2018 | In vivo In vitro | miR-145a was downregulated; upregulation reduced inflammation |
| Zhao et al. [120] | 2020 | In vivo | miR-125b was upregulated; it is a marker of sepsis and risk of mortality |
| Sisti et al. [141] | 2018 | In vivo In vitro | miR-125b was upregulated; it is a marker of sepsis and risk of mortality |
| Zhu et al. [142] | 2020 | In vivo | miR-125b was upregulated; it is a marker of sepsis and risk of mortality |
| Sun et al. [143] | 2021 | In vivo In vitro | miR-27b was downregulated; upregulation reduced levels of inflammation |
| Sisti et al. [141] | 2018 | In vivo In vitro | miR-203b was downregulated; its expression reduced inflammation |
| Zheng et al. [144] | 2020 | In vivo In vitro | miR-10 a was downregulated; its expression was associated with disease’s severity scores |
| Liu et al. [145] | 2015 | In vivo | miR-155 was upregulated; upregulation was associated with severity of disease |
| Zhang et al. [146] | 2019 | In vivo | miR-7110-5p was upregulated, representing a good biomarker for sepsis |
| Jiang et al. [147] | 2015 | In vivo In vitro | miR-19a was upregulated, upregulation determined an increase in BCR signaling. Downregulation promoted the expression of CD22 |
| Xu et al. [148] | 2020 | In vivo In vitro | miR-19b-3p was downregulated; upregulation reduced inflammation |
| Liu et al. [149] | 2021 | In vivo In vitro | miR-150 was downregulated; upregulation reduced inflammation |
| Sheng et al. [150] | 2017 | In vivo | miR-375 was downregulated; upregulation reduced inflammation |
| McClure et al. [151] | 2014 | In vivo | miR-21 and miR-181b were upregulated; downregulation reduced immunosuppression and improved bacterial clearance |
| McClure et al. [152] | 2017 | In vivo | miR-21 and miR-181b were upregulated; downregulation reduced immunosuppression and improved bacterial clearance |
| Han et al. [153] | 2016 | In vivo | miR-143 was upregulated, representing a good marker for sepsis |
| Liu et al. [154] | 2020 | In vivo In vitro | miR-20b-5p was upregulated; downregulation reduced inflammation |
| Ji et al. [155] | 2019 | In vivo In vitro | miR-17-5p was downregulated; upregulation reduced inflammation |
| Szilagyi et al. [156] | 2020 | In vivo | miR-26b was downregulated; downregulation increased inflammation and mortality |
| Chen et al. [157] | 2020 | In vivo In vitro | miR-96-5p was downregulated; upregulation reduced inflammation |
| Wang et al. [158] | 2014 | In vivo In vitro | miR-27a was upregulated; downregulation reduced inflammation and promoted survival |
| Yang et al. [159] | 2018 | In vivo | miR-27a was upregulated; downregulation reduced inflammation and promoted survival |
| Wang et al. [43] | 2012 | In vivo | miR-574-5p was upregulated; upregulation increased mortality rate |
| Sun et al. [45] | 2012 | In vivo | miR-181b was downregulated; upregulation reduced inflammation and mortality |
| Yang et al. [160] | 2019 | In vivo In vitro | miR-346 upregulation promoted proliferation |
| Bai et al. [161] | 2020 | In vivo In vitro | miR-148a-3p was upregulated; upregulation induced inflammation |
| Zhang et al. [162] | 2019 | In vivo In vitro | miR-218-5p was upregulated; downregulation improved survival |
| Ma et al. [163] | 2021 | In vivo | miR-1298-5p was upregulated; upregulation reduced inflammation |
| Gu et al. [164] | 2020 | In vitro | miR-608 was upregulated; upregulation reduced inflammation |
| Zhang et al. [165] | 2018 | In vivo | miR-124 was downregulated; upregulation reduced inflammation |
| Huo et al. [166] | 2017 | In vivo | miR-29a was upregulated, and is considered an independent risk factor for mortality in sepsis |
| Cao et al. [167] | 2021 | In vivo In vitro | miR-155 was upregulated; upregulation induced inflammation |
| Ma et al. [168] | 2017 | In vivo In vitro | miR-155 was upregulated; upregulation induced inflammation |
| Wang et al. [169] | 2014 | In vivo | miR-30a was downregulated; upregulation reduced inflammation |
| Mei et al. [170] | 2019 | In vivo In vitro | miR-339-5p was downregulated; upregulation reduced inflammation |
| Wang et al. [171] | 2016 | In vivo | miR-99b was upregulated |
