CXCL14 Attenuates Triple-Negative Breast Cancer Progression by Regulating Immune Profiles of the Tumor Microenvironment in a T Cell-Dependent Manner
Abstract
:1. Introduction
2. Results
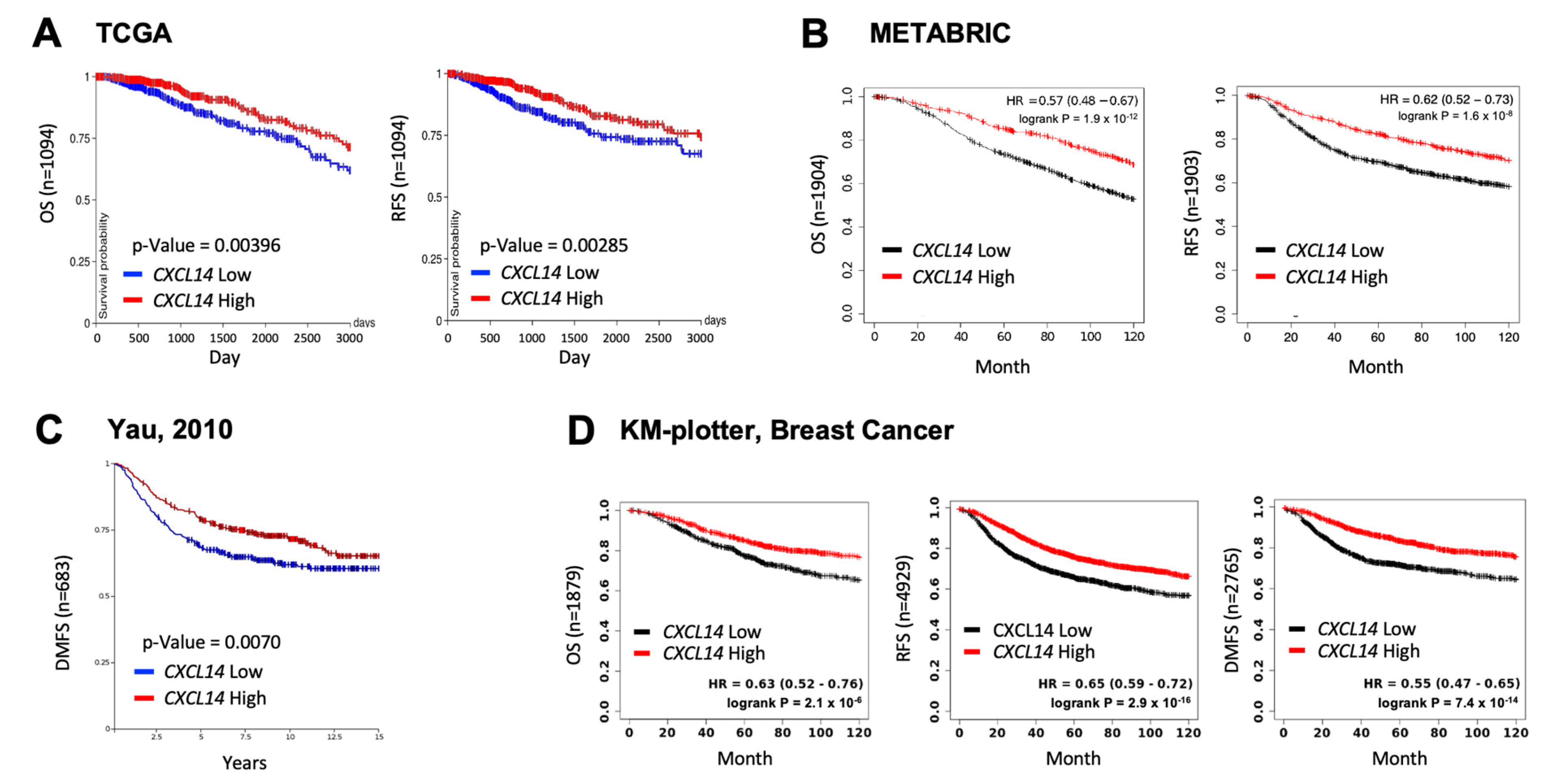
2.1. High Levels of CXCL14 Correlated with Better Survivals in Patients with Breast Cancer
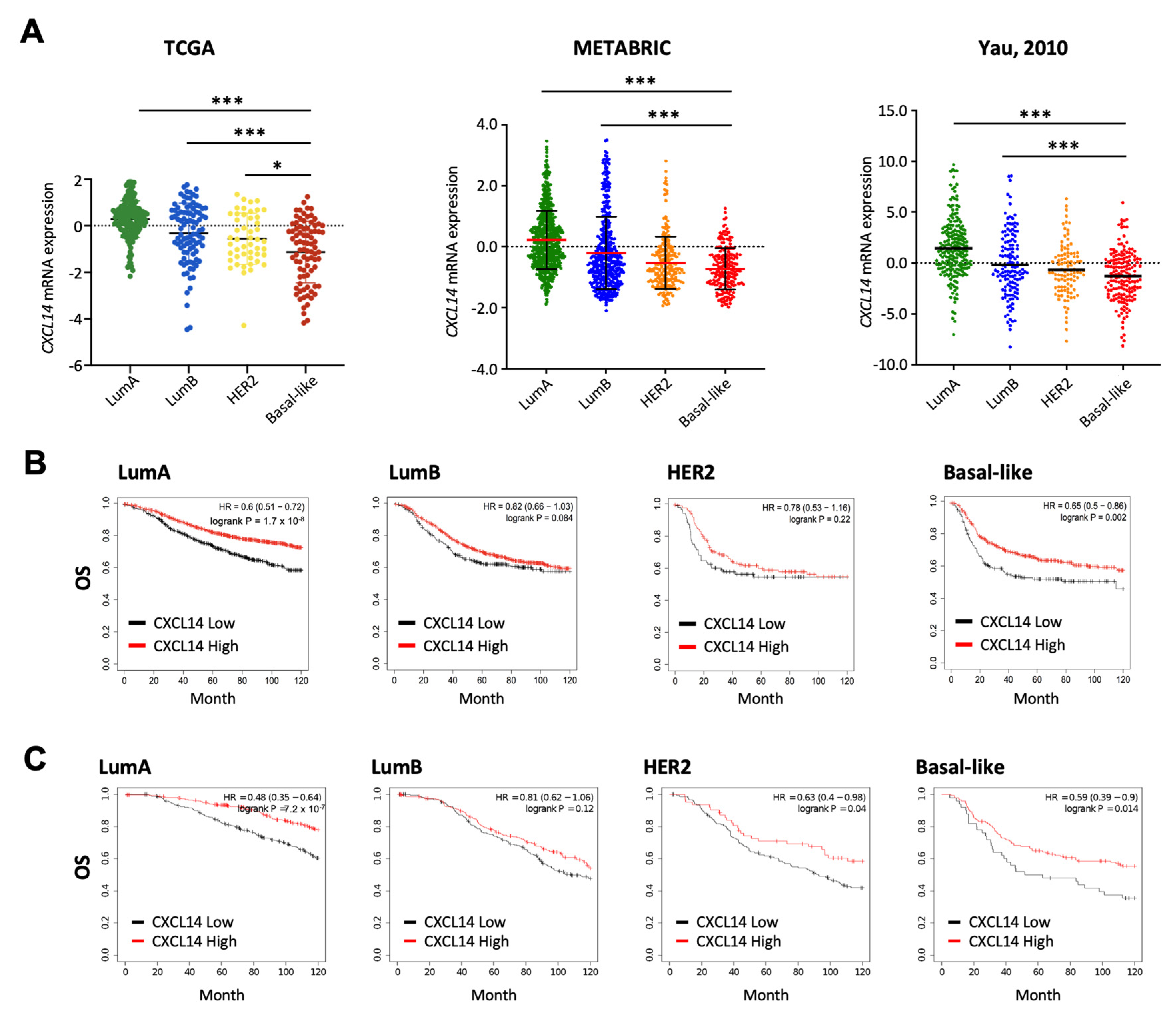
2.2. Comparison of CXCL14 Expression Levels and CXCL14-Contributed Survivals in Breast Cancer Subtypes
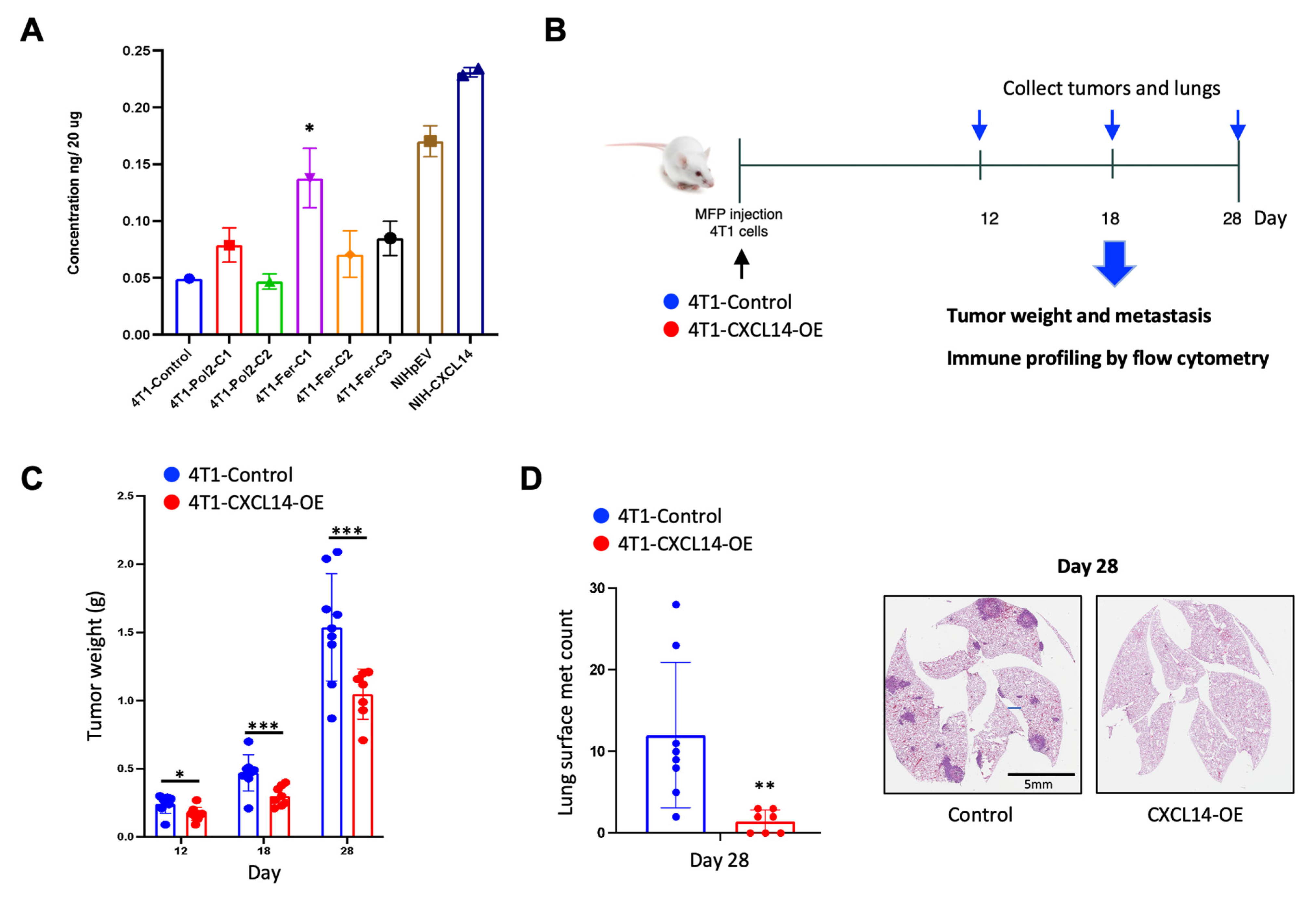
2.3. Overexpression of CXCL14 Decreased Breast Tumor Growth and Metastasis
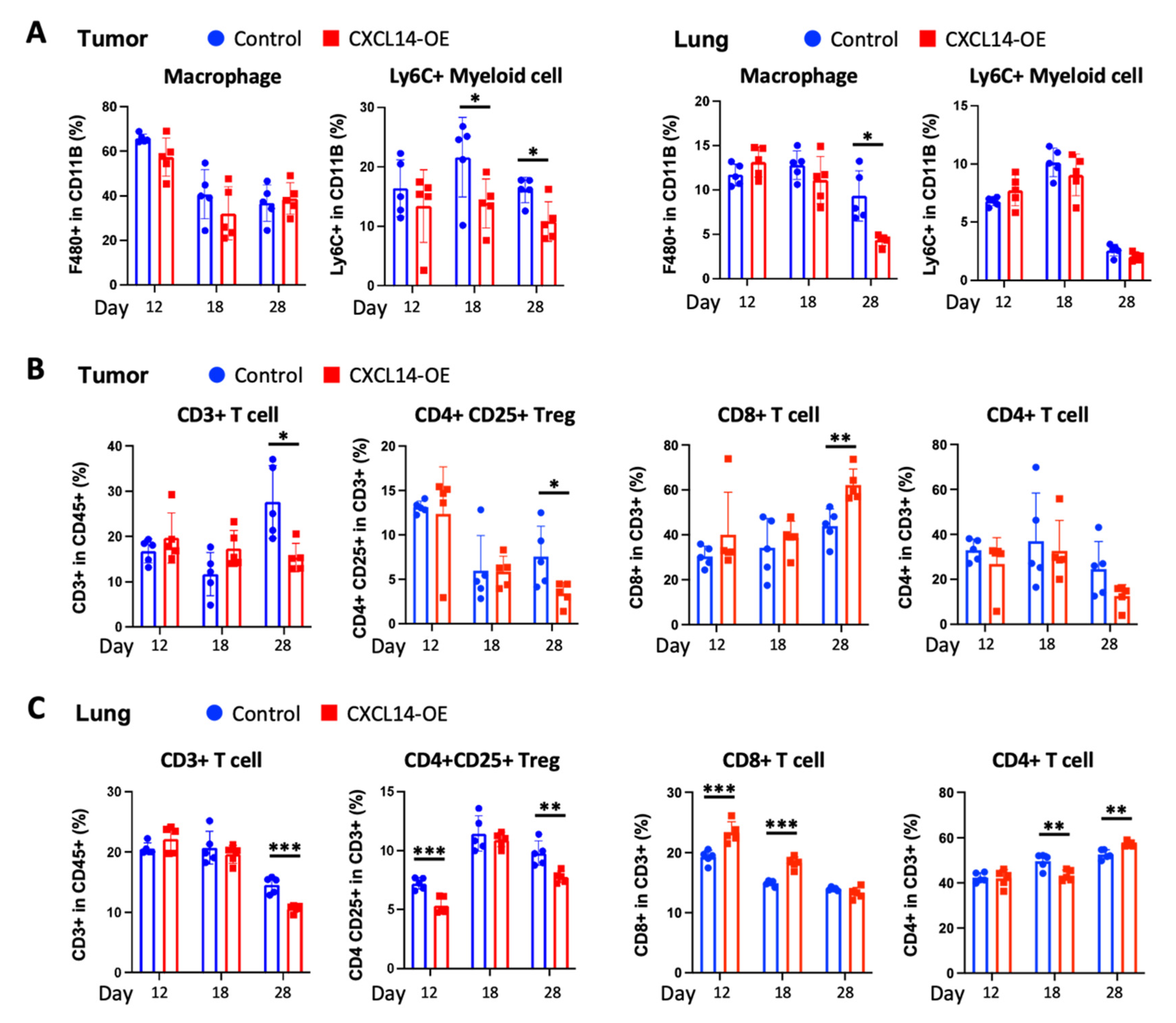
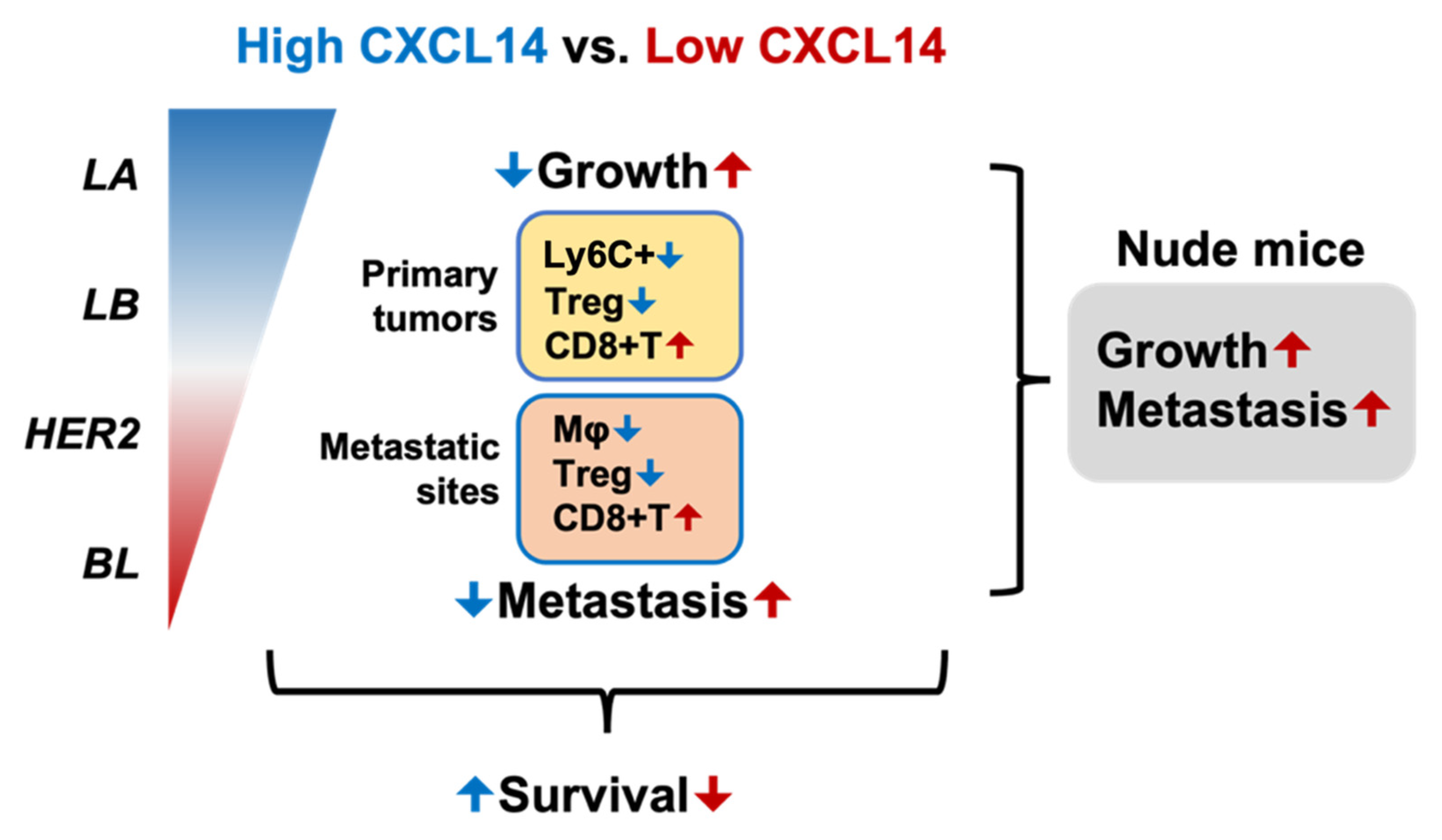
2.4. CXCL14 Modulates Immune Profiles in the Tumor Microenvironment in Primary Tumors and Metastatic Sites
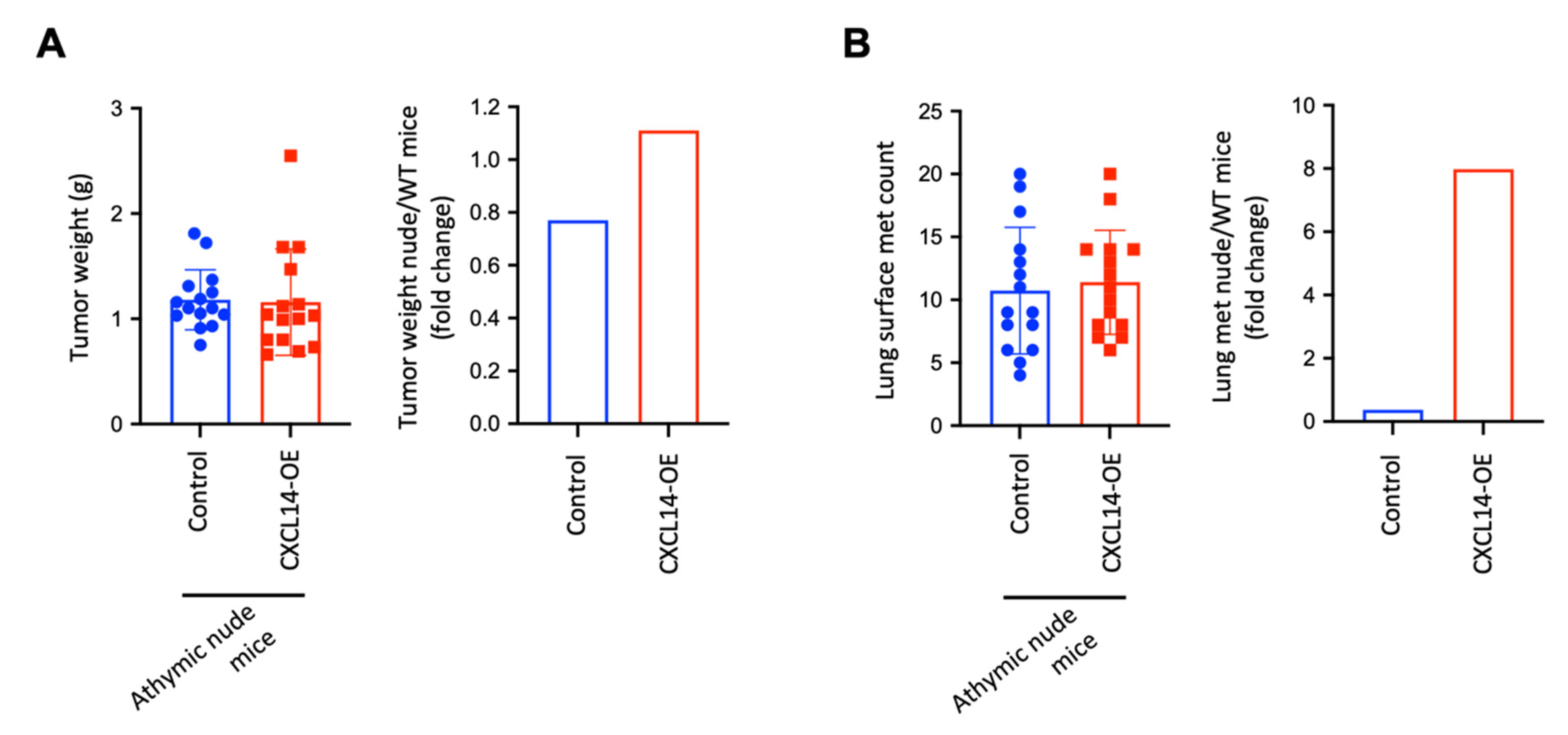
2.5. CXCL14 Attenuates Tumor Growth and Metastasis in a T Cell-Dependent Manner
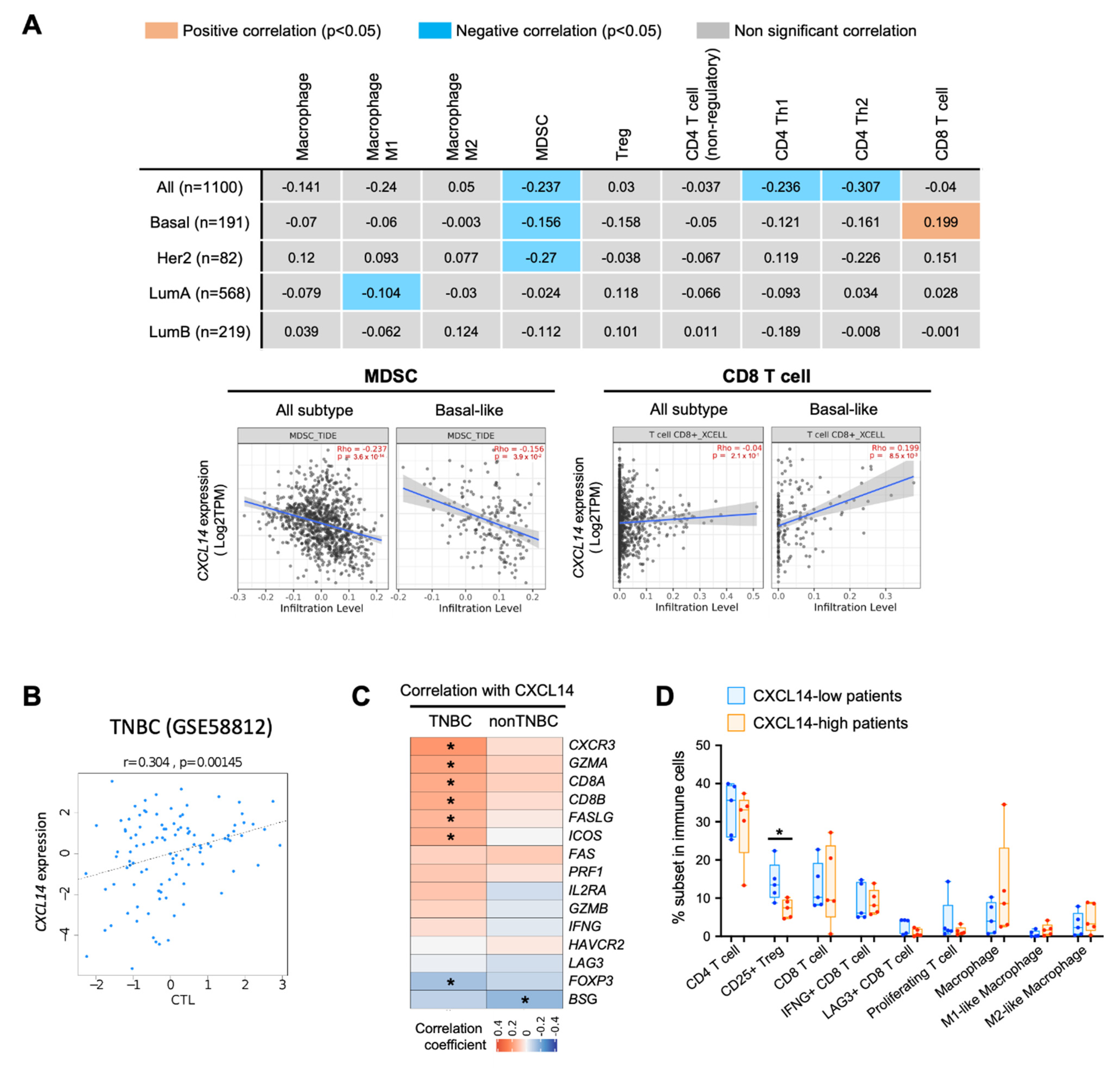
2.6. Correlation of CXCL14 Expression with Immune Modulators in Breast Cancer Subtypes
3. Discussion
4. Materials and Methods
4.1. Data Analysis of CXCL14 Correlation with Patient Survival and Breast-Cancer Subtypes
4.2. Cell Line and Cell Culture
4.3. Generation of CXCL14 Overexpressed Cell Line
4.4. Single Cell Preparation from Tissue Samples
4.5. Flow Cytometry Analysis
4.6. In Vivo Animal Study
4.7. H&E Staining
