Enhancer-Mediated Formation of Nuclear Transcription Initiation Domains
Abstract
:1. Introduction
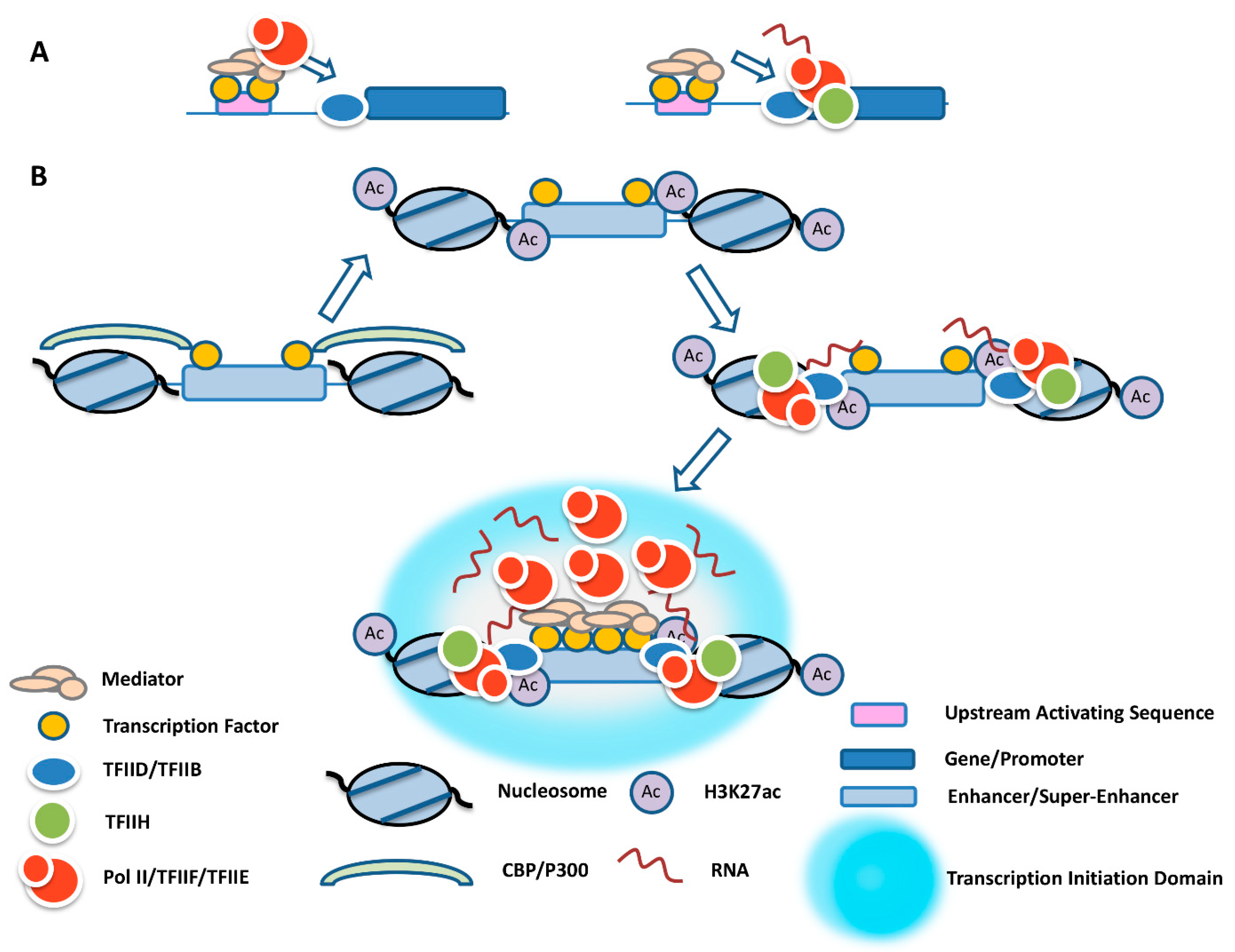
2. Promoter and Formation of a Pol II Transcription Complex
3. Enhancers and Super-Enhancers
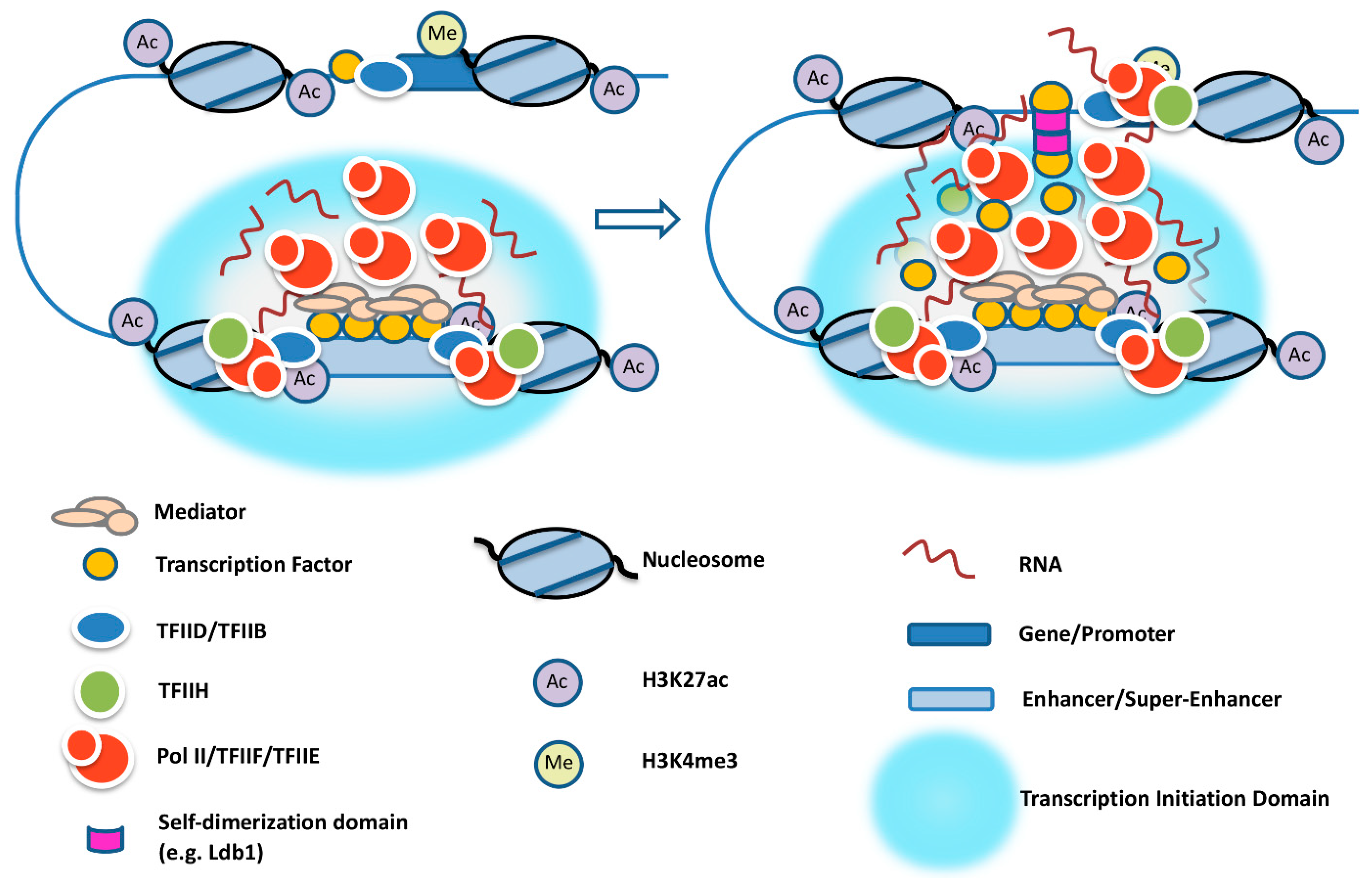
4. Transcription Complex Recruitment at Enhancers, Super-Enhancers, and Yeast Upstream Activating Sequences
5. Enhancer-Mediated Transcription Bursting
6. The Role of Enhancer Transcription and eRNA
7. Establishment of Promoter–Enhancer Contacts
8. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rank, G.; Prestel, M.; Paro, R. Transcription through Intergenic Chromosomal Memory Elements of the Drosophila Bithorax Complex Correlates with an Epigenetic Switch. Mol. Cell. Biol. 2002, 22, 8026–8034. [Google Scholar] [CrossRef]
- Tuan, D.; Kong, S.; Hu, K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc. Natl. Acad. Sci. USA 1992, 89, 11219–11223. [Google Scholar] [CrossRef]
- Ashe, H.L.; Monks, J.; Wijgerde, M.; Proudfoot, N.J. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997, 11, 2494–2509. [Google Scholar] [CrossRef]
- Szutorisz, H.; Canzonetta, C.; Georgiou, A.; Chow, C.-M.; Tora, L.; Dillon, N. Formation of an Active Tissue-Specific Chromatin Domain Initiated by Epigenetic Marking at the Embryonic Stem Cell Stage. Mol. Cell. Biol. 2005, 25, 1804–1820. [Google Scholar] [CrossRef]
- Levings, P.P.; Bungert, J. The human beta-globin locus control region: A center of attraction. Eur. J. Biochem. 2002, 269, 1589–1599. [Google Scholar] [CrossRef]
- Szutorisz, H.; Dillon, N.; Tora, L. The role of enhancers as centers for general transcription factor recruitment. Trends Biochem. Sci. 2005, 30, 593–599. [Google Scholar] [CrossRef]
- Johnson, K.D.; Christensen, H.M.; Zhao, B.; Bresnick, E.H. Distinct Mechanisms Control RNA Polymerase II Recruitment to a Tissue-Specific Locus Control Region and a Downstream Promoter. Mol. Cell 2001, 8, 465–471. [Google Scholar] [CrossRef]
- Leach, K.M.; Nightingale, K.; Igarashi, K.; Levings, P.P.; Engel, J.D.; Becker, P.B.; Bungert, J. Reconstitution of Human β-Globin Locus Control Region Hypersensitive Sites in the Absence of Chromatin Assembly. Mol. Cell. Biol. 2001, 21, 2629–2640. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef]
- Koch, F.; Fenouil, R.; Gut, M.; Cauchy, P.; Albert, T.K.; Zacarias-Cabeza, J.; Spicuglia, S.; De La Chapelle, A.L.; Heidemann, M.; Hintermair, C.; et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 2011, 18, 956–963. [Google Scholar] [CrossRef]