| Yao et al. [172] | 2020 | In vitro | miR-215-5p was downregulated; upregulation reduced inflammatory response |
| Wang et al. [173] | 2012 | In vivo In vitro | miR-15a was upregulated, and is useful to distinguish sepsis from SIRS |
| Liang et al. [174] | 2020 | In vivo In vitro | miR-206 was upregulated, and is considered a good biomarker for sepsis |
| Wang et al. [175] | 2020 | In vivo | miR-92a-3p was downregulated; upregulation induced inflammation |
| Author and Reference | Year of Publication | Sepsis Model | Brief Description of miRNAs in Lung Tissue during Sepsis |
|---|---|---|---|
| Liu et al. [191] | 2017 | In vivo In vitro | miR-155 was upregulated; upregulation attenuated inflammation in vivo and in vitro |
| Jiang et al. [192] | 2019 | In vivo | miR-155 was upregulated; upregulation induced inflammation and macrophage activation |
| Yuan et al. [193] | 2016 | In vivo | miR-155 was upregulated; upregulation induced inflammation. It was targeted by TREM-1 |
| Li et al. [195] | 2020 | In vivo | miR-155 was upregulated; upregulation induced inflammation. Downregulation reduced inflammation |
| Han et al. [196] | 2016 | In vivo | miR-155 was downregulated |
| Vergadi et al. [197] | 2014 | In vivo | miR-146 was upregulated; upregulation protected against LPS-induced lung injury |
| Zeng et al. [198] | 2013 | In vivo | miR-146 was upregulated; upregulation protected against LPS-induced lung injury |
| Xu et al. [199] | 2021 | In vivo | miR-144-3p was upregulated; upregulation induced inflammation and apoptosis |
| Ren et al. [200] | 2021 | In vivo | miR-19a-3p was upregulated; upregulation induced inflammation |
| Liu et al. [201] | 2018 | In vivo | miR-199a was downregulated; downregulation reduced inflammation |
| Chen et al. [202] | 2020 | In vivo | miR-34a was downregulated; downregulation reduced inflammation |
| Qiu et al. [221] | 2020 | In vivo In vitro | miR-34b-5p was downregulated; downregulation reduced inflammation and lung injury |
| Zhu et al. [203] | 2021 | In vivo In vitro | miR-132 was upregulated; upregulation induced LPS-induced lung injury |
| Jiao et al. [204] | 2021 | In vivo | miR-30d-5p was upregulated; upregulation induced lung injury |
| Ma et al. [226] | 2021 | In vivo In vitro | miR-1298-5p was upregulated; upregulation induced inflammation. Downregulation reduced inflammatory response |
| Wang et al. [205] | 2022 | In vivo | miR-92a-3p was upregulated; upregulation induced LPS-induced lung injury |
| Yang et al. [206] | 2018 | In vivo In vitro | miR-23a was upregulated; upregulation reduced inflammation and lung injury |
| Zhou et al. [227] | 2018 | In vivo In vitro | miR-218 was upregulated; upregulation reduced inflammation and lung injury |
| Meng et al. [207] | 2019 | In vivo | miR-539-5p was upregulated; upregulation reduced inflammation and apoptosis |
| Cao et al. [139] | 2019 | In vivo In vitro | miR-145 was downregulated; downregulation induced LPS-induced inflammation |
| Pan et al. [208] | 2019 | In vivo | miR-124 was downregulated, downregulation induced LPS-induced inflammation |
| Leng et al. [209] | 2020 | In vivo In vitro | miR-483-5p was upregulated; upregulation induced inflammation. Downregulation reduced inflammation and apoptosis |
| Lou et al. [210] | 2021 | In vivo In vitro | miR-497-5p was upregulated; downregulation reduced inflammation and apoptosis |
| You et al. [211] | 2020 | In vivo In vitro | miR-802 was downregulated; upregulation reduced inflammation and apoptosis |
| Wang et al. [212] | 2020 | In vivo In vitro | miR-326 was downregulated, upregulation reduced inflammation and lung injury |
| Yang et al. [213] | 2020 | In vivo In vitro | miR-129-5p was upregulated; upregulation reduced inflammation and lung injury |
| Yao et al. [215] | 2021 | In vitro | miR-129 was downregulated; upregulation reduced inflammation and lung injury |
| Lin et al. [214] | 2021 | In vivo | miR-490 was downregulated; upregulation reduced inflammation and lung injury |
| Xie et al. [216] | 2012 | In vitro | miR-127 was upregulated; upregulation reduced inflammation and apoptosis |
| Jiang et al. [217] | 2021 | In vivo | miR-125-5p was downregulated; upregulation reduced inflammation and apoptosis |
