Phylogeography of Sub-Saharan Mitochondrial Lineages Outside Africa Highlights the Roles of the Holocene Climate Changes and the Atlantic Slave Trade
Abstract
1. Introduction
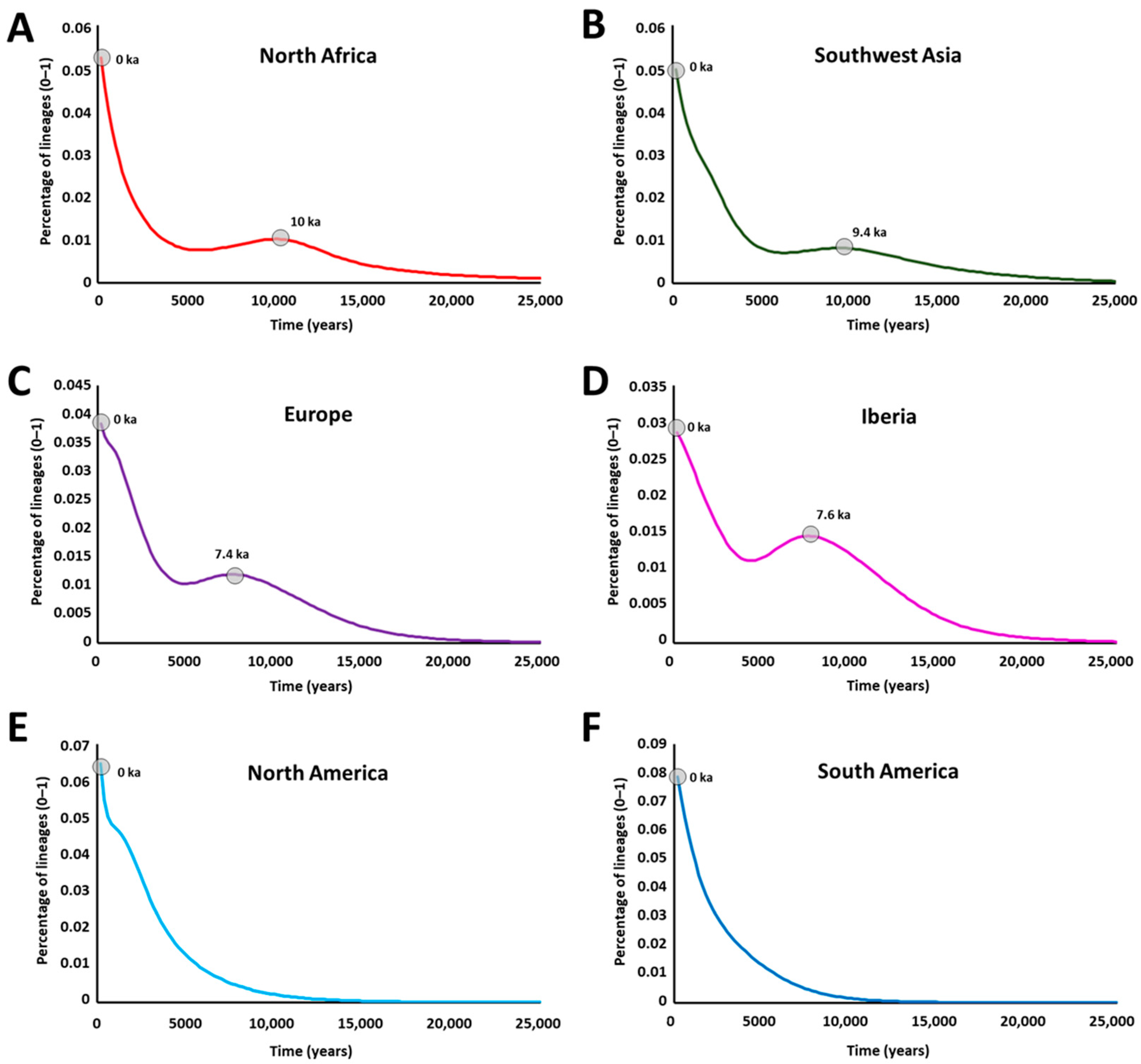
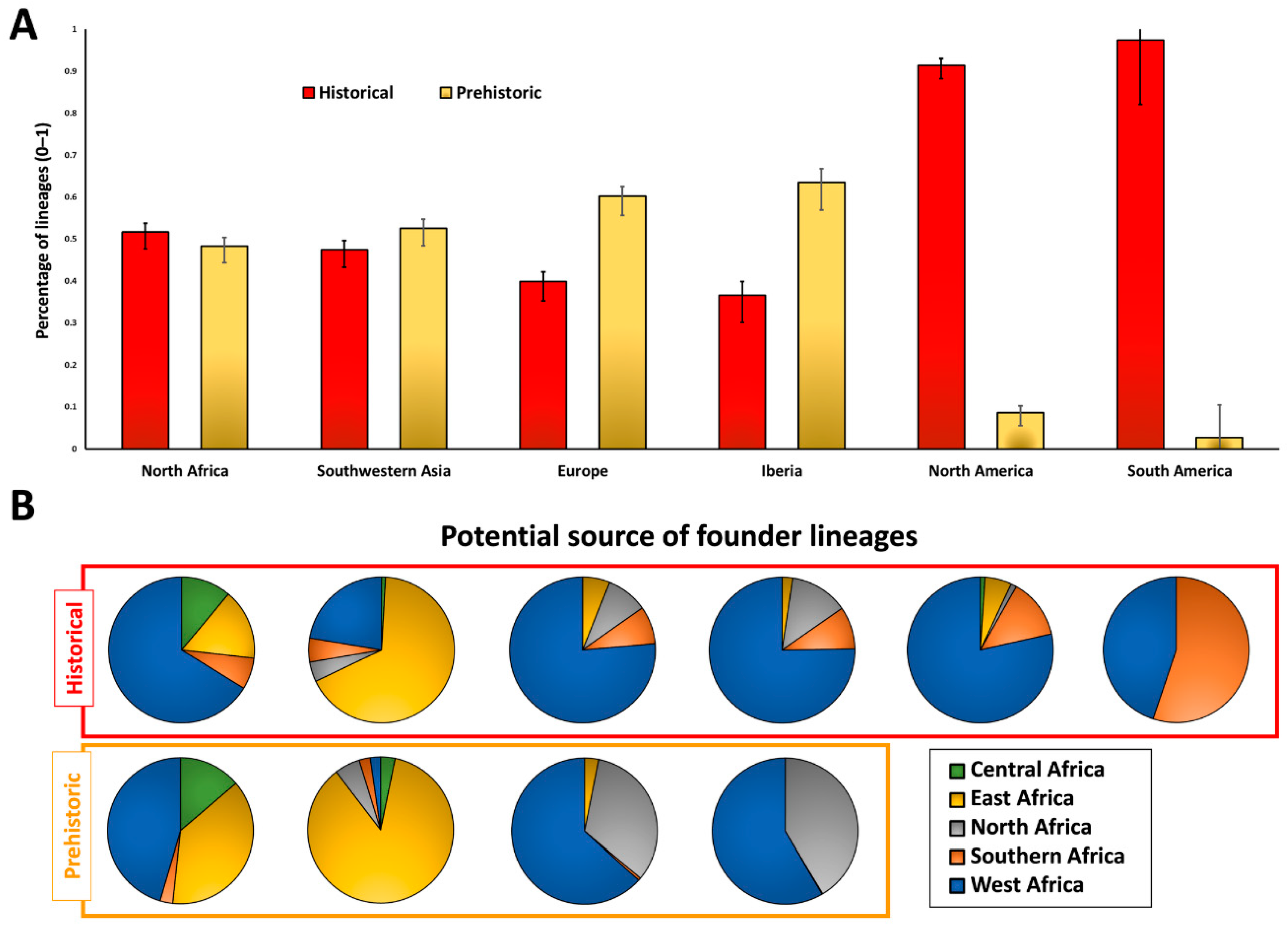
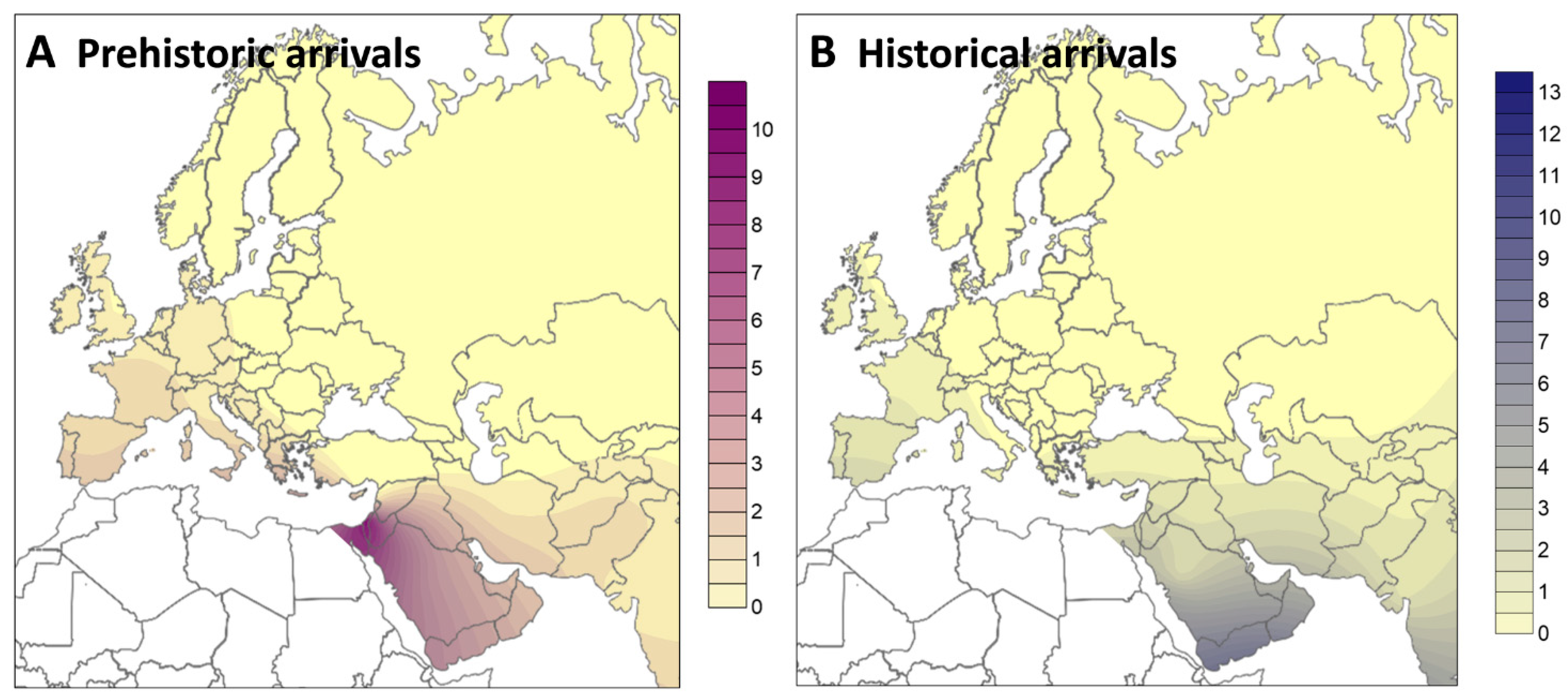
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scerri, E.M.L. The North African Middle Stone Age and Its Place in Recent Human Evolution. Evol. Anthropol. Issues News Rev. 2017, 26, 119–135. [Google Scholar] [CrossRef]
- Scerri, E.M.L.; Thomas, M.G.; Manica, A.; Gunz, P.; Stock, J.T.; Stringer, C.; Grove, M.; Groucutt, H.S.; Timmermann, A.; Rightmire, G.P.; et al. Did Our Species Evolve in Subdivided Populations across Africa, and Why Does It Matter? Trends Ecol. Evol. 2018, 33, 582–594. [Google Scholar] [CrossRef]
- Bergström, A.; Stringer, C.; Hajdinjak, M.; Scerri, E.M.L.; Skoglund, P. Origins of Modern Human Ancestry. Nature 2021, 590, 229–237. [Google Scholar] [CrossRef]
- Soares, P.; Rito, T.; Pereira, L.; Richards, M.B. A Genetic Perspective on African Prehistory. In Africa from MIS 6-2; Springer: Berlin/Heidelberg, Germany, 2016; pp. 383–405. [Google Scholar]
- Rito, T.; Richards, M.B.; Fernandes, V.; Alshamali, F.; Cerny, V.; Pereira, L.; Soares, P. The First Modern Human Dispersals across Africa. PLoS ONE 2013, 8, e80031. [Google Scholar] [CrossRef]
- Rito, T.; Vieira, D.; Silva, M.; Conde-Sousa, E.; Pereira, L.; Mellars, P.; Richards, M.B.; Soares, P. A Dispersal of Homo Sapiens from Southern to Eastern Africa Immediately Preceded the Out-of-Africa Migration. Sci. Rep. 2019, 9, 4728. [Google Scholar] [CrossRef]
- Gunz, P.; Bookstein, F.L.; Mitteroecker, P.; Stadlmayr, A.; Seidler, H.; Weber, G.W. Early Modern Human Diversity Suggests Subdivided Population Structure and a Complex Out-of-Africa Scenario. Proc. Natl. Acad. Sci. USA 2009, 106, 6094–6098. [Google Scholar] [CrossRef]
- Skoglund, P.; Thompson, J.C.; Prendergast, M.E.; Mittnik, A.; Sirak, K.; Hajdinjak, M.; Salie, T.; Rohland, N.; Mallick, S.; Peltzer, A.; et al. Reconstructing Prehistoric African Population Structure. Cell 2017, 171, 59–71.e21. [Google Scholar] [CrossRef]