- Baek, I.; Friedman, L.J.; Gelles, J.; Buratowski, S. Single-molecule studies reveal branched pathways for activator-dependent assembly of RNA polymerase II pre-initiation complexes. Mol. Cell 2021, 81, 3576–3588. [Google Scholar] [CrossRef]
- Roeder, R.G. 50+ years of eukaryotic transcription: An expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 2019, 26, 783–791. [Google Scholar] [CrossRef]
- Patel, A.B.; Louder, R.K.; Greber, B.J.; Grünberg, S.; Luo, J.; Fang, J.; Liu, Y.; Ranish, J.; Hahn, S.; Nogales, E. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 2018, 362, eaau8872. [Google Scholar] [CrossRef]
- Chen, X.; Qi, Y.; Wu, Z.; Wang, X.; Li, J.; Zhao, D.; Hou, H.; Li, Y.; Yu, Z.; Liu, W.; et al. Structural insights into preinitiation complex assembly on core promoters. Science 2021, 372, eaba8490. [Google Scholar] [CrossRef]
- Harlen, K.M.; Churchman, L.S. The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Dollinger, R.; Gilmour, D.S. Regulation of Promoter Proximal Pausing of RNA Polymerase II in Metazoans. J. Mol. Biol. 2021, 33, 166897. [Google Scholar] [CrossRef]
- Bhuiyan, T.; Timmers, H.M. Promoter Recognition: Putting TFIID on the Spot. Trends Cell Biol. 2019, 29, 752–763. [Google Scholar] [CrossRef]
- Schaffner, W. Enhancers, enhancers-from their discovery to today’s universe of transcription enhancers. Biol. Chem. 2015, 396, 311–327. [Google Scholar] [CrossRef]
- Field, A.; Adelman, K. Evaluating Enhancer Function and Transcription. Annu. Rev. Biochem. 2020, 89, 213–234. [Google Scholar] [CrossRef]
- Bose, D.A.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 2017, 168, 135–149. [Google Scholar] [CrossRef]
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Prakash, C.; Sum, C.; Gong, Y.; Li, Y.; Kwok, J.J.T.; Thiessen, N.; Pettersson, S.; Jones, S.; Knapp, S.; et al. Bromodomain-containing Protein 4 (BRD4) Regulates RNA Polymerase II Serine 2 Phosphorylation in Human CD4+ T Cells. J. Biol. Chem. 2012, 287, 43137–43155. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G.; Groudine, M. Controlling the double helix. Nature 2003, 421, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Pott, S.; Lieb, J.D. What are super-enhancers. Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Dębek, S.; Juszczyński, P. Super enhancers as master gene regulators in the pathogenesis of hematologic malignancies. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188697. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef]
