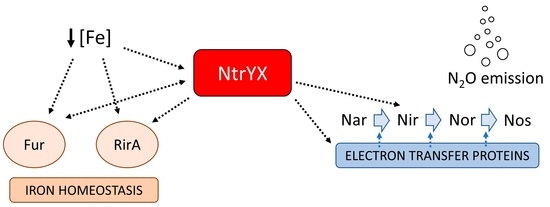
The NtrYX Two-Component System of Paracoccus denitrificans Is Required for the Maintenance of Cellular Iron Homeostasis and for a Complete Denitrification under Iron-Limited Conditions
Abstract
1. Introduction
2. Results
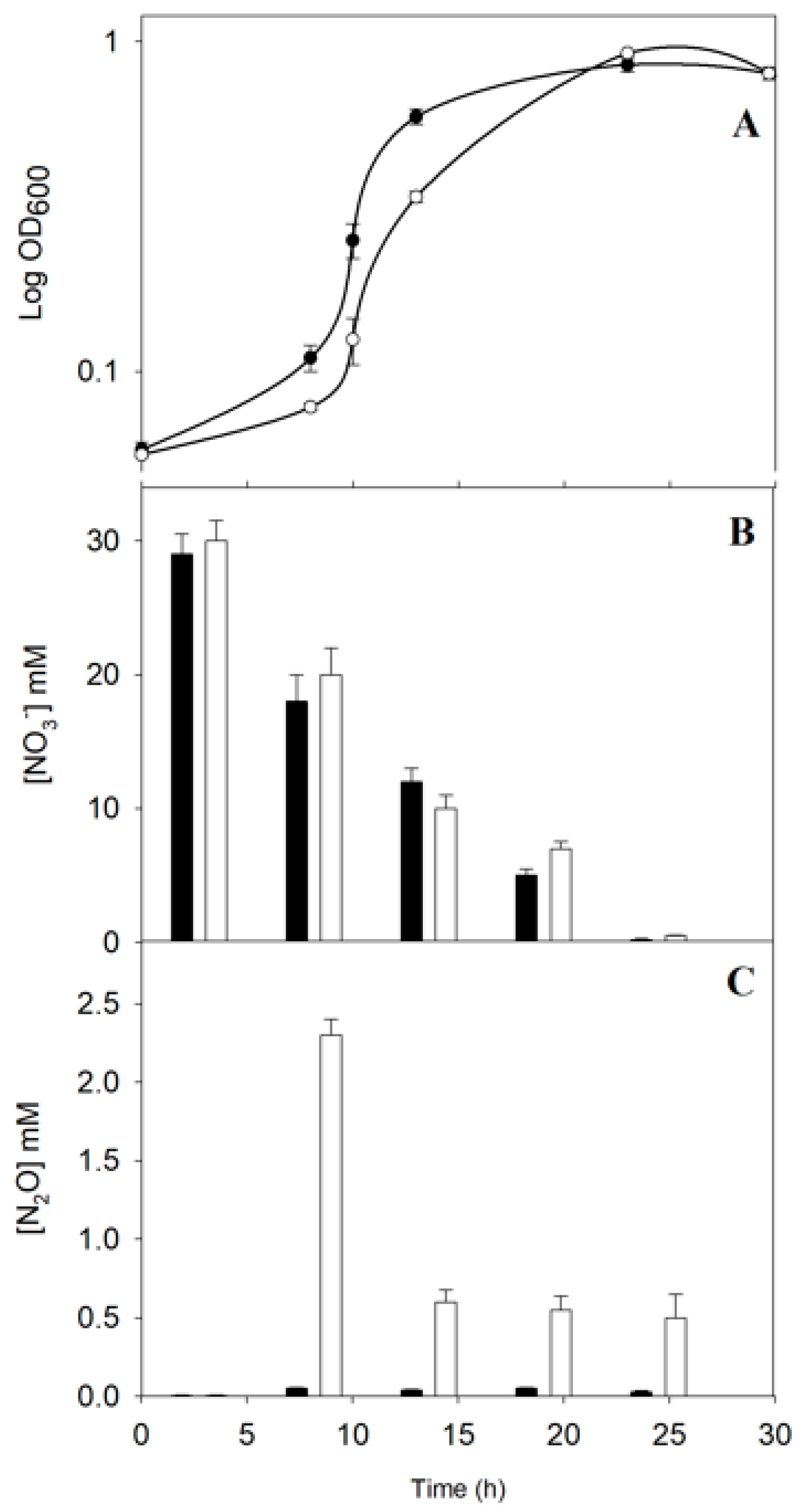
2.1. Physiological Characterization of the P. denitrificans NtrY Mutant under Denitrifying Conditions
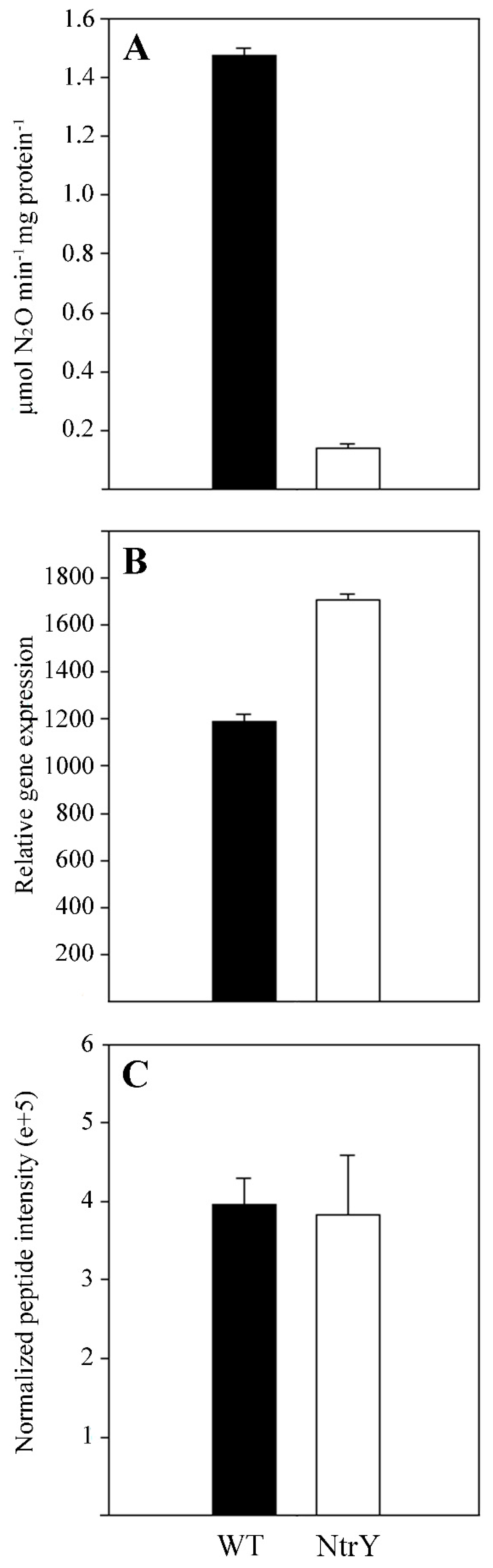
2.2. Quantitative Proteomic Analysis of the P. denitrificans NtrY Mutant under Denitrifying Conditions
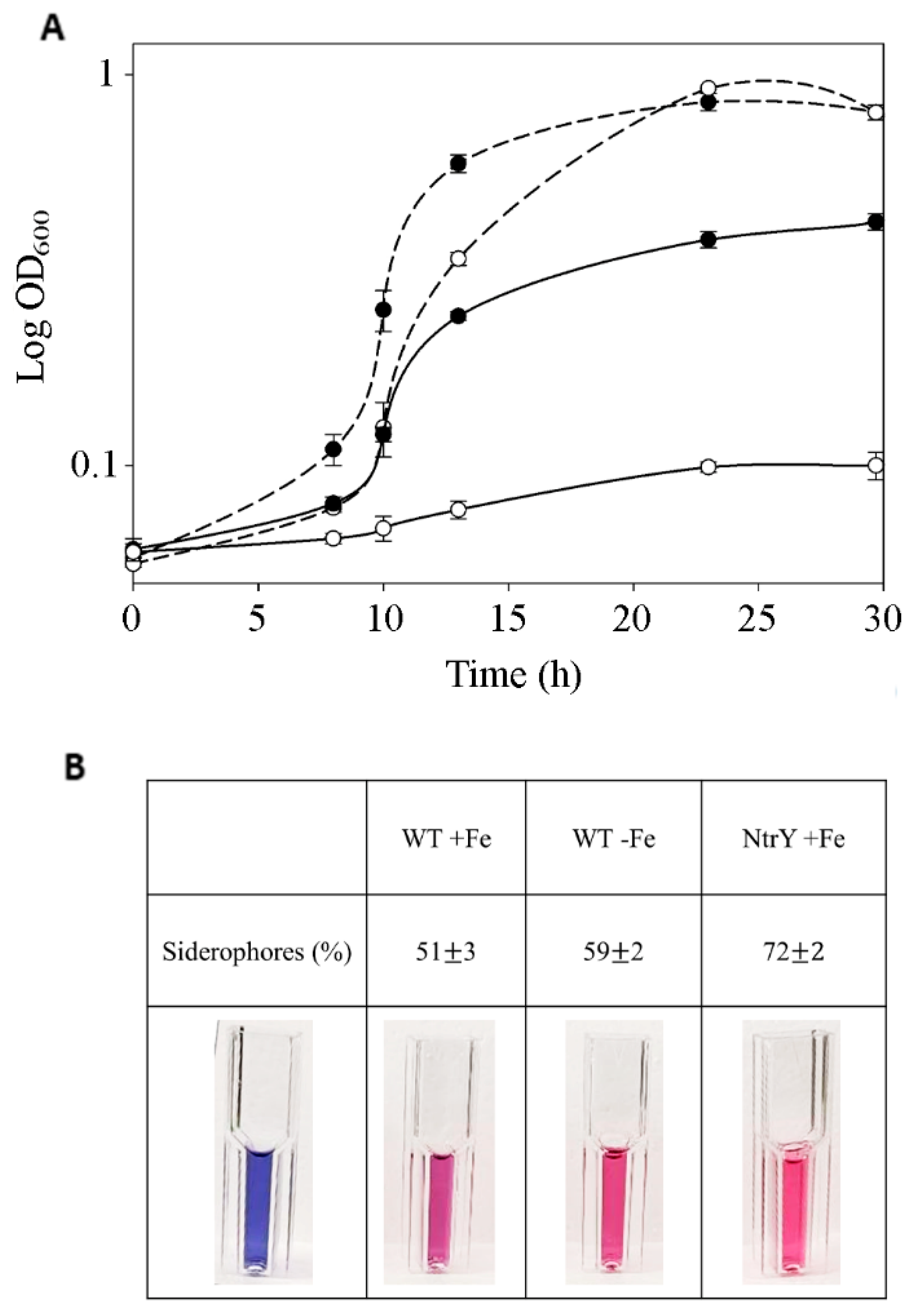
2.3. Intracellular Iron Content, Siderophore Production, and Gene Expression Analysis of P. denitrificans Iron-Responsive Regulators
2.4. Identification of NtrX Binding Boxes and Target Genes in the Genome of P. denitrificans
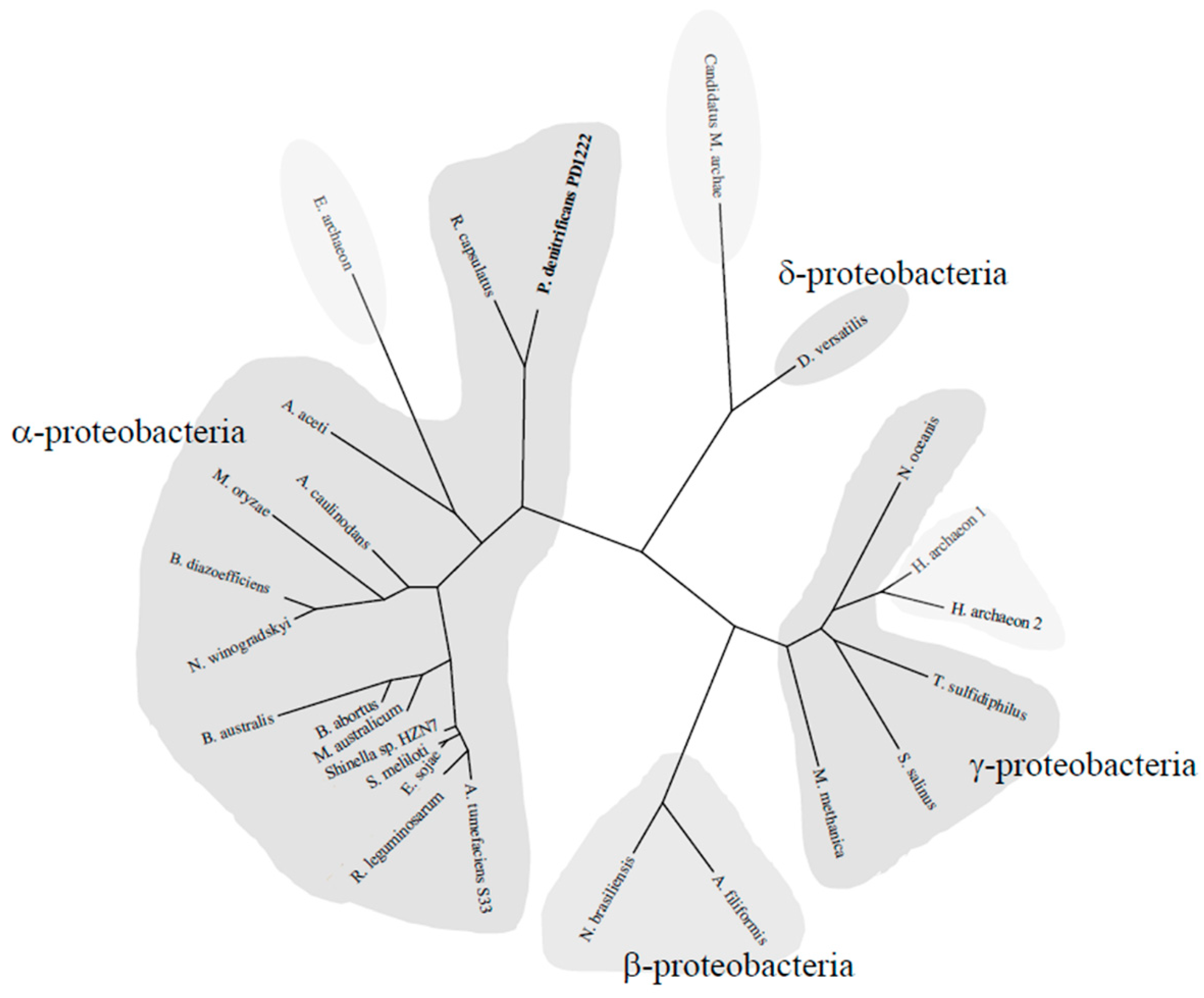
2.5. Phylogenetic Tree of the NtrYX System
3. Discussion
3.1. Physiological Characterization of the P. denitrificans NtrY Mutant under Denitrifying Conditions
3.2. Proteomic Analysis, Iron Content and Siderophore Production of the P. denitrificans NtrY Mutant
3.3. Iron-Responsive Regulators of P. denitrificans under Denitrifying Conditions
3.4. NtrX Targets Involved in Denitrification of P. denitrificans
3.5. Phylogenetic Distribution of the NtrYX Regulon
4. Materials and Methods
4.1. Bacterial Strains, Media, and Growth Conditions
4.2. In Vivo Determination of Nitrous Oxide Production
4.3. Determination of Intracellular Metals Concentration by ICP-MS
4.4. Detection of Siderophore Production
4.5. In Vitro Assays of Denitrification Enzymes
4.6. Quantitative Proteomic Analysis by LC-MS/MS
4.7. Gene Expression Analysis by qRT-PCR
4.8. Bioinfomatic Analisis of the NtrX Binding Sequence in the Genome of P. denitrificans
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pawlowski, K.; Klosse, U.; de Bruijn, F.J. Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol. Gen. Genet. 1991, 231, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Carrica, M.C.; Fernandez, I.; Martí, M.A.; Paris, G.; Goldbaum, F.A. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol. Microbiol. 2012, 85, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Luque-Almagro, V.M.; Manso, I.; Sullivan, M.J.; Rowley, G.; Ferguson, S.J.; Moreno-Vivián, C.; Richardson, D.J.; Gates, A.J.; Roldán, M.D. Transcriptional and translational adaptation to aerobic nitrate anabolism in the denitrifier Paracoccus denitrificans. Biochem. J. 2017, 474, 1769–1787. [Google Scholar] [CrossRef] [PubMed]