4.8. Human Correlation of CXCL14 with the Immune Microenvironment
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Kaljee, L.M. Health Disparities and Triple-Negative Breast Cancer in African American Women: A Review. JAMA Surg. 2017, 152, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Hossain, F.; Danos, D.; Lassak, A.; Scribner, R.; Miele, L. Racial Disparities in Triple Negative Breast Cancer: A Review of the Role of Biologic and Non-biologic Factors. Front. Public Health 2020, 8, 576964. [Google Scholar] [CrossRef] [PubMed]
- Foy, K.C.; Fisher, J.L.; Lustberg, M.B.; Gray, D.M.; DeGraffinreid, C.R.; Paskett, E.D. Disparities in breast cancer tumor characteristics, treatment, time to treatment, and survival probability among African American and white women. NPJ Breast Cancer 2018, 4, 7. [Google Scholar] [CrossRef]
- Kumar, P.; Aggarwal, R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 2016, 293, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Picon-Ruiz, M.; Morata-Tarifa, C.; Valle-Goffin, J.J.; Friedman, E.R.; Slingerland, J.M. Obesity and adverse breast cancer risk and outcome: Mechanistic insights and strategies for intervention. CA Cancer J. Clin. 2017, 67, 378–397. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Brumec, M.; Sobocan, M.; Takac, I.; Arko, D. Clinical Implications of Androgen-Positive Triple-Negative Breast Cancer. Cancers 2021, 13, 1642. [Google Scholar] [CrossRef]