| Zhang et al. [218] | 2021 | In vivo | miR-499-5p was downregulated; upregulation reduced inflammation and apoptosis |
| Lu et al. [219] | 2021 | In vitro | miR-942-5p was downregulated; upregulation reduced inflammation and apoptosis |
| Yin et al. [220] | 2021 | In vivo | miR-16-5p was downregulated; upregulation reduced inflammation and lung injury |
| Wang et al. [222] | 2020 | In vitro | miR-195-5p was upregulated; upregulation induced inflammation. Downregulation reduced inflammation and apoptosis |
| Zhu et al. [223] | 2021 | In vitro | miR-152-3p was upregulated; downregulation reduced inflammation |
| Xie et al. [224] | 2021 | In vivo In vitro | miR-128-3p was upregulated; upregulation induced inflammation and macrophage activation |
| Chen et al. [225] | 2020 | In vivo In vitro | miR-424 was downregulated; downregulation increased LPS-induced inflammation |
| Author and Reference | Year of Publication | Sepsis Model | Brief Description of miRNAs in Liver during Sepsis |
|---|---|---|---|
| Yang et al. [229] | 2018 | In vivo | miR-155 is upregulated in septic liver; miR-155 antagomir is linked to reduced septic liver injury |
| Ling et al. [230] | 2018 | In vivo | Low miR-30a levels are linked to an increased expression of FOSL2 and the JAK/STAT pathway |
| Yuan et al. [231] | 2019 | In vivo | High miR-30a levels and diminished SOCS-1 are linked to increased hepatocyte apoptosis |
| Zhu et al. [86] | 2019 | In vivo | Low miR-98 levels are described in septic liver, cardiac and lung tissue. High miR-98 levels seem to protect from septic injury. |
| Zhou and Xia. [232] | 2019 | In vivo | High miR-103a-3p levels are reported in septic mice liver and also in the serum of septic mice and human |
| Chen et al. [233] | 2020 | In vivo | Low miR-103a-3p levels in septic live tissue compared to controls; miR-103a-3p agomiR diminished septic liver damage |
| Li et al. [234] | 2021 | In vivo | Low miR-103a-3p levels in liver and lung tissue of septic mice; diminished levels also in blood samples of human septic samples |
| Gu et al. [235] | 2020 | In vivo | miR-425-5p downregulation is linked to high liver damage in septic mice |
| Zhou et al. [236] | 2020 | In vivo | miR-10a upregulated in mimics group and downregulated in inhibitor group |
| Xu et al. [237] | 2021 | In vivo In vitro | miR-142-5p upregulation increases apoptosis and hepatocyte inflammation |
| Kim et al. [238] | 2021 | In vivo | High miR-147 levels described in liver but also in lung, kidney and stomach; miR-147 could be involved in inflammatory response |
| Li et al. [239] | 2020 | In vitro | miR-373-3p is downregulated in septic liver models; miR-373-3p reduces apoptosis of LPS THLE cells. |
| Li et al. [240] | 2020 | In vivo In vitro | miR-126-5p upregulation is linked to augmented apoptosis and low cell viability |
| Chen et al. [241] | 2020 | In vivo In vitro | miR-204-5p downregulation is associated with high LPS liver injury |
| Han et al. [242] | 2019 | In vivo | miR-9 is downregulated with high levels of MCPIP1 target, which plays a protective role in septic liver injury |
| Yang et al. [243] | 2018 | In vivo | miR-27a upregulation ameliorates liver injury |
| Wang et al. [244] | 2020 | In vivo | Inhibition of miR-640 is linked to reduction in liver damage |
| Author and Reference | Year of Publication | Sepsis Model | Brief Description of miRNAs in Renal Tissue during Sepsis |
|---|---|---|---|
| Xu, G. et al. [254] | 2019 | In vivo | LPS-induced inflammation stimulated the downregulation of miR-15a-5p and the upregulation of XIST and CUL3 with the inhibition of podocyte growth in animals. |
| Fu, D. et al. [255] | 2017 | In vivo | Overexpression of miRNA-21 during sepsis-induced renal cell injury may regulate PTEN/PI3K/AKT signaling and inhibit apoptosis. |
| Wei, W. et al. [256] | 2020 | In vivo | Suppression of miR-21 inhibits cell apoptosis in septic mice and LPS-stimulated human renal tubule cells. miR-21 targets the CDK6 gene inducing cellular apoptosis and leading to septic renal dysfunction |
| Lin Z., et al. [257] | 2018 | In vivo | miR-21-3p upregulated FOXO1, which enhanced cell apoptosis and the cell cycle arrest of tubule epithelial cells during sepsis-induced kidney injury. |