- Albright, L.M.; Huala, E.; Ausubel, F.M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu. Rev. Genet. 1989, 23, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Kustu, S.; Santero, E.; Keener, J.; Popham, D.; Weiss, D. Expression of σ54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 1989, 53, 367–376. [Google Scholar] [CrossRef]
- Roop, R.M.; Caswell, C.C. Redox-responsive regulation of denitrification genes in Brucella. Mol. Microbiol. 2012, 85, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Carrica, M.C.; Fernández, I.; Sieira, R.; Paris, G.; Goldbaum, F.A. The two-component systems PrrBA and NtrYX co-ordinately regulate the adaptation of Brucella abortus to an oxygen-limited environment. Mol. Microbiol. 2013, 88, 222–233. [Google Scholar] [CrossRef]
- Atack, J.M.; Srikhanta, Y.N.; Djoko, K.Y.; Welch, J.P.; Hasri, N.H.; Steichen, C.T.; Vanden-Hoven, R.N.; Grimmond, S.M.; Othman, D.S.; Kappler, U.; et al. Characterization of a ntrX mutant of Neisseria gonorrhoeae reveals a response regulator that controls expression of respiratory enzymes in oxidase-positive proteobacteria. J. Bacteriol. 2013, 195, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- López, M.F.; Hegel, V.A.; Torres, M.J.; Hidalgo-García, A.; Delgado, M.J.; López-García, S.L. The Bradyrhizobium diazoefficiens two-component system NtrYX has a key role in symbiotic nitrogen fixation of soybean plants and cbb3 oxidase expression in bacteroids. Plant Soil 2019, 440, 167–183. [Google Scholar] [CrossRef]
- Lemmer, K.C.; Alberge, F.; Myers, K.S.; Dohnalkova, A.C.; Schau, R.E.; Lenz, J.D.; Imam, S.; Dillard, J.P.; Noguera, D.R.; Donohue, T.J. The NtrYX two-component system regulates the bacterial cell envelope. mBio 2020, 11, e00957-20. [Google Scholar] [CrossRef] [PubMed]
- Schulz, A.; Hermann, L.; Freibert, S.A.; Bönig, T.; Hoffmann, T.; Riclea, R.; Dickschat, J.S.; Heider, J.L.; Bremer, E. Transcriptional regulation of ectoine catabolism in response to multiple metabolic and environmental cues. Environ. Microbiol. 2017, 19, 4599–4619. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.C.; Ferguson, S.J.; Ludwig, B.; Page, M.D.; Richter, O.M.H.; van Spanning, R.J.M. Molecular genetics of the genus Paracoccus: Metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 1998, 62, 1046–1078. [Google Scholar] [CrossRef] [PubMed]
- Gates, A.J.; Luque-Almagro, V.M.; Goddard, A.D.; Ferguson, S.J.; Roldán, M.D.; Richardson, D.J. A composite biochemical system for bacterial nitrate and nitrite assimilation as exemplified by Paracoccus denitrificans. Biochem. J. 2011, 435, 743–753. [Google Scholar] [CrossRef]