- Cho, W.-K.; Spille, J.-H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef]
- Morinière, J.; Rousseaux, S.; Steuerwald, U.; Soler-López, M.; Curtet, S.; Vitte, A.-L.; Govin, J.; Gaucher, J.; Sadoul, K.; Hart, D.J.; et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 2009, 461, 664–668. [Google Scholar] [CrossRef]
- Dey, A.; Chitsaz, F.; Abbasi, A.; Misteli, T.; Ozato, K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 2003, 100, 8758–8763. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, A.; O’Malley, B.W. Mechanisms of enhancer action: The known and the unknown. Genome Biol. 2021, 22, 108. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Rupon, J.W.; Krivega, I.; Breda, L.; Motta, I.; Jahn, K.S.; Reik, A.; Gregory, P.D.; Rivella, S.; Dean, A.; et al. Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell 2014, 158, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Benabdallah, N.S.; Williamson, I.; Illingworth, R.S.; Kane, L.; Boyle, S.; Sengupta, D.; Grimes, G.R.; Therizols, P.; Bickmore, W.A. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 2019, 76, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Gurumurthy, A.; Shen, Y.; Gunn, E.M.; Bungert, J. Phase separation and transcription regulation: Are super-enhancers and locus control regions primary sites of transcription complex assembly? Bioessays 2019, 41, e1800164. [Google Scholar] [CrossRef]
- Fukaya, T.; Lim, B.; Levine, M. Enhancer Control of Transcriptional Bursting. Cell 2016, 166, 358–368. [Google Scholar] [CrossRef]
- Lim, B. Imaging transcriptional dynamics. Curr. Opin. Biotechnol. 2018, 52, 49–55. [Google Scholar] [CrossRef]
- Henninger, J.E.; Oksuz, O.; Shrinivas, K.; Sagi, I.; LeRoy, G.; Zheng, M.M.; Andrews, J.O.; Zamudio, A.V.; Lazaris, C.; Hannett, N.M.; et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 2021, 184, 207–225. [Google Scholar] [CrossRef]
- Donovan, B.T.; Huynh, A.; Ball, D.A.; Patel, H.P.; Poirier, M.G.; Larson, D.R.; Ferguson, M.L.; Lenstra, T.L. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019, 38, e100809. [Google Scholar] [CrossRef]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2022, 28, 52–57. [Google Scholar] [CrossRef]