- Lipson, M.; Ribot, I.; Mallick, S.; Rohland, N.; Olalde, I.; Adamski, N.; Broomandkhoshbacht, N.; Lawson, A.M.; López, S.; Oppenheimer, J.; et al. Ancient West African Foragers in the Context of African Population History. Nature 2020, 577, 665–670. [Google Scholar] [CrossRef]
- Lipson, M.; Sawchuk, E.A.; Thompson, J.C.; Oppenheimer, J.; Tryon, C.A.; Ranhorn, K.L.; de Luna, K.M.; Sirak, K.A.; Olalde, I.; Ambrose, S.H.; et al. Ancient DNA and Deep Population Structure in Sub-Saharan African Foragers. Nature 2022, 603, 290–296. [Google Scholar] [CrossRef]
- Pala, M.; Chaubey, G.; Soares, P.; Richards, M.B. The Archaeogenetics of European Ancestry. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; ISBN 978-0-470-01590-2. [Google Scholar]
- Torroni, A.; Achilli, A.; Macaulay, V.; Richards, M.; Bandelt, H.-J. Harvesting the Fruit of the Human MtDNA Tree. Trends Genet. TIG 2006, 22, 339–345. [Google Scholar] [CrossRef]
- Henn, B.M.; Gignoux, C.R.; Jobin, M.; Granka, J.M.; Macpherson, J.M.; Kidd, J.M.; Rodríguez-Botigué, L.; Ramachandran, S.; Hon, L.; Brisbin, A.; et al. Hunter-Gatherer Genomic Diversity Suggests a Southern African Origin for Modern Humans. Proc. Natl. Acad. Sci. USA 2011, 108, 5154–5162. [Google Scholar] [CrossRef]
- Silva, M.; Alshamali, F.; Silva, P.; Carrilho, C.; Mandlate, F.; Jesus Trovoada, M.; Černý, V.; Pereira, L.; Soares, P. 60,000 Years of Interactions between Central and Eastern Africa Documented by Major African Mitochondrial Haplogroup L2. Sci. Rep. 2015, 5, 12526. [Google Scholar] [CrossRef]
- Blome, M.W.; Cohen, A.S.; Tryon, C.A.; Brooks, A.S.; Russell, J. The Environmental Context for the Origins of Modern Human Diversity: A Synthesis of Regional Variability in African Climate 150,000–30,000 Years Ago. J. Hum. Evol. 2012, 62, 563–592. [Google Scholar] [CrossRef]
- Soares, P.; Alshamali, F.; Pereira, J.B.; Fernandes, V.; Silva, N.M.; Afonso, C.; Costa, M.D.; Musilová, E.; Macaulay, V.; Richards, M.B.; et al. The Expansion of MtDNA Haplogroup L3 within and out of Africa. Mol. Biol. Evol. 2012, 29, 915–927. [Google Scholar] [CrossRef]