- Shu, C.J.; Zhulin, I.B. ANTAR: An RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 2002, 27, 3–5. [Google Scholar] [CrossRef]
- Luque-Almagro, V.M.; Lyall, V.J.; Ferguson, S.J.; Roldán, M.D.; Richardson, D.J.; Gates, A.J. Nitrogen oxyanion-dependent dissociation of a two-component complex that regulates bacterial nitrate assimilation. J. Biol. Chem. 2013, 288, 29692–29702. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Luque-Almagro, V.M.; Manso, I.; Gates, A.J.; Moreno-Vivián, C.; Richardson, D.J.; Roldán, M.D. Poly(3-hydroxybutyrate) hyperproduction by a global nitrogen regulator NtrB mutant strain of Paracoccus denitrificans PD1222. FEMS Microbiol. Lett. 2018, 365, fnx251. [Google Scholar] [CrossRef]
- Richardson, D.J. Bacterial respiration: A flexible process for a changing environment. Microbiology 2000, 146, 551–571. [Google Scholar] [CrossRef]
- Philippot, L.; Hallin, S.; Schloter, M. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 2007, 96, 249–305. [Google Scholar]
- Olaya-Abril, A.; Hidalgo-Carrillo, J.; Luque-Almagro, V.M.; Fuentes-Almagro, C.; Urbano, F.J.; Moreno-Vivián, C.; Richardson, D.J.; Roldán, M.D. Exploring the denitrification proteome of Paracoccus denitrificans PD1222. Front. Microbiol. 2018, 29, 1137. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robison, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Berks, B.C.; Ferguson, S.J.; Moir, J.W.B.; Richardson, D.J. Enzymes and associated electron transport systems that catalyse their respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1995, 1232, 97–173. [Google Scholar] [CrossRef]
- Giannopoulos, G.; Sullivan, M.J.; Hartop, K.R.; Rowley, G.; Gates, A.J.; Watmough, N.J.; Richardson, D.J. Tuning the modular Paracoccus denitrificans respirome to adapt from aerobic respiration to anaerobic denitrification. Environ. Microbiol. 2017, 19, 4953–4964. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A.; Gelfand, M.S.; Todd, J.D.; Curson, A.R.J.; Johnston, A.W.B. Computational reconstruction of iron and manganese-responsive transcriptional networks in α-Proteobacteria. PLoS Comput. Biol. 2006, 2, e163. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 1, 36. [Google Scholar] [CrossRef] [PubMed]
- Gaimster, H.; Chalklen, L.; Alston, M.; Munnoch, J.T.; Richardson, D.J.; Gates, A.J.; Rowley, G. Genome-wide discovery of putative sRNAs in Paracoccus denitrificans expressed under nitrous oxide emitting conditions. Front. Microbiol. 2016, 7, 1806. [Google Scholar] [CrossRef]
- Moreno-Vivián, C.; Flores, E. Nitrate assimilation in bacteria. In Biology of the Nitrogen Cycle; Bothe, H., Ferguson, S.J., Newton, W.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 263–282. ISBN 9780444528575. [Google Scholar]