- Zhou, Z.; Giles, K.E.; Felsenfeld, G. DNA·RNA triple helix formation can function as a cis-acting regulatory mechanism at the human β-globin locus. Proc. Natl. Acad. Sci. USA 2019, 116, 6130–6139. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.W.; Li, S.; Franco, H.L. Transcriptional control by enhancers and enhancer RNAs. Transcription 2019, 10, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Andersson, R.; Refsing Andersen, P.; Valen, E.; Core, L.J.; Bornholdt, J.; Boyd, M.; Jensen, T.H.; Sandelin, A. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat. Commun. 2014, 5, 5226. [Google Scholar] [CrossRef]
- Core, L.J.; Martins, A.L.; Danko, C.G.; Waters, C.T.; Siepel, A.; Lis, J.T. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014, 46, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef]
- Henriques, T.; Scruggs, B.S.; Inouye, M.O.; Muse, G.W.; Williams, L.H.; Burkholder, A.; Lavender, C.; Fargo, D.C.; Adelman, K. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018, 32, 26–41. [Google Scholar] [CrossRef]
- Tippens, N.D.; Vihervaara, A.; Lis, J.T. Enhancer transcription: What, where, when, and why? Genes Dev. 2018, 32, 1–3. [Google Scholar] [CrossRef]
- Gurumurthy, A.; Yu, D.T.; Stees, J.R.; Chamales, P.; Gavrilova, E.; Wassel, P.; Li, L.; Stribling, D.; Chen, J.; Brackett, M.; et al. Super-enhancer mediated regulation of adult β-globin gene expression: The role of eRNA and Integrator. Nucleic Acids Res. 2021, 49, 1383–1396. [Google Scholar] [CrossRef]
- Kouno, T.; Moody, J.; Kwon, A.T.-J.; Shibayama, Y.; Kato, S.; Huang, Y.; Böttcher, M.; Motakis, E.; Mendez, M.; Severin, J.; et al. C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution. Nat. Commun. 2019, 10, 360. [Google Scholar] [CrossRef]
- Rahman, S.; Zorca, C.E.; Traboulsi, T.; Noutahi, E.; Krause, M.R.; Mader, S.; Zenklusen, D. Single-cell profiling reveals that eRNA accumulation at enhancer–promoter loops is not required to sustain transcription. Nucleic Acids Res. 2017, 45, 3017–3030. [Google Scholar] [CrossRef]
- Bulger, M.; Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011, 3, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Ito, S.; Higashijima, Y.; Chu, W.K.; Neumann, K.; Walter, J.; Satpathy, S.; Liebner, T.; Hamilton, W.B.; Maskey, E.; et al. Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol. Cell 2021, 81, 2166–2182. [Google Scholar] [CrossRef]
- Chan, J.; Kumar, A.; Kono, H. RNAPII driven post-translational modifications of nucleosomal histones. Trends Genet. 2022; in press. [Google Scholar] [CrossRef]
- Bae, H.J.; Dubarry, M.; Jeon, J.; Soares, L.M.; Dargemont, C.; Kim, J.; Geli, V.; Buratowski, S. The Set1 N-terminal domain and Swd2 interact with RNA polymerase II CTD to recruit COMPASS. Nat. Commun. 2020, 11, 2181. [Google Scholar] [CrossRef] [PubMed]