- Locatelli, M.A.; Curigliano, G.; Eniu, A. Extended Adjuvant Chemotherapy in Triple-Negative Breast Cancer. Breast Care 2017, 12, 152–158. [Google Scholar] [CrossRef]
- Nedeljkovic, M.; Damjanovic, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Deepak, K.G.K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; S, N.; Rao, D.N.; Malla, R.R. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020, 153, 104683. [Google Scholar] [CrossRef] [PubMed]
- Wein, L.; Loi, S. Mechanisms of resistance of chemotherapy in early-stage triple negative breast cancer (TNBC). Breast 2017, 34 (Suppl. 1), S27–S30. [Google Scholar] [CrossRef]
- Stone, M.J.; Hayward, J.A.; Huang, C.; Z, E.H.; Sanchez, J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int. J. Mol. Sci. 2017, 18, 342. [Google Scholar] [CrossRef] [PubMed]
- Ignacio, R.M.C.; Gibbs, C.R.; Lee, E.S.; Son, D.S. The TGFalpha-EGFR-Akt signaling axis plays a role in enhancing proinflammatory chemokines in triple-negative breast cancer cells. Oncotarget 2018, 9, 29286–29303. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chatterjee, M.; Schmid, H.; Beck, S.; Gawaz, M. CXCL14 as an emerging immune and inflammatory modulator. J. Inflamm. 2016, 13, 1. [Google Scholar] [CrossRef]