- Luque-Almagro, V.M.; Gates, A.J.; Moreno-Vivián, C.; Ferguson, S.J.; Richardson, D.J.; Roldán, M.D. Bacterial nitrate assimilation: Gene distribution and regulation. Biochem. Soc. Trans. 2011, 39, 838–1843. [Google Scholar] [CrossRef]
- Vitorino, J.C.; Steffens, M.B.; Machado, H.B.; Yates, M.G.; Souza, E.M.; Pedrosa, F.O. Potential roles for the glnB and ntrYX genes in Azospirillum brasilense. FEMS Microbiol. Lett. 2001, 201, 199–204. [Google Scholar] [CrossRef][Green Version]
- Ishida, M.L.; Assumpção, M.C.; Machado, H.B.; Benelli, E.M.; Souza, E.M.; Pedros, F.O. Identification and characterization of the two-component NtrY/NtrX regulatory system in Azospirillum brasilense. Braz. J. Med. Biol. Res. 2002, 35, 651–661. [Google Scholar] [CrossRef]
- Assumpção, M.C.; de Souza, E.M.; Yates, M.G.; de Oliviera-Pedrosa, F.; Benelli, E.M. Purification and characterization of Azospirillum brasilense N-truncated NtrX protein. Prot. Express. Purif. 2007, 53, 302–308. [Google Scholar] [CrossRef]
- Bonato, P.; Alves, L.R.; Osaki, J.H.; Rigo, L.U.; Pedrosa, F.O.; Souza, E.M.; Zhang, N.; Schumacher, J.; Buck, M.; Wassem, R.; et al. The NtrY/NtrX two-component system is involved in controlling nitrate assimilation in Herbaspirillum seropedicae strain SmR1. FEBS J. 2016, 283, 3919–3930. [Google Scholar] [CrossRef]
- Drepper, T.; Wiethaus, J.; Giaourakis, D.; Groβ, S.; Schubert, B.; Vogt, M.; Wiencek, Y.; McEwan, A.G.; Masepohl, B. Cross-talk towards response regulator NtrC controlling nitrogen metabolism in Rhodobacter capsulatus. FEMS Microbiol. Lett. 2006, 258, 250–256. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bergaust, L.; van Spanning, R.J.M.; Frostegård, A.; Bakken, L.R. Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 2012, 158, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, 44, 655–683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chen, J.; Zheng, P.; Gong, W.; Sun, J.; Liu, H. Crystal structure of 5-aminolevulinate synthase HemA from Rhodopseudomonas palustris presents multiple conformations. Biochem. Biophys. Res. Commun. 2022, 609, 100–104. [Google Scholar] [CrossRef]
- Huertas, M.J.; Luque-Almagro, V.M.; Martínez-Luque, M.; Blasco, R.; Moreno-Vivián, C.; Castillo, F.; Roldán, M.D. Cyanide metabolism of Pseudomonas pseudoalcaligenes CECT5344: Role of siderophores. Biochem. Soc. Trans. 2006, 34, 152–155. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef]
- Bergeron, R.J.; Weimar, W.R.; Dionis, J.B. Demonstration of ferric L-parabactin-binding activity in the outer membrane of Paracoccus denitrificans. J. Bacteriol. 1988, 170, 3711–3717. [Google Scholar] [CrossRef]