- Marean, C.W. When the Sea Saved Humanity. Sci. Am. 2010, 303, 54–61. [Google Scholar] [CrossRef]
- Miller, J.M.; Wang, Y.V. Ostrich Eggshell Beads Reveal 50,000-Year-Old Social Network in Africa. Nature 2022, 601, 234–239. [Google Scholar] [CrossRef]
- Macaulay, V.; Hill, C.; Achilli, A.; Rengo, C.; Clarke, D.; Meehan, W.; Blackburn, J.; Semino, O.; Scozzari, R.; Cruciani, F.; et al. Single, Rapid Coastal Settlement of Asia Revealed by Analysis of Complete Mitochondrial Genomes. Science 2005, 308, 1034–1036. [Google Scholar] [CrossRef]
- Aumassip, G. Milieux, Hommes et Techniques du Sahara Préhistorique: Problèmes Actuels; L’Harmattan: Paris, France, 1994; ISBN 978-2-7384-2889-9. [Google Scholar]
- Triska, P.; Soares, P.; Patin, E.; Fernandes, V.; Cerny, V.; Pereira, L. Extensive Admixture and Selective Pressure Across the Sahel Belt. Genome Biol. Evol. 2015, 7, 3484–3495. [Google Scholar] [CrossRef]
- Černý, V.; Pereira, L.; Musilová, E.; Kujanová, M.; Vašíková, A.; Blasi, P.; Garofalo, L.; Soares, P.; Diallo, I.; Brdička, R.; et al. Genetic Structure of Pastoral and Farmer Populations in the African Sahel. Mol. Biol. Evol. 2011, 28, 2491–2500. [Google Scholar] [CrossRef]
- Pereira, L.; Černý, V.; Cerezo, M.; Silva, N.M.; Hájek, M.; Vašíková, A.; Kujanová, M.; Brdička, R.; Salas, A. Linking the Sub-Saharan and West Eurasian Gene Pools: Maternal and Paternal Heritage of the Tuareg Nomads from the African Sahel. Eur. J. Hum. Genet. 2010, 18, 915–923. [Google Scholar] [CrossRef]
- Cerezo, M.; Achilli, A.; Olivieri, A.; Perego, U.A.; Gómez-Carballa, A.; Brisighelli, F.; Lancioni, H.; Woodward, S.R.; López-Soto, M.; Carracedo, Á.; et al. Reconstructing Ancient Mitochondrial DNA Links between Africa and Europe. Genome Res. 2012, 22, 821–826. [Google Scholar] [CrossRef]
- Prendergast, M.E.; Lipson, M.; Sawchuk, E.A.; Olalde, I.; Ogola, C.A.; Rohland, N.; Sirak, K.A.; Adamski, N.; Bernardos, R.; Broomandkhoshbacht, N.; et al. Ancient DNA Reveals a Multistep Spread of the First Herders into Sub-Saharan Africa. Science 2019, 365, eaaw6275. [Google Scholar] [CrossRef]