| Wei, W. et al. [258] | 2021 | In vitro | The expression of miR-21e5p is significantly increased in LPS-stimulated HK-2 cells. hsa_circ_0068,888 has a protective effect in LPS-induced AKI by sponging miR-21e5p. |
| Zou Z., et al. [259] | 2020 | In vivo In vitro | Upregulation of miR-21a-3p in plasma and TECs during sepsis is promoted by the internalization of plasmatic Ago2. |
| Zhang P. et al. [261] | 2020 | In vivo In vitro | Overexpression of miR-22-3p inhibits renal cell apoptosis and reduces the accumulation of cleaved caspase-3 by targeting the AIFM1 gene. |
| Wang X. et al. [262] | 2020 | In vivo In vitro | Overexpression of miR-22-3p might have a beneficial effect by attenuating sepsis- or LPS-induced inflammation and apoptosis by targeting PTEN. |
| Ye J. et al. [263] | 2021 | In vivo In vitro | Overexpression of miR-23a-3p inhibits hyperuricemia-induced renal tubular injury and renal cell apoptosis by targeting the WNT5A gene. |
| Chen Y. et al. [264] | 2022 | In vivo | The miR-26a-5p/ IL-6 axis can ameliorate sepsis-induced acute kidney injury by inhibiting renal inflammation. |
| Ha ZL. et al. [265] | 2021 | In vivo In vitro | Downregulation of miR-29b-3p causes podocyte damage by targeting the HDAC4 gene in LPS-induced AKI. |
| Ding G. et al. [266] | 2022 | In vivo In vitro | Downregulation of miR-103a-3p during sepsis is associated with the overexpression of pro-inflammatory CXCL12 gene. |
| Shen, Y. et al. [267] | 2019 | In vivo | Upregulation of miR-106a exacerbated LPS-induced the inflammation and apoptosis of TCMK-1 cells via regulating THBS2 expression |
| Whang, S. et al. [268] | 2017 | In vivo | Upregulation of miR-107 induces the secretion of TNF-α by targeting DUSP7 in endothelial cells, leading to tubule cell injury in septic AKI |
| Zhang H. et al. [269] | 2022 | In vitro | Downregulation of miR-124-3p.1 is directly related to the increased activity of LPCAT3, a key enzyme of phospholipid metabolism involved in cellular ferroptosis |
| Shi L. et al. [270] | 2021 | In vivo In vitro | Downregulation of miR-150-5p by Stat-3 activation in septic models. miR-150-5p can attenuate apoptosis in HK-2 cells, renal inflammatory response, and oxidative stress |
| Ren Y., et al. [272] | 2017 | In vivo | Downregulation of miR-155 in the kidney reduced mortality of septic mice, improved renal function, and suppressed expression of the JAK /STAT pathway |
| Lin Y., et al. [273] | 2019 | In vivo | miR-210, miR-494, and miR-205 have predictive value for the prognosis and survival of patients with sepsis-induced AKI. MiR-205 appears to be an independent risk factor for sepsis-induced AKI and its decreased expression is associated with shorter patient survival |
| Sang Z. et al. [273] | 2020 | In vivo | miR-214 could alleviate AKI in septic models by inhibiting renal autophagy through silencing PTEN expression |
| Guo C., et al. [275] | 2022 | In vivo In vitro | Downregulation of miR-214-5p dramatically reduced renal inflammation and oxidative stress, preventing septic AKI |
| Liu Z. et al. [276] | 2020 | In vivo | Overexpression of miR-452 stimulated the pro-inflammatory NF-κB pathway |
| Qi Y. et al. [277] | 2022 | In vivo | Downregulation of miR-665 was able to inhibit LPS-induced renal inflammation and apoptosis and improve renal function |
| Ge QM. et al. [278] | 2017 | In vivo | Overexpression of these miRNAs is associated with oxidative stress miR-4321 is involved in the regulation of AKT1, mTOR, and NOX5 genes miR-4270 is involved in the regulation of PPARGC1A, AKT3, NOX5, PIK3C3, and WNT1 genes |
| miRNA | Target Gene/Pathway | Upregulation/Downregulation and Its Effect on Brain Function | Author and Reference |
|---|---|---|---|
| miR-181b | - | ↓miR-181b ↓Brain function | Dong et al. [19] |
| miR-200a-3p | NLRP3 | ↑miR-200a-3p ↑NLRP3 ↓Brain function | Yu et al. [21] |
| miR-181b | S1PR1 NCALD | ↑miR-181b ↓S1PR1 and NCALD ↓Brain function | Chen et al. [20] |
| miR-370-3p | - | ↑ miR-370-3p ↓Brain function | Visitchanakun et al. [22] |
| miR-147 | - | ↓ miR-147 ↓Brain function | Kim et al. [23] |
| miR-126 | - | ↓miR-126 - ↓Brain function | Nong et al. [24] |