- Lycus, P.; Lovise-Bøthun, K.; Bergaust, L.; Shapleigh, J.P.; Bakken, L.R.; Frostegård, A. Phenotypic and genotypic richness of denitrifiers revealed by a novel isolation strategy. ISME J. 2017, 11, 2219–2232. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Hidalgo-Carrillo, J.; Luque-Almagro, V.M.; Fuentes-Almagro, C.; Urbano, F.J.; Moreno-Vivián, C.; Richardson, D.J.; Roldán, M.D. Effect of pH on the denitrification proteome of the soil bacterium Paracoccus denitrificans PD1222. Sci. Rep. 2021, 11, 17276. [Google Scholar] [CrossRef]
- Spiro, S. Nitrous oxide production and consumption: Regulation of gene expression by gas-sensitive transcription factors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Spiro, S. Regulation of denitrification. In Metalloenzymes in Denitrification: Applications and Environmental Impacts; Moura, I., Moura, J.J.G., Pauleta, S.R., Maia, L.B., Eds.; Royal Society of Chemistry: London, UK, 2016; pp. 312–330. [Google Scholar]
- Van Spanning, R.J.M.; de Boer, A.P.N.; Reijnders, W.N.M.; Westerhoff, H.V.; Stouthamer, A.H.; der Oost, J.V. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol. Microbiol. 1997, 23, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Vaccaro, B.J.; Thorgersen, M.P.; Lancaster, W.A.; Price, M.N.; Wetmore, K.M.; Poole, F.L.; Deutschbauer, I.A.; Arkin, A.P.; Adams, M.W.W. Determining roles of accessory genes in denitrification by mutant fitness analyses. Appl. Environ. Microbiol. 2016, 82, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Harms, N.; de Vries, G.E.; Maurer, K.; Veltkamp, E.; Stouthamer, A.H. Isolation and characterization of Paracoccus denitrificans mutants with defects in the metabolism of one-carbon compounds. J. Bacteriol. 1985, 164, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Frunzke, K.; Zumft, W.G. Rapid, single sample analysis of H2, O2, N2, NO, CO, N2O and CO2 by isothermal gas chromatography: Applications to the study of bacterial denitrification. J. Chromatogr. 1984, 299, 477–483. [Google Scholar] [CrossRef]
- Cawse, P.A. The determination of nitrate in soil solutions by ultraviolet spectrophotometry. Analyst 1967, 92, 311–315. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Snell, F.D.; Snell, C.T. Colorimetric Methods of Analysis; Van Nostrand: New York, NY, USA, 1949; pp. 802–807. [Google Scholar]
- Ridley, H.; Watts, C.A.; Richardson, D.J.; Butler, C.S. Development of a viologen-based microtiter plate assay for the analysis of oxyanion activity: Application to the membrane-bound selenate reductase from Enterobacter cloacae SLD1a-1. Anal. Biochem. 2006, 358, 289–294. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–256. [Google Scholar] [CrossRef]