- Inlamea, O.F.; Soares, P.; Ikuta, C.Y.; Heinemann, M.B.; Achá, S.J.; Machado, A.; Ferreira Neto, J.S.; Correia-Neves, M.; Rito, T. Evolutionary Analysis of Mycobacterium Bovis Genotypes across Africa Suggests Co-Evolution with Livestock and Humans. PLoS Negl. Trop. Dis. 2020, 14, e0008081. [Google Scholar] [CrossRef]
- Beleza, S.; Gusmão, L.; Amorim, A.; Carracedo, A.; Salas, A. The Genetic Legacy of Western Bantu Migrations. Hum. Genet. 2005, 117, 366–375. [Google Scholar] [CrossRef]
- Plaza, S.; Salas, A.; Calafell, F.; Corte-Real, F.; Bertranpetit, J.; Carracedo, A.; Comas, D. Insights into the Western Bantu Dispersal: MtDNA Lineage Analysis in Angola. Hum. Genet. 2004, 115, 439–447. [Google Scholar] [CrossRef]
- Gomes, V.; Pala, M.; Salas, A.; Álvarez-Iglesias, V.; Amorim, A.; Gómez-Carballa, A.; Carracedo, Á.; Clarke, D.J.; Hill, C.; Mormina, M.; et al. Mosaic Maternal Ancestry in the Great Lakes Region of East Africa. Hum. Genet. 2015, 134, 1013–1027. [Google Scholar] [CrossRef]
- De Filippo, C.; Bostoen, K.; Stoneking, M.; Pakendorf, B. Bringing Together Linguistic and Genetic Evidence to Test the Bantu Expansion. Proc. R. Soc. B Biol. Sci. 2012, 279, 3256–3263. [Google Scholar] [CrossRef]
- Salas, A.; Richards, M.; De la Fe, T.; Lareu, M.-V.; Sobrino, B.; Sánchez-Diz, P.; Macaulay, V.; Carracedo, Á. The Making of the African MtDNA Landscape. Am. J. Hum. Genet. 2002, 71, 1082–1111. [Google Scholar] [CrossRef]
- Tishkoff, S.A.; Reed, F.A.; Friedlaender, F.R.; Ehret, C.; Ranciaro, A.; Froment, A.; Hirbo, J.B.; Awomoyi, A.A.; Bodo, J.-M.; Doumbo, O.; et al. The Genetic Structure and History of Africans and African Americans. Science 2009, 324, 1035–1044. [Google Scholar] [CrossRef]
- Rosa, A.; Brehm, A. African Human MtDNA Phylogeography At-a-Glance. J. Anthropol. Sci. Riv. Antropol. JASS 2011, 89, 25–58. [Google Scholar] [CrossRef]
- Fage, J.D. A History of Africa; Hutchinson: London, UK, 1978; ISBN 978-0-09-132850-4. [Google Scholar]