- Kouzeli, A.; Collins, P.J.; Metzemaekers, M.; Meyrath, M.; Szpakowska, M.; Artinger, M.; Struyf, S.; Proost, P.; Chevigne, A.; Legler, D.F.; et al. CXCL14 Preferentially Synergizes With Homeostatic Chemokine Receptor Systems. Front. Immunol. 2020, 11, 561404. [Google Scholar] [CrossRef]
- Lee, H.T.; Liu, S.P.; Lin, C.H.; Lee, S.W.; Hsu, C.Y.; Sytwu, H.K.; Hsieh, C.H.; Shyu, W.C. A Crucial Role of CXCL14 for Promoting Regulatory T Cells Activation in Stroke. Theranostics 2017, 7, 855–875. [Google Scholar] [CrossRef]
- Lv, J.; Wu, Z.L.; Gan, Z.; Gui, P.; Yao, S.L. CXCL14 Overexpression Attenuates Sepsis-Associated Acute Kidney Injury by Inhibiting Proinflammatory Cytokine Production. Mediat. Inflamm. 2020, 2020, 2431705. [Google Scholar] [CrossRef]
- Westrich, J.A.; Vermeer, D.W.; Silva, A.; Bonney, S.; Berger, J.N.; Cicchini, L.; Greer, R.O.; Song, J.I.; Raben, D.; Slansky, J.E.; et al. CXCL14 suppresses human papillomavirus-associated head and neck cancer through antigen-specific CD8(+) T-cell responses by upregulating MHC-I expression. Oncogene 2019, 38, 7166–7180. [Google Scholar] [CrossRef]
- Hasegawa, T.; Feng, Z.; Yan, Z.; Ngo, K.H.; Hosoi, J.; Demehri, S. Reduction in Human Epidermal Langerhans Cells with Age Is Associated with Decline in CXCL14-Mediated Recruitment of CD14(+) Monocytes. J. Investig. Dermatol. 