- Shakir, F.K.; Audilet, D.; Drake, A.J.; Shakir, K.M. A rapid protein determination by modification of the Lowry procedure. Anal. Biochem. 1994, 216, 232–233. [Google Scholar] [CrossRef]
- Bache, N.; Geyer, P.E.; Bekker-Jensen, D.B.; Hoerning, O.; Falkenby, L.; Treit, P.; Doll, S.; Paron, I.; Müller, J.B.; Meier, F.; et al. A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Mol. Cell. Proteom. 2018, 17, 2284–2296. [Google Scholar] [CrossRef]
- Meier, F.; Brunner, A.-D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M.A.; et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteom. 2018, 17, 2534–2545. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Brunner, A.D.; Frank, M.; Ha, A.; Bludau, I.; Voytik, E.; Kaspar-Schoenefeld, S.; Lubeck, M.; Raether, O.; Aebersold, R.; et al. DiaPASEF: Parallel accumulation–serial fragmentation combined with data-independent acquisition. Nat. Methods 2020, 17, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, G.; Bludau, I.; Schmitt, U.; Heusel, M.; Hunter, C.L.; Liu, Y.; MacCoss, M.J.; MacLean, B.X.; Nesvizhskii, A.I.; Pedrioli, P.G.A.; et al. Statistical control of peptide and protein error rates in large-scale targeted data-independent acquisition analyses. Nat. Methods 2017, 14, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Fruzangohar, M.; Ebrahimie, E.; Ogunniyi, A.D.; Mahdi, L.K.; Paton, J.C.; Adelsonet, D.L. Comparative GO: A web application for comparative gene ontology and gene ontology-based gene selection in bacteria. PLoS ONE 2013, 8, e58759. [Google Scholar] [CrossRef]
- Mrázek, J.; Xie, S. Pattern locator: A new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 2006, 22, 3099–3100. [Google Scholar] [CrossRef] [PubMed]


| Locus 1 | Protein ID 2 | Name | NtrX Binding Sequence (Distance from Start Codon) | Fur Binding Sequence (Distance from Start Codon) | qRT-PCR | ||
|---|---|---|---|---|---|---|---|
| WT | NtrY | ||||||
| +Fe | −Fe | +Fe | |||||
| Pden_4128 | A1B9J8 | PAS/PAC sensor signal transduction histidine kinase (nrtY) | CACAGAACGGCCGC (-33) GTGGCCGTTCAGCG (-44) | TGACACGGCGCCGCAA (-92) | 0.3 ± 0.1 | 3.1 ± 0.1 | - |
| Pden_4127 | A1B9J7 | Two-component, σ54-specific, transcriptional regulator, Fis family (ntrX) | - | - | 1.1 ± 0.6 | 3.7 ± 0.9 | 2.3 ± 0.4 |
| Pden_1260 | A1B1H1 | Manganese uptake regulator, Fur family (mur/irr) | CTTGTGCTCTGGGC (-82) | TGAGATCGTCCGCCAC (-29) | 0.1 ± 0.1 | 0.7 ± 0.2 | 1.0 ± 0.4 |