- Barry, B. La Sénégambie du XVe au XIXe Siècle: Traite Négrière, Islam et Conquête Coloniale; L’Harmattan: Paris, France, 1988; ISBN 978-2-85802-670-8. [Google Scholar]
- Klein, H.S. The Atlantic Slave Trade; Cambridge University Press: Cambridge, UK, 1999; ISBN 978-0-521-46588-5. [Google Scholar]
- Ki-Zerbo, J. Histoire de l’Afrique Noire: D’hier à Demain; A. Hatier: Paris, France, 1978; ISBN 978-2-218-04176-1. [Google Scholar]
- Salas, A.; Richards, M.; Lareu, M.-V.; Scozzari, R.; Coppa, A.; Torroni, A.; Macaulay, V.; Carracedo, A. The African Diaspora: Mitochondrial DNA and the Atlantic Slave Trade. Am. J. Hum. Genet. 2004, 74, 454–465. [Google Scholar] [CrossRef]
- Salas, A.; Carracedo, A.; Richards, M.; Macaulay, V. Charting the Ancestry of African Americans. Am. J. Hum. Genet. 2005, 77, 676–680. [Google Scholar] [CrossRef][Green Version]
- Heinz, T.; Pala, M.; Gómez-Carballa, A.; Richards, M.B.; Salas, A. Updating the African Human Mitochondrial DNA Tree: Relevance to Forensic and Population Genetics. Forensic Sci. Int. Genet. 2017, 27, 156–159. [Google Scholar] [CrossRef]
- Schroeder, H.; Ávila-Arcos, M.C.; Malaspinas, A.-S.; Poznik, G.D.; Sandoval-Velasco, M.; Carpenter, M.L.; Moreno-Mayar, J.V.; Sikora, M.; Johnson, P.L.F.; Allentoft, M.E.; et al. Genome-Wide Ancestry of 17th-Century Enslaved Africans from the Caribbean. Proc. Natl. Acad. Sci. USA 2015, 112, 3669–3673. [Google Scholar] [CrossRef]
- Fortes-Lima, C.; Verdu, P. Anthropological Genetics Perspectives on the Transatlantic Slave Trade. Hum. Mol. Genet. 2021, 30, R79–R87. [Google Scholar] [CrossRef]
- Richards, M.B.; Torroni, A.; Soares, P. Phylogeography. In The International Encyclopedia of Biological Anthropology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 1–3. ISBN 978-1-118-58453-8. [Google Scholar]
- Pereira, J.B.; Costa, M.D.; Vieira, D.; Pala, M.; Bamford, L.; Harich, N.; Cherni, L.; Alshamali, F.; Hatina, J.; Rychkov, S.; et al. Reconciling Evidence from Ancient and Contemporary Genomes: A Major Source for the European Neolithic within Mediterranean Europe. Proc. R. Soc. B Biol. Sci. 2017, 284, 20161976. [Google Scholar] [CrossRef]