2020, 140, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Kleer, C.G.; Bloushtain-Qimron, N.; Chen, Y.H.; Carrasco, D.; Hu, M.; Yao, J.; Kraeft, S.K.; Collins, L.C.; Sabel, M.S.; Argani, P.; et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin. Cancer Res. 2008, 14, 5357–5367. [Google Scholar] [CrossRef]
- Kumar, A.; Mohamed, E.; Tong, S.; Chen, K.; Mukherjee, J.; Lim, Y.; Wong, C.M.; Boosalis, Z.; Shai, A.; Pieper, R.O.; et al. CXCL14 promotes a robust brain tumor-associated immune response in glioma. Clin. Cancer Res. 2022. [Google Scholar] [CrossRef]
- Lin, K.; Zou, R.; Lin, F.; Zheng, S.; Shen, X.; Xue, X. Expression and effect of CXCL14 in colorectal carcinoma. Mol. Med. Rep. 2014, 10, 1561–1568. [Google Scholar] [CrossRef]
- Ignacio, R.M.C.; Lee, E.S.; Wilson, A.J.; Beeghly-Fadiel, A.; Whalen, M.M.; Son, D.S. Chemokine Network and Overall Survival in TP53 Wild-Type and Mutant Ovarian Cancer. Immune Netw. 2018, 18, e29. [Google Scholar] [CrossRef]
- Zeng, J.; Yang, X.; Cheng, L.; Liu, R.; Lei, Y.; Dong, D.; Li, F.; Lau, Q.C.; Deng, L.; Nice, E.C.; et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J. Transl. Med. 2013, 11, 6. [Google Scholar] [CrossRef]
- Li, X.; Zhao, L.; Meng, T. Upregulated CXCL14 is associated with poor survival outcomes and promotes ovarian cancer cells proliferation. Cell Biochem. Funct. 2020, 38, 613–620. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, Q.; Wu, X.; Yu, Y.; Zhang, H. Effect of chemokine CXCL14 on in vitro angiogenesis of human hepatocellular carcinoma cells. Arch. Physiol. Biochem. 2020, 1–7. [Google Scholar] [CrossRef]