- Francette, A.M.; Tripplehorn, S.A.; Arndt, K.M. The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin. J. Mol. Biol. 2021, 433, 166979. [Google Scholar] [CrossRef] [PubMed]
- DiFiore, J.V.; Ptacek, T.S.; Wang, Y.; Li, B.; Simon, J.M.; Strahl, B.D. Unique and Shared Roles for Histone H3K36 Methylation States in Transcription Regulation Functions. Cell Rep. 2020, 31, 107751. [Google Scholar] [CrossRef]
- Jha, R.K.; Levens, D.; Kouzine, F. Mechanical determinants of chromatin topology and gene expression. Nucleus 2022, 13, 94–115. [Google Scholar] [CrossRef]
- Gosh, R.P.; Shi, Q.; Yang, L.; Reddick, M.P.; Nikitina, T.; Zhurkin, V.B.; Fordyce, P.; Stasevich, T.J.; Chang, H.Y.; Greenleaf, W.J.; et al. Sat1b integrates DNA binding site geometry and torsional stress to differentially target nucleosome-dense regions. Nat. Commun. 2019, 10, 3221. [Google Scholar] [CrossRef]
- Tan-Wong, S.M.; Dhir, S.; Proudfoot, N.J. R-Loops Promote Antisense Transcription across the Mammalian Genome. Mol. Cell 2019, 76, 600–616. [Google Scholar] [CrossRef]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.-P.; et al. RNA Exosome-Regulated Long Non-Coding RNA Transcription Controls Super-Enhancer Activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef]
- Ishov, A.M.; Gurumurthy, A.; Bungert, J. Coordination of transcription, processing, and export of highly expressed RNAs by distinct molecular condensates. Emerg. Top. Life Sci. 2020, 4, 281–291. [Google Scholar]
- Nair, S.J.; Yang, L.; Meluzzi, D.; Oh, S.; Yang, F.; Friedman, M.J.; Wang, S.; Suter, T.; Alshareedah, I.; Gamliel, A.; et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019, 26, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Choi, J.-M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Laghmach, R.; Alshareedah, I.; Pham, M.; Raju, M.; Banerjee, P.R.; Potoyan, D.A. RNA chain length and stoichiometry govern surface tension and stability of protein-RNA condensates. iScience 2022, 25, 104105. [Google Scholar] [CrossRef]
- Trojanowski, J.; Frank, L.; Rademacher, A.; Mücke, N.; Grigaitis, P.; Rippe, K. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 2022, 82, 1878–1893.e10. [Google Scholar] [CrossRef]
- Azofeifa, J.G.; Allen, M.A.; Hendrix, J.R.; Read, T.; Rubin, J.D.; Dowell, R.D. Enhancer RNA profiling predicts transcription factor activity. Genome Res. 2018, 28, 334–344. [Google Scholar] [CrossRef]