- Macaulay, V.; Richards, M.B. Mitochondrial Genome Sequences and Their Phylogeographic Interpretation. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; ISBN 978-0-470-01590-2. [Google Scholar]
- Richards, M.; Macaulay, V.; Hickey, E.; Vega, E.; Sykes, B.; Guida, V.; Rengo, C.; Sellitto, D.; Cruciani, F.; Kivisild, T.; et al. Tracing European Founder Lineages in the Near Eastern MtDNA Pool. Am. J. Hum. Genet. 2000, 67, 1251–1276. [Google Scholar] [CrossRef]
- Silva, M.; Oteo-García, G.; Martiniano, R.; Guimarães, J.; von Tersch, M.; Madour, A.; Shoeib, T.; Fichera, A.; Justeau, P.; Foody, M.G.B.; et al. Biomolecular Insights into North African-Related Ancestry, Mobility and Diet in Eleventh-Century Al-Andalus. Sci. Rep. 2021, 11, 18121. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstätter, A.; Forer, L.; Specht, G.; Bandelt, H.-J.; Kronenberg, F.; Salas, A.; Schönherr, S. HaploGrep 2: Mitochondrial Haplogroup Classification in the Era of High-Throughput Sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Sykes, B.C.; Richards, M.B. Mitochondrial Portraits of Human Populations Using Median Networks. Genetics 1995, 141, 743–753. [Google Scholar] [CrossRef]
- Soares, P.; Ermini, L.; Thomson, N.; Mormina, M.; Rito, T.; Röhl, A.; Salas, A.; Oppenheimer, S.; Macaulay, V.; Richards, M.B. Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock. Am. J. Hum. Genet. 2009, 84, 740–759. [Google Scholar] [CrossRef]
- Van Oven, M. PhyloTree Build 17: Growing the Human Mitochondrial DNA Tree. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e392–e394. [Google Scholar] [CrossRef]
- Vieira, D.; Almeida, M.; Richards, M.B.; Soares, P. An Efficient and User-Friendly Implementation of the Founder Analysis Methodology. In Proceedings of the 13th International Conference on Practical Applications of Computational Biology and Bioinformatics, Ávila, Spain, 26–28 June 2019; Fdez-Riverola, F., Rocha, M., Mohamad, M.S., Zaki, N., Castellanos-Garzón, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 121–128. [Google Scholar]