| miR-190a-3p, miR-3085-3p | - | ↑ miR-190a-3p, ↓miR-383-5p ↑ Brain function | Rani et al. [25] |
| miR-146a | - | ↓ miR-146 a ↑ Brain function | Zou et al. [26] |
| miRNA | Target Gene/Pathway | Upregulation/Downregulation and Its Effect on Heart Function | Author and Reference |
|---|---|---|---|
| miR-223 | STAT3 Sema3A | ↓miR-223 ↑STAT3 and Sema3A ↓Cardiac function | Wang et al. [54] |
| miR-223 | FOXO1 | ↑miR-223 ↓FOXO1 ↑Cardiac function | Liu et al. [56] |
| miR-223 | NF-κB | ↑miR-223 ↑NF-κB - | M’baya-Moutoula et al. [57] |
| miR-27a | Nrf2 | ↑miR-27a ↑Nrf2 ↓Cardiac function | Xue et al. [58] |
| miR-146a | ICAM1 VCAM1 | ↑miR-146a ↓ICAM1 and VCAM1 ↑Cardiac function | Gao et al. [60] |
| miR-146a | TRAF6 IRAK1 | ↑miR-146a ↓ TRAF6 and IRAK1 ↑Cardiac function | An et al. [61] |
| miR-146a | TRAF6 IRAK1 | ↑miR-146a ↓TRAF6 and IRAK1 ↑Cardiac function | Xie et al. [62] |
| miR-146b | Notch1 | ↑miR-146b ↓Notch1 ↑Cardiac function | Wang et al. [63] |
| miR-125b | TRAF6 | ↓miR-125b ↑TRAF6 ↑Cardiac function | Ma et al. [64] |
| miR-150-5p | - | ↓miR-150-5p - ↓Cardiac function | Wei et al. [65] |
| miR-150-5p | Akt2 | ↓miR-150-5p ↑Akt2 ↓Cardiac function | Zhu et al. [66] |
| miR-21-3p | SORBS2 | ↑miR-21-3p ↓SORBS2 ↓Cardiac function | Wang et al. [67] |
| miR-155 | Pea15a | ↓miR-155 ↑Pea15a ↑Cardiac function | Wang et al. [68] |
| miR-155 | Arrb2 | ↑miR-155 ↓Arrb2 ↑Cardiac function | Zhou et al. [71] |
| miR-124a | STX2 | ↑miR-124 ↓STX2 ↑Cardiac function | Diao et al.. [69] |
| miR-135a | - | ↑miR-135a - ↓Cardiac function | Zheng et al. [70] |
| miR-214 | PTEN | ↑miR-214 ↑PTEN ↑Cardiac function | Ge et al. [72] |
| miR-214-3p | PTEN | ↑miR-214-3p ↓PTEN ↑Cardiac function | Sang et al. [73] |
| miR-874 | AQP1 | ↑miR-874 ↓AQP1 ↓Cardiac function | Fang et al. [74] |
| miR-93-3p | TLR4 | ↑miR-93-3p ↓TLR4 ↑Cardiac function | Tang et al. [78] |
| miR-25 | PTEN | ↑miR-25 ↓PTEN ↑Cardiac function | Yao et al. [79] |
| miR-494-3p | PTEN | ↑miR-494-3p ↓PTEN ↑Cardiac function | Wu et al. [80] |
| miR-23b | TGIF1 PTEN | ↑miR-23b ↓TGIF1 and PTEN ↑Cardiac function | Zhang et al. [81] |
| miR-23b | TRAF6 IKKβ | ↑miR-23b ↓TRAF6 and IKKβ ↑Cardiac function | Cao et al. [84] |
| miR-495 | - | ↑miR-495- ↑Cardiac function | Guo et al. [85] |
| miR-98 | HMGA2 | ↑miR-98 ↓HMGA2 ↑Cardiac function | Zhu et al. [86] |
| miR-208-5p | SOCS2 | ↑miR-208-5p ↓SOCS2 ↓Cardiac function | Ouyang et al. [87] |
| miR-328 | - | ↑miR-328 - ↓Cardiac function | Sun et al. [88] |
| miR-29a | SIRT1 | ↑miR-29a ↑Cardiac function | Zhu et al. [91] |
| miR-29a | - | ↑miR-29a ↑Cardiac function | Song et al. [92] |
| miR-24 | XIAP | ↑miR-24 ↓XIAP ↓Cardiac function | Chen et al. [93] |
| miR-192-5p | XIAP | ↓miR-192-5p ↑XIAP ↑Cardiac function | Sun et al. [94] |
| miR-144-3p | - | ↑miR-144-3p ↑Cardiac function | Wei et al. [95] |
| miR-330-5p | TRAF6 | ↑miR-330-5p ↓TRAF6 ↑Cardiac function | Xing et al. [96] |
| miR-1-5p | HSPA4 | ↓miR-1-5p ↑HSPA4 ↑Cardiac function | Han et al. [97] |
| miR-29b-3p | FOXO3A | ↑miR-29b-3p ↓FOXO3A ↑Cardiac function | Li et al. [98] |
| miR-101-3p | DUSP1 | ↓miR-101-3p ↑DUSP1 ↑Cardiac function | Xin et al. [99] |
| miR-106b-5p | PTENP1 | ↑miR-106b-5p ↓PTENP1 ↑Cardiac function | Liu et al. [100] |
| miR-29c-3p | - | ↑miR-29c-3p ↓Cardiac function | Zhang et al. [101] |
| miR-499a-5p | EIF4E | ↑miR-499a-5p ↓EIF4E ↑Cardiac function | Yang et al. [102] |
| miRNA | Target Gene/Pathway | Upregulation/Downregulation and Its Effect on Inflammation Function | Author and Reference |
|---|---|---|---|
| miR-494-3p | TLR6 | ↓miR-494-3p ↓NF-κB ↑Inflammation | Wang et al. [113] |
| miR-218 | VOPP1 | ↓miR-218 ↓VOPP1 ↑Inflammation | Li et al. [114] |
| miR-218 | RUNX2 | ↓miR-218 ↑RUNX2 ↑Inflammation | Zhou et al. [115] |
| miR-122 | - | ↑miR-122 ↑Inflammation | Abou El-Khier NT et al. [116] |
| miR-328 | - | ↑miR-328 ↑Inflammation | Sun et al. [117] |
| miR-21 | - | ↓miR-21 ↑Inflammation | Na et al. [118] |
| miR-126 | EGFL7 | ↑miR-126 ↑Inflammation | Lin et al. [119] |
| miR-125a | - | ↑miR-125a ↑Inflammation | Li et al. [120] |
| miR-146a | - | ↓miR-146a ↑Inflammation | Song et al. [121] |
| miR-146a | NF-κB | ↑miR-146a ↓NF-κB ↓Inflammation | Chen et al. [122] |