- Schaukowitch, K.; Joo, J.-Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.-K. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Gorbovytska, V.; Kim, S.-K.; Kuybu, F.; Götze, M.; Um, D.; Kang, K.; Pittroff, A.; Brennecke, T.; Schneider, L.-M.; Leitner, A.; et al. Enhancer RNAs stimulate Pol II pause release by harnessing multivalent interactions to NELF. Nat. Commun. 2022, 13, 2429. [Google Scholar] [CrossRef]
- Hou, T.Y.; Kraus, W.L. Analysis of estrogen-regulated enhancer RNAs identifies a functional motif required for enhancer assembly and gene expression. Cell Rep. 2022, 39, 110944. [Google Scholar] [CrossRef]
- Qi, C.; Zhu, Y.T.; Chang, J.; Yeldandi, A.V.; Rao, M.S.; Zhu, Y.-J. Potentiation of estrogen receptor transcriptional activity by breast cancer amplified sequence 2. Biochem. Biophys. Res. Commun. 2005, 328, 393–398. [Google Scholar] [CrossRef]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Greifenstein, A.A.; Jo, S.; Bierhoff, H. RNA:DNA triple helices: From peculiar structures to pervasive chromatin regulators. Essays Biochem. 2021, 65, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, S.; Zhang, M.; Wu, Q. Systematic functional characterization of antisense eRNA of protocadherin α composite enhancer. Genes Dev. 2021, 35, 1383–1394. [Google Scholar] [CrossRef] [PubMed]
- Trembinski, D.J.; Bink, D.I.; Theodoru, K.; Sommer, J.; Fischer, A.; van Bergen, A.; Kuo, C.-C.; Costa, I.G.; Schürmann, C.; Leisegang, M.S.; et al. Aging-related anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat. Commun. 2020, 11, 2039. [Google Scholar] [CrossRef] [PubMed]
- Preker, P.; Almvig, K.; Christensen, M.S.; Valen, E.; Mapendano, C.K.; Sandelin, A.; Jensen, T.H. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011, 39, 7179–7193. [Google Scholar] [CrossRef]
- Cai, Z.; Cao, C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z.; Hu, N.; Yu, X.; et al. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature 2020, 582, 432–437. [Google Scholar] [CrossRef]
- Deforzh, E.; Uhlmann, E.J.; Das, E.; Galitsyna, A.; Arora, R.; Saravanan, H.; Rabinovsky, R.; Wirawan, A.D.; Teplyuk, N.M.; El Fatimy, R.; et al. Promoter and enhancer RNAs regulate chromatin reorganization and activation of miR-10b/HOXD locus, and neoplastic transformation in glioma. Mol. Cell 2022, 82, 1894–1908. [Google Scholar] [CrossRef]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588. [Google Scholar] [CrossRef]
- El Khattabi, L.; Zhao, H.; Kalchschmidt, J.; Young, N.; Jung, S.; Van Blerkom, P.; Kieffer-Kwon, P.; Kieffer-Kwon, K.-R.; Park, S.; Wang, X.; et al. A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 2022, 178, 1145–1158. [Google Scholar] [CrossRef]