- Gu, X.L.; Ou, Z.L.; Lin, F.J.; Yang, X.L.; Luo, J.M.; Shen, Z.Z.; Shao, Z.M. Expression of CXCL14 and its anticancer role in breast cancer. Breast Cancer Res. Treat 2012, 135, 725–735. [Google Scholar] [CrossRef]
- Schwarze, S.R.; Luo, J.; Isaacs, W.B.; Jarrard, D.F. Modulation of CXCL14 (BRAK) expression in prostate cancer. Prostate 2005, 64, 67–74. [Google Scholar] [CrossRef]
- Hata, R.; Izukuri, K.; Kato, Y.; Sasaki, S.; Mukaida, N.; Maehata, Y.; Miyamoto, C.; Akasaka, T.; Yang, X.; Nagashima, Y.; et al. Suppressed rate of carcinogenesis and decreases in tumour volume and lung metastasis in CXCL14/BRAK transgenic mice. Sci. Rep. 2015, 5, 9083. [Google Scholar] [CrossRef] [PubMed]
- Takiguchi, S.; Korenaga, N.; Inoue, K.; Sugi, E.; Kataoka, Y.; Matsusue, K.; Futagami, K.; Li, Y.J.; Kukita, T.; Teramoto, N.; et al. Involvement of CXCL14 in osteolytic bone metastasis from lung cancer. Int. J. Oncol. 2014, 44, 1316–1324. [Google Scholar] [CrossRef]
- Gowhari Shabgah, A.; Haleem Al-Qaim, Z.; Markov, A.; Valerievich Yumashev, A.; Ezzatifar, F.; Ahmadi, M.; Mohammad Gheibihayat, S.; Gholizadeh Navashenaq, J. Chemokine CXCL14; a double-edged sword in cancer development. Int. Immunopharmacol. 2021, 97, 107681. [Google Scholar] [CrossRef]