| miR-146a | Notch1 | ↑miR-146a ↓Notch1 ↓Inflammation | Bai et al. [123] |
| miR-146a | p38 MAPK | ↑miR-146a ↓p38 MAPK ↓Inflammation | Brudecki et al. [124] |
| miR-146a | IRAK1 TRAF6 | ↑miR-146a ↓IRAK1/TRAF6 ↓Inflammation | Banarjee et al. [125] |
| miR-146a | STAT4 | ↓miR-146a ↑PRKCε/STAT4 ↑Inflammation | Mohnle et al. [126] |
| miR-146a | - | ↑miR-146a ↓Inflammation | Paik et al. [127] |
| miR-146b | p38 MAPK | ↑miR-146b ↓p38 MAPK ↓Inflammation | Chen et al. [122] |
| miR-146b | - | ↑miR-146b ↑Inflammation | Zhang et al. [128] |
| miR-146b | NF-κB | ↓miR-146b ↑NF-κB ↑Inflammation | Gao et al. [129] |
| miR-126 | CASP3 | ↑miR-126 ↑CASP3 ↓Mortality | Zou et al. [130] |
| miR-223 | HIF-1α | ↑miR-223 ↓HIF-1α ↓Inflammation | Dang et al. [131] |
| miR-25 | NOX4 | ↓miR-25 ↑NOX4 ↑Inflammation | Yao et al. [132] |
| miR-21 | NLRP3 NF-κB TNFAIP3 | ↑miR-21 ↑NLRP3 ↑NF-κB ↓TNFAIP3 ↑Inflammation | Xue et al. [133] |
| miR-223 | FOXO1 | ↑miR-223 ↓FOXO1 ↑Inflammation | Liu et al. [56] |
| miR-223 | - | ↑miR-223 ↑Inflammation | Wu et al. [135] |
| miR-205 | HMGB1 | ↑miR-205 ↓HMGB1 ↓Inflammation | Zhou et al. [136] |
| miR-145a | Fli-1/ NF-κB | ↑miR-145a ↓Fli-1/ NF-κB ↓Inflammation | Wu et al. [137] |
| miR-145a | TGF-β2 | ↓miR-145a ↑TGF-β2 ↑Inflammation | Ma et al. [138] |
| miR-145a | TGF-β2 | ↓miR-145a ↑TGF-β2 ↑Inflammation | Cao et al. [139] |
| miR-145a | - | ↑JMJD3 ↓miR-145a ↑Inflammation | Pan et al. [140] |
| miR-125b | - | ↑miR-125b ↑Inflammation | Zhao et al. [120] |
| miR-125b | PTEN/MyD88 | ↑miR-125b ↑PTEN ↓MyD88 ↓Inflammation | Sisti et al. [141] |
| miR-125b | - | ↑miR-125b ↑Inflammation | Zhu et al. [142] |
| miR-27b | NF-κB | ↑miR-27b ↓NF-κB ↓Inflammation | Sun et al. [143] |
| miR-203b | PTEN/MyD88 | ↑miR-203b ↑PTEN ↓MyD88 ↓Inflammation | Sisti et al. [141] |
| miR-10a | MAP3K7/ NF-κB | ↓miR-10a ↑MAP3K7/ NF-κB ↑Inflammation | Zheng et al. [144] |
| miR-155 | - | ↑miR-155 ↑Inflammation | Liu et al. [145] |
| miR-7110-5p | - | ↑miR-7110-5p ↑Inflammation | Zhang et al. [146] |
| miR-19a | - | ↑miR-19a ↑Inflammation | Jiang et al. [147] |
| miR-19b-3p | - | ↑miR-19b-3p ↓Inflammation | Xu et al. [148] |
| miR-150 | ARG1 | ↑miR-150 ↓ARG1 ↓Inflammation | Liu et al. [149] |
| miR-375 | JAK2-STAT3 | ↑miR-375 ↓JAK2-STAT3 ↓Inflammation | Sheng et al. [150] |
| miR-21 miR-181b | NFI-A | ↓miR-21 and miR-181b ↓NFI-A ↓Inflammation | McClure et al. [151] |
| miR-21 miR-181b | STAT3 and C/EBPβ | ↓miR-21 and miR-181b ↓STAT3 and C/EBPβ ↓Inflammation | McClure et al. [152] |
| miR-143 | - | ↑miR-143 ↑Inflammation | Han et al. [153] |
| miR-20b-5p | - | ↑circDNMT3B ↓miR-20b-5p ↓Inflammation | Liu et al. [154] |
| miR-17-5p | TLR4 | ↑miR-17-5p ↓TLR4 ↓Inflammation | Ji et al. [155] |
| miR-26b | SELP | ↓miR-26b ↑SELP ↑Inflammation | Szilagyi et al. [156] |
| miR-96-5p | NAMPT | ↑miR-96-5p ↓NAMPT ↓Inflammation | Chen et al. [157] |
| miR-27a | PPAR γ | ↓miR-27a ↑PPAR γ ↓Inflammation | Wang et al. [158] |
| miR-27a | TAB3 | ↓miR-27a ↑TAB3 ↓Inflammation | Yang et al. [159] |
| miR-574-5p | - | ↑miR-574-5p ↑Inflammation | Wang et al. [43] |
| miR-181b | Importin-α3 NFkβ | ↑miR-181b ↓Importin-α3 and NFkβ ↓Inflammation function | Sun et al. [45] |
| miR-346 | SMAD3 | ↑miR-346 ↓SMAD3 ↑Inflammation | Yang et al. [160] |
| miR-148a-3p | Notch1 | ↑miR-148a-3p ↑Notch1 ↑Inflammation | Bai et al. [161] |
| miR-218-5p | HO-1 | ↓miR-218-5p ↑HO-1 ↓Inflammation | Zhang et al. [162] |
| miR-1298-5p | SOCS6 | ↓miR-1298-5p ↓STAT3 ↓inflammation | Ma et al. [163] |
| miR-608 | ELANE | ↑miR-608 ↓Inflammation | Gu et al. [164] |
| miR-124 | - | ↓miR-124 ↑Inflammation | Zhang et al. [165] |
| miR-29a | - | ↑miR-29a ↑Inflammation | Huo et al. [166] |
| miR-155 | NF-κB | ↑miR-155 ↓NF-κB ↑Inflammation | Cao et al. [167] |
| miR-155 | - | ↑miR-155 ↑Inflammation | Ma et al. [168] |
| miR-30a | MD-2 | ↑miR-30a ↓MD-2 ↑Inflammation | Wang et al. [169] |
| miR-339-5p | HMGB1 | ↑miR-339-5p ↓HMGB1 ↓Inflammation | Mei et al. [170] |
| miR-99b | MFG-E8 | ↑miR-99b ↓MFG-E8 ↑Inflammation | Wang et al. [171] |
| miR-215-5p | ILF3 and LRRFIP1 | ↓miR-215-5p ↑ILF3 and LRRFIP1 ↑Inflammation | Yao et al. [172] |
| miR-15a | - | ↑miR-15a ↑Inflammation | Wang et al. [173] |