- Wang, H.L.V.; Corces, V.G. The cupid shuffle: Do enhancers prefer specific promoters? Mol. Cell 2022, 82, 2357–2359. [Google Scholar] [CrossRef]
- Luo, H.; Zhu, G.; Eshelman, M.A.; Fung, T.K.; Lai, Q.; Wang, F.; Zeisig, B.B.; Lesperance, J.; Ma, X.; Chen, S.; et al. HOTTIP-dependent R-loop formation regulates CTCF boundary activity and TAD integrity in leukemia. Mol. Cell 2022, 82, 833–851. [Google Scholar] [CrossRef]
- Papantonis, A.; Cook, P.R. Transcription Factories: Genome Organization and Gene Regulation. Chem. Rev. 2013, 113, 8683–8705. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Vähärautio, A.; Taipale, J.; Zhang, G.; Jarvis, E.D.; Gilbert, M.T.P. Cancer by super-enhancer. Science 2014, 346, 1291–1292. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef]
- Menzel, S.; Garner, C.; Gut, I.; Matsuda, F.; Yamaguchi, M.; Heath, S.; Foglio, M.; Zelenika, D.; Boland, A.; Rooks, H.; et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 2007, 39, 1197–1199. [Google Scholar] [CrossRef]
- Canver, M.C.; Smith, E.C.; Sher, F.; Pinello, L.; Sanjana, N.E.; Shalem, O.; Chen, D.D.; Schupp, P.G.; Vinjamur, D.S.; Garcia, S.P.; et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 2015, 527, 192–197. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibbons, M.D.; Fang, Y.; Spicola, A.P.; Linzer, N.; Jones, S.M.; Johnson, B.R.; Li, L.; Xie, M.; Bungert, J. Enhancer-Mediated Formation of Nuclear Transcription Initiation Domains. Int. J. Mol. Sci. 2022, 23, 9290. https://doi.org/10.3390/ijms23169290
Gibbons MD, Fang Y, Spicola AP, Linzer N, Jones SM, Johnson BR, Li L, Xie M, Bungert J. Enhancer-Mediated Formation of Nuclear Transcription Initiation Domains. International Journal of Molecular Sciences. 2022; 23(16):9290. https://doi.org/10.3390/ijms23169290
Chicago/Turabian StyleGibbons, Matthew D., Yu Fang, Austin P. Spicola, Niko Linzer, Stephen M. Jones, Breanna R. Johnson, Lu Li, Mingyi Xie, and Jörg Bungert. 2022. "Enhancer-Mediated Formation of Nuclear Transcription Initiation Domains" International Journal of Molecular Sciences 23, no. 16: 9290. https://doi.org/10.3390/ijms23169290
APA StyleGibbons, M. D., Fang, Y., Spicola, A. P., Linzer, N., Jones, S. M., Johnson, B. R., Li, L., Xie, M., & Bungert, J. (2022). Enhancer-Mediated Formation of Nuclear Transcription Initiation Domains. International Journal of Molecular Sciences, 23(16), 9290. https://doi.org/10.3390/ijms23169290