- Rito, T.; Richards, M.B.; Pala, M.; Correia-Neves, M.; Soares, P.A. Phylogeography of 27,000 SARS-CoV-2 Genomes: Europe as the Major Source of the COVID-19 Pandemic. Microorganisms 2020, 8, 1678. [Google Scholar] [CrossRef]
- Macaulay, V.; Soares, P.; Richards, M.B. Rectifying Long-Standing Misconceptions about the ρ Statistic for Molecular Dating. PLoS ONE 2019, 14, e0212311. [Google Scholar] [CrossRef]
- Saillard, J.; Forster, P.; Lynnerup, N.; Bandelt, H.-J.; Nørby, S. MtDNA Variation among Greenland Eskimos: The Edge of TheBeringian Expansion. Am. J. Hum. Genet. 2000, 67, 718–726. [Google Scholar] [CrossRef]
- Gandini, F.; Achilli, A.; Pala, M.; Bodner, M.; Brandini, S.; Huber, G.; Egyed, B.; Ferretti, L.; Gómez-Carballa, A.; Salas, A.; et al. Mapping Human Dispersals into the Horn of Africa from Arabian Ice Age Refugia Using Mitogenomes. Sci. Rep. 2016, 6, 25472. [Google Scholar] [CrossRef]
- Font-Porterias, N.; Solé-Morata, N.; Serra-Vidal, G.; Bekada, A.; Fadhlaoui-Zid, K.; Zalloua, P.; Calafell, F.; Comas, D. The Genetic Landscape of Mediterranean North African Populations through Complete MtDNA Sequences. Ann. Hum. Biol. 2018, 45, 98–104. [Google Scholar] [CrossRef]
- Hernández, C.L.; Soares, P.; Dugoujon, J.M.; Novelletto, A.; Rodríguez, J.N.; Rito, T.; Oliveira, M.; Melhaoui, M.; Baali, A.; Pereira, L.; et al. Early Holocenic and Historic MtDNA African Signatures in the Iberian Peninsula: The Andalusian Region as a Paradigm. PLoS ONE 2015, 10, e0139784. [Google Scholar] [CrossRef]
- Wilson, A. Saharan Trade in the Roman Period: Short-,Medium-and Long-Distance Trade Networks. Azania Archaeol. Res. Afr. 2012, 47, 409–449. [Google Scholar] [CrossRef]
- Richards, M.; Rengo, C.; Cruciani, F.; Gratrix, F.; Wilson, J.F.; Scozzari, R.; Macaulay, V.; Torroni, A. Extensive Female-Mediated Gene Flow from Sub-Saharan Africa into near Eastern Arab Populations. Am. J. Hum. Genet. 2003, 72, 1058–1064. [Google Scholar] [CrossRef][Green Version]
- Gouveia, M.H.; Borda, V.; Leal, T.P.; Moreira, R.G.; Bergen, A.W.; Kehdy, F.S.G.; Alvim, I.; Aquino, M.M.; Araujo, G.S.; Araujo, N.M.; et al. Origins, Admixture Dynamics, and Homogenization of the African Gene Pool in the Americas. Mol. Biol. Evol. 2020, 37, 1647–1656. [Google Scholar] [CrossRef]