| miR-206 | - | ↑miR-206 ↑Inflammation | Liang et al. [174] |
| miR-92a-3p | LCN2 | ↓miR-92a-3p ↓LCN2 ↓Inflammation | Wang et al. [175] |
| miRNA | Target Gene/Pathway | Upregulation/Downregulation and Its Effect on Lung Function | Author and Reference |
|---|---|---|---|
| miR-155 | TAB2 | ↑miR-155 ↓TAB2 ↑Pulmonary function | Liu et al. [191] |
| miR-155 | SHIP1 SOCS1 | ↑miR-155 ↓SHIP1 and SOCS1 ↓Pulmonary function | Jiang et al. [192] |
| miR-155 | TREM1 | ↑miR-155 ↑TREM1 ↓Pulmonary function | Yuan et al. [193] |
| miR-155 | IRF2BP2 | ↑miR-155 ↓IRF2BP2 ↓Pulmonary function | Li et al. [195] |
| miR-155 | - | ↓miR-155 - - | Han et al. [196] |
| miR-146 | Akt2 | ↑miR-146 ↓Akt2 ↑Pulmonary function | Vergadi et al. [197] |
| miR-146 | IRAK1 TRAF6 | ↑miR-146 ↓IRAK1 and TRAF6 ↑Pulmonary function | Zeng et al. [198] |
| miR-144-3p | CAV2 | ↑miR-144-3p ↓CAV2 ↓Pulmonary function | Xu et al. [199] |
| miR-19a-3p | USP13 | ↑miR-19a-3p ↓USP13 ↓Pulmonary function | Ren et al. [200] |
| miR-199a | SIRT1 | ↓miR-199a ↑SIRT1 ↑Pulmonary function | Liu et al. [201] |
| miR-34a | SIRT1 ATG4B | ↓miR-34a ↑SIRT1 and ATG4B ↑Pulmonary function | Chen et al. [202] |
| miR-34b-5p | TUG1 | ↓miR-34b-5p ↑TUG1 ↑Pulmonary function | Qiu et al. [221] |
| miR-132 | SIRT1 | ↑miR-132 ↓SIRT1 ↓Pulmonary function | Zhu et al. [203] |
| miR-30d-5p | SIRT1 SOCS1 | ↑miR-30d-5p ↑SIRT1 and SOCS1 ↓Pulmonary function | Jiao et al. [204] |
| miR-1298-5p | SOCS6 | ↑miR-1298-5p ↓SOCS6 ↓Pulmonary function | Ma et al. [226] |
| miR-92a-3p | AKAP1 | ↑miR-92a-3p ↓AKAP1 ↓Pulmonary function | Wang et al. [205] |
| miR-23a | PTEN | ↑miR-23a ↓PTEN ↑Pulmonary function | Yang et al. [206] |
| miR-218 | RUNX2 | ↑miR-218 ↓RUNX2 ↑Pulmonary function | Zhou et al. [227] |
| miR-539-5p | ROCK1 | ↑miR-539-5p ↓ROCK1 ↑Pulmonary function | Meng et al. [207] |
| miR-145 | TGF-β2 | ↓miR-145 ↑TGF-β2 ↓Pulmonary function | Cao et al. [139] |
| miR-124 | MAPK14 | ↓miR-124 ↑MAPK14 ↓Pulmonary function | Pan et al. [208] |
| miR-483-5p | PIAS1 | ↑miR-483-5p ↓PIAS1 ↓Pulmonary function | Leng et al. [209] |
| miR-497-5p | IL2RB | ↓miR-497-5p ↑IL2RB ↑Pulmonary function | Lou et al. [210] |
| miR-802 | Peli2 | ↑miR-802 ↓Peli2 ↑Pulmonary function | You et al. [211] |
| miR-326 | TLR4 | ↑miR-326 ↓TLR4 ↑Pulmonary function | Wang et al. [212] |
| miR-129-5p | HMGB1 | ↑miR-129-5p ↓HMGB1 ↑Pulmonary function | Yang et al. [213] |
| miR-129 | TAK1 | ↑miR-129 ↓TAK1 ↑Pulmonary function | Yao et al. [215] |
| miR-490 | MRP4 | ↑miR-490 ↓MRP4 ↑Pulmonary function | Lin et al. [214] |
| miR-127 | CD64 | ↑miR-127 ↓CD64 ↑Pulmonary function | Xie et al. [216] |
| miR-125-5p | TOP2A | ↑miR-125-5p ↓TOP2A ↑Pulmonary function | Jiang et al. [217] |
| miR-499-5p | SOX6 | ↑miR-499-5p ↓SOX6 ↑Pulmonary function | Zhang et al. [218] |
| miR-942-5p | TRIM37 | ↑miR-942-5p ↓TRIM37 ↑Pulmonary function | Lu et al. [219] |
| miR-16-5p | BRD4 | ↑miR-16-5p ↓BRD4 ↑Pulmonary function | Yin et al. [220] |
| miR-195-5p | PDK4 | ↑miR-195-5p ↓PDK4 ↓Pulmonary function | Wang et al. [222] |
| miR-152-3p | PDK4 | ↓miR-152-3p ↑PDK4 ↑Pulmonary function | Zhu et al. [223] |
| miR-128-3p | SIRT1 | ↑miR-128-3p ↓SIRT1 ↓Pulmonary function | Xie et al. [224] |
| miR-424 | ROCK2 | ↓miR-424 ↑ROCK2 ↓Pulmonary function | Chen et al. [225] |
| miRNA | Target Gene/Pathway | Upregulation/Downregulation and Its Effect on Liver Function | Author and Reference |
|---|---|---|---|
| miR-155 | Nrf-2 | ↑miR-155 ↓Nrf-2 ↓Liver function | Yang et al. [229] |
| miR-30a | FOSL2 | ↓miR-30a ↑FOSL2 ↓Liver function | Ling et al. [230] |
| miR-30a | SOCS-1 | ↑ miR-30a ↓SOCS-1 ↓Liver function | Yuan et al. [231] |
| miR-98 | HMGA2 | ↓miR-98 ↑HMGA2 ↓Liver function | Zhu et al. [86] |
| miR-103a-3p | FBXW7 | ↑miR-103a-3p ↓FBXW7 ↓Liver function | Zhou and Xia. [232] |
| miR-103a-3p | HMGB1 | ↓miR-103a-3p ↓HMGB1 with miR-103a-3p agomiR ↓Liver function | Chen et al. [233] |
| miR-103a-3p | HMGB1 | ↓miR-103a-3p ↑HMGB1 ↓Liver function | Li et al. [234] |
| miR-425-5p | RIP1 | ↓miR-425-5p ↑RIP1 ↓Liver function | Gu et al. [235] |
| miR-10a | CYP2E1, TGF-β1, and Smad2 | ↑miR-10a in mimics and↓in inhibitor ↑CYP2E1, TGF-β1, and Smad2 in mimics ↓Liver function with miR-10a silencing | Zhou et al. [236] |