- Veglia, F.; Sanseviero, E.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021, 21, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.; Jiang, J.; Pang, Y.; Achyut, B.R.; Lizardo, M.; Liang, X.; Hunter, K.; Khanna, C.; Hollander, C.; Yang, L. CCL9 Induced by TGFbeta Signaling in Myeloid Cells Enhances Tumor Cell Survival in the Premetastatic Organ. Cancer Res. 2015, 75, 5283–5298. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression—Implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Peterson, F.C.; Thorpe, J.A.; Harder, A.G.; Volkman, B.F.; Schwarze, S.R. Structural determinants involved in the regulation of CXCL14/BRAK expression by the 26 S proteasome. J. Mol. Biol. 2006, 363, 813–822. [Google Scholar] [CrossRef]
- Liu, K.; Wu, L.; Yuan, S.; Wu, M.; Xu, Y.; Sun, Q.; Li, S.; Zhao, S.; Hua, T.; Liu, Z.J. Structural basis of CXC chemokine receptor 2 activation and signalling. Nature 2020, 585, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Yau, C.; Esserman, L.; Moore, D.H.; Waldman, F.; Sninsky, J.; Benz, C.C. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010, 12, R85. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Colaprico, A.; Silva, T.C.; Chen, J.; An, H.; Ban, Y.; Huang, H.; Wang, L.; James, J.L.; Balko, J.M.; et al. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun. 2021, 12, 6276. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibbs, C.; So, J.Y.; Ahad, A.; Michalowski, A.M.; Son, D.-S.; Li, Y. CXCL14 Attenuates Triple-Negative Breast Cancer Progression by Regulating Immune Profiles of the Tumor Microenvironment in a T Cell-Dependent Manner. Int. J. Mol. Sci. 2022, 23, 9314. https://doi.org/10.3390/ijms23169314
Gibbs C, So JY, Ahad A, Michalowski AM, Son D-S, Li Y. CXCL14 Attenuates Triple-Negative Breast Cancer Progression by Regulating Immune Profiles of the Tumor Microenvironment in a T Cell-Dependent Manner. International Journal of Molecular Sciences. 2022; 23(16):9314. https://doi.org/10.3390/ijms23169314
Chicago/Turabian StyleGibbs, Carla, Jae Young So, Abdul Ahad, Aleksandra M. Michalowski, Deok-Soo Son, and Yang Li. 2022. "CXCL14 Attenuates Triple-Negative Breast Cancer Progression by Regulating Immune Profiles of the Tumor Microenvironment in a T Cell-Dependent Manner" International Journal of Molecular Sciences 23, no. 16: 9314. https://doi.org/10.3390/ijms23169314
APA StyleGibbs, C., So, J. Y., Ahad, A., Michalowski, A. M., Son, D.-S., & Li, Y. (2022). CXCL14 Attenuates Triple-Negative Breast Cancer Progression by Regulating Immune Profiles of the Tumor Microenvironment in a T Cell-Dependent Manner. International Journal of Molecular Sciences, 23(16), 9314. https://doi.org/10.3390/ijms23169314