- Micheletti, S.J.; Bryc, K.; Ancona Esselmann, S.G.; Freyman, W.A.; Moreno, M.E.; Poznik, G.D.; Shastri, A.J.; 23andMe Research Team; Beleza, S.; Mountain, J.L. Genetic Consequences of the Transatlantic Slave Trade in the Americas. Am. J. Hum. Genet. 2020, 107, 265–277. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sá, L.; Almeida, M.; Azonbakin, S.; Matos, E.; Franco-Duarte, R.; Gómez-Carballa, A.; Salas, A.; Laleye, A.; Rosa, A.; Brehm, A.; et al. Phylogeography of Sub-Saharan Mitochondrial Lineages Outside Africa Highlights the Roles of the Holocene Climate Changes and the Atlantic Slave Trade. Int. J. Mol. Sci. 2022, 23, 9219. https://doi.org/10.3390/ijms23169219
Sá L, Almeida M, Azonbakin S, Matos E, Franco-Duarte R, Gómez-Carballa A, Salas A, Laleye A, Rosa A, Brehm A, et al. Phylogeography of Sub-Saharan Mitochondrial Lineages Outside Africa Highlights the Roles of the Holocene Climate Changes and the Atlantic Slave Trade. International Journal of Molecular Sciences. 2022; 23(16):9219. https://doi.org/10.3390/ijms23169219
Chicago/Turabian StyleSá, Luísa, Mafalda Almeida, Simon Azonbakin, Erica Matos, Ricardo Franco-Duarte, Alberto Gómez-Carballa, Antonio Salas, Anatóle Laleye, Alexandra Rosa, António Brehm, and et al. 2022. "Phylogeography of Sub-Saharan Mitochondrial Lineages Outside Africa Highlights the Roles of the Holocene Climate Changes and the Atlantic Slave Trade" International Journal of Molecular Sciences 23, no. 16: 9219. https://doi.org/10.3390/ijms23169219
APA StyleSá, L., Almeida, M., Azonbakin, S., Matos, E., Franco-Duarte, R., Gómez-Carballa, A., Salas, A., Laleye, A., Rosa, A., Brehm, A., Richards, M. B., Soares, P., & Rito, T. (2022). Phylogeography of Sub-Saharan Mitochondrial Lineages Outside Africa Highlights the Roles of the Holocene Climate Changes and the Atlantic Slave Trade. International Journal of Molecular Sciences, 23(16), 9219. https://doi.org/10.3390/ijms23169219