| miR-142-5p | - | ↑miR-142-5p ↓Liver function | Xu et al. [237] |
| miR-147 | - | ↑miR-147 ↓Liver function | Kim et al. [238] |
| miR-373-3p | TRIM8 | ↓miR-373-3p ↑TRIM8 ↑Liver function (protective role against apoptosis) | Li et al. [239] |
| miR-126-5p | - | ↑miR-126-5p - ↓Liver function | Li et al. [240] |
| miR-204-5p | TRPM7 | ↓miR-204-5p ↑TRPM7 ↓Liver function | Chen et al. [241] |
| miR-9 | MCPIP1 | ↓miR-9 ↑MCPIP1 ↑Liver function | Han et al. [242] |
| miR-27a | TAB3 | ↑miR-27a ↓TAB3 ↑Liver function | Yang et al. [243] |
| miR-640 | LRP1 | ↑miR-640 ↓LRP1 ↓Liver function | Wang et al. [244] |
| miRNA | Target Gene/Pathway | Upregulation/Downregulation and Its Effect on Renal Function | Author and Reference |
|---|---|---|---|
| miR-15a-5p | XIST CUL3 | ↓miR-15a-5p ↑XIST and CUL3 ↓Renal function | Xu, G. et al. [254] |
| miR-21 | PTEN/PI3K/AKT | ↑miR-21 ↓PTEN/PI3K/AKT ↑Renal function | Fu, D. et al. [255] |
| miR-21 | CDK6 | ↓miR-21 ↓CDK6 ↑Renal function | Wei, W. et al. [256] |
| miR-21-3p | FOXO1 | ↑miR-21-3p ↑FOXO1 ↓Renal function | Lin Z., et al. [257] |
| miR-21e5p | - | ↑hsa_circ_0068,888 ↓miR-21e5p ↑Renal function | Wei, W. et al. [258] |
| miR-21a-3p | - | ↑Ago2 ↑miR-21a-3p ↓Renal function | Zou Z., et al. [259] |
| miR-22-3p | AIFM1 | ↑miR-22-3p ↓AIFM1 ↑Renal function | Zhang P. et al. [261] |
| miR-22-3p | PTEN | ↑miR-22-3p ↓PTEN ↑Renal function | Wang X. et al. [262] |
| miR-23a-3p | WNT5A | ↑miR-23a-3p ↓WNT5A ↑Renal function | Ye J. et al. [263] |
| miR-26a-5p | IL-6 | ↑miR-26a-5p ↓IL-6 ↑Renal function | Chen Y. et al. [264] |
| miR-29b-3p | HDAC4 | ↓miR-29b-3p ↑HDAC4 ↓Renal function | Ha ZL. et al. [265] |
| miR-103a-3p | CXCL12 | ↓miR-103a-3p ↑CXCL12 ↓Renal function | Ding G. et al. [266] |
| miR-106a | THBS2 | ↑miR-106a ↑THBS2 ↓Renal function | Shen, Y. et al. [267] |
| miR-107 | DUSP7 | ↑miR-107 ↓DUSP7 ↓Renal function | Whang, S. et al. [268] |
| miR-124-3p.1 | LPCAT3 | ↓miR-124-3p.1 ↑LPCAT3 ↓Renal function | Zhang H. et al. [269] |
| miR-150-5p | MEKK3/JNK | ↓miR-150-5p ↑MEKK3/JNK ↓Renal function | Shi L. et al. [270] |
| miR-155 | JAK /STAT | ↓miR-155 ↓JAK /STAT ↑Renal function | Ren Y., et al. [272] |
| miR-210 miR-494 miR-205 | - - - | ↑miR-210 ↑Renal function ↑miR-494 ↑Renal function ↓miR-205 ↓Renal function | Lin Y., et al. [273] |
| miR-214 | PTEN | ↑miR-214 ↓PTEN ↑Renal function | Sang Z. et al. [273] |
| miR-214-5p | - | ↓miR-214-5p ↑Renal function | Guo C., et al. [275] |
| miR-452 | - | ↑miR-452 ↑NF-κB pathway ↓Renal function | Liu Z. et al. [276] |
| miR-665 | BCL-2 | ↑miR-665 ↓BLC-2 ↓Renal function | Qi Y. et al. [277] |
| miR-4321 miR-4270 | AKT1, mTOR, NOX5 PPARGC1A, AKT3, NOX5, PIK3C3, WNT1 | ↑miR-4321 ↑AKT1, mTOR, NOX5 ↓Renal function ↑miR-21 ↑PPARGC1A, AKT3, NOX5, PIK3C3, WNT1 ↓Renal function | Ge QM. et al. [278] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiese, A.; Scatena, A.; Costantino, A.; Chiti, E.; Occhipinti, C.; La Russa, R.; Di Paolo, M.; Turillazzi, E.; Frati, P.; Fineschi, V. Expression of MicroRNAs in Sepsis-Related Organ Dysfunction: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 9354. https://doi.org/10.3390/ijms23169354
Maiese A, Scatena A, Costantino A, Chiti E, Occhipinti C, La Russa R, Di Paolo M, Turillazzi E, Frati P, Fineschi V. Expression of MicroRNAs in Sepsis-Related Organ Dysfunction: A Systematic Review. International Journal of Molecular Sciences. 2022; 23(16):9354. https://doi.org/10.3390/ijms23169354
Chicago/Turabian StyleMaiese, Aniello, Andrea Scatena, Andrea Costantino, Enrica Chiti, Carla Occhipinti, Raffaele La Russa, Marco Di Paolo, Emanuela Turillazzi, Paola Frati, and Vittorio Fineschi. 2022. "Expression of MicroRNAs in Sepsis-Related Organ Dysfunction: A Systematic Review" International Journal of Molecular Sciences 23, no. 16: 9354. https://doi.org/10.3390/ijms23169354
APA StyleMaiese, A., Scatena, A., Costantino, A., Chiti, E., Occhipinti, C., La Russa, R., Di Paolo, M., Turillazzi, E., Frati, P., & Fineschi, V. (2022). Expression of MicroRNAs in Sepsis-Related Organ Dysfunction: A Systematic Review. International Journal of Molecular Sciences, 23(16), 9354. https://doi.org/10.3390/ijms23169354