| CTTCGTTCTGCTGC (-69) | |||||||
| CTGCCGTTATCTGC (-59) | |||||||
| CTCAACTGTCAAGC (-37) | |||||||
| CTTCGCATCTGGGC (-11) | |||||||
| Pden_4139 | A1B9K9 | Putative ferric uptake regulator, Fur family (fur) | CTTGGCGTTTCCGC (-53) | - | 5.3 ± 0.4 | 28.0 ± 1.3 | 8.4 ± 0.3 |
| Pden_1690 | A1B2P3 | Transcriptional regulator, BadM/Rrf2famil (rirA) | CAAGGTTGCGCGGC (-98) | TGCAATCAGGATGCAT (-19) | 1.8 ± 0.2 | 8.9 ± 0.7 | 12.7 ± 1.2 |
| CTGCAAGGTTGCGC (-101) | TGTATTCTGGATACAT (-56) | ||||||
| Pden_2958 | A1B698 | Transcriptional regulator, BadM/Rrf2family (iscR) | CTTGCCTATGTCGC (-78) | TGCCGCGGCCCCTCAC (-51) | 1.0 ± 0.5 | 4.3 ± 0.7 | 2.8 ± 0.7 |
| GATCTAGAGCCGCG (-111) | |||||||
| Locus 1 | Protein ID 2 | Name | NtrX Binding Sequences (Position from Start Codon) | qRT-PCR | ||
|---|---|---|---|---|---|---|
| WT | NtrY | |||||
| +Fe | −Fe | +Fe | ||||
| Pden_0893 | A1B0G0 | Cytochrome c peroxidase (Ccp) | GTGGGCGCGTCTCG (-78) GTCAGATGTTTTCG (-133) GTTCTGCCTTGCCG (-149) GACGGTCCGTCGCG (-108) | 0.4 ± 0.2 | 1.1 ± 0.3 | 8.4 ± 0.9 |
| Pden_1355 | A1B1R3 | Methionine synthase B12-independent (MetE) | CATGCCACTGGCGC (-201) CAAGGTGACATCGC (-77) CATCGCCGCTTCGC (-69) CTGTTTCCTCAGGC (-19) CTCAGAAGGCATGC (-3) GTTTCCTCAGGCCG (-17) GACCGCGGTGGGCG (-111) GAGCGGCCCATCCG (-91) GATATGCAAGGACG (-7) | 0.8 ± 0.4 | 4.3 ± 0.9 | 36.5 ± 2.3 |
| Pden_1848 | Q51679 | Cytochrome c oxidase, cbb3-type, subunit I | CTTAAATCCTGCGC (-11) GTCACACGGTTTCG (-86) GACTTTGATCTGCG (-103) CAATCTGTCATTGC (-118) CAGGATGTCGCAGC (-163) CAGGTGAAACTTGC (-211) | 0.6 ± 0.3 | 1.86 ± 0.3 | 7.5 ± 0.5 |
| Pden_1850 | A1B353 | Putative transcriptional regulator, Crp/Fnr family (FnrP) | CAAGGTTCCAGCGC (-125) CTCATCGCCTTCGC (-18) CTGCTGCGTCGCGC (-78) | 0.3 ± 0.1 | 1.1 ± 0.2 | 4.1 ± 0.9 |
| Pden_1937 | P00096 | Cytochrome c550 | CACAATGATCTTGC (-61) CATGATCCGCAGGC (-39) | 0.4 ± 0.1 | 1.2 ± 0.1 | 6.9 ± 0.2 |
| Pden_2046 | A1B3P4 | TonB-dependent receptor | GATCCCTTGTCCCG (-105) CTCGCCCTCTCGGC (-37) | 0.5 ± 0.1 | 13.2 ± 1.1 | 5.4 ± 0.1 |
| Pden_2305 | A1B4F2 | Ubiquinol-cytochrome c reductase iron-sulfur subunit (cytochrome bc1) | CTGCGGCGATTTGC (-119) GTTCCGTCGTATCG (-20) GTCGTATCGCCCCG (-15) GATCGCTAGAACCG (-64) | 0.6 ± 0.2 | 1.1 ± 0.1 | 8.9 ± 0.6 |
| Pden_2484 | Q51662 | Nitric oxide reductase subunit C (NorC) | CAAGCGTGAGTCGC (-47) GACCTCACTGTCCG (-32) | 0.2 ± 0.1 | 3.8 ± 0.3 | 12.5 ± 1.2 |
| Pden_2486 | O33432 | Protein NirI | GTCAAAGCCCCGCG (-59) GAACGGCGTGAACG (-89) | 3.2 ± 0.6 | 3.4 ± 0.1 | 3.4 ± 0.6 |
| Pden_2487 | Q2HPX3 | Nitrite reductase NirS | GTCAAAGCCCCGCG (-59) GAACGGCGTGAACG (-89) | 4.4 ± 0.9 | 5.0 ± 0.9 | 24.6 ± 2.2 |
| Pden_2610 | A1B5A3 | TonB-dependent receptor | GTCGGCAGGCTGCG (-155) | 0.2 ± 0.1 | 1.4 ± 0.4 | 5.0 ± 0.9 |
| Pden_2832 | A1B5X2 | Rieske [2Fe-2S] domain protein (Stc2) | CAGCCGAATGTCGC (-176) CTGCCGTAACTTGC (-54) CTCCGTCCGGTCGC (-286) | 0.3 ± 0.9 | 3.6 ± 0.2 | 5.2 ± 0.9 |
| Pden_3027 | A1B6G6 | Alkylhydroperoxide reductase (AhpD) | GACGCTTGCCGCCG (-58) | 0.5 ± 0.1 | 1.7 ± 0.4 | 5.2 ± 0.4 |
| Pden_4044 | A1B9B6 | NnrS family protein | GAGCCGGTGCCACG (-30) GAGGGGCCGCATCG (-10) | 0.2 ± 0.1 | 0.3 ± 0.1 | 6.7 ± 0.7 |
| Pden_4213 | A1B9T3 | Acetyl-coenzyme A synthetase (Acs) | GTACGGGACATGCG (-221) GAAAACCGATTGCG (-75) | 0.4 ± 0.1 | 0.6 ± 0.1 | 2.1 ± 0.7 |
| Pden_4221 | A1B9U1 | NosC protein | GTTTATGGATCGCG (-74) GTCCCGACCCTGCG (-194) GAAGGAGAATCGCG (-35) GAGTTTTTTCCTCG (-231) | 1.6 ± 0.8 | 3.1 ± 0.2 | 12.5 ± 0.9 |
| Pden_4222 | Q71RW5 | Pseudoazurin | GAAGGAGAATCGCG (-287) GAGTTTTTTCCTCG (-91) GTTTATGGATCGCG (-248) GTCCCGACCCTGCG (-128) | 0.5 ± 0.1 | 0.7 ± 0.1 | 20.0 ± 1.3 |
| Pden_4237 | A1B9V7 | Nitrate/nitrite transporter NarK | CTCAAATCGTCAGC (-150) GTCCGGCCGGCCCG (-89) GATTGGGACTTTCG (-12) GACTTCTCAAATCG (-145) GATTTTTGCAAGCG (-221) | 6.8 ± 0.8 | 8.8 ± 0.13 | 45.3 ± 3.5 |
| Pden_4238 | A1B9V8 | Putative transcriptional regulator, Crp/Fnr family (NarR) | CTCAAATCGTCAGC (-150) GTCCGGCCGGCCCG (-89) GATTGGGACTTTCG (-12) GACTTCTCAAATCG (-145) GATTTTTGCAAGCG (-221) | 0.4 ± 0.1 | 1.2 ± 0.4 | 6.1 ± 0.3 |
| Pden_4453 | A1BAH3 | Major facilitator superfamily MFS_1, nitrate transporter NasA | CTGATGGCGAAGGC (-18) CTGTCGGAAAGCGC (-31) CTGGGTCAGGACGC (-176) CTCCGCCCGAAAGC (-235) GTCGCGGGAATCCG (-104) | 1.9 ± 0.2 | 53.4 ± 4.3 | 14.7 ± 0.1 |
| Pden_4455 | A1BAH5 | Response regulator receiver ANTAR domain protein NasT | GTCCCGGTTTTGCG (-82) GAACCCCGCGCCCG (-38) | 1.0 ± 0.3 | 7.5 ± 1.6 | 2.9 ± 0.9 |
| Pden_4719 | A1BB86 | Periplasmic nitrate reductase subunit (NapE) | GAGGCGTTTTGACG (-16) | 0.2 ± 0.1 | 2.2 ± 0.1 | 2.1 ± 0.3 |
| Pden_5108 | A1BCC4 | Cytochrome ba3 quinoloxidase subunit 2 | CAGGTGCGGACGGC (-25) CAAGCTCGCCATGC (-103) CTGAGTCGCAGGGC (-38) GATGTCGGTGAGCG (-10) GAGCGACAGGTGCG (-19) GACAGGTGCGGACG (-23) GAAACGCCGCGGCG (-154) | 0.3 ± 0.1 | 1.1 ± 0.7 | 6.9 ± 0.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olaya-Abril, A.; Luque-Almagro, V.M.; Hidalgo-Carrillo, J.; Chicano-Gálvez, E.; Urbano, F.J.; Moreno-Vivián, C.; Richardson, D.J.; Roldán, M.D. The NtrYX Two-Component System of Paracoccus denitrificans Is Required for the Maintenance of Cellular Iron Homeostasis and for a Complete Denitrification under Iron-Limited Conditions. Int. J. Mol. Sci. 2022, 23, 9172. https://doi.org/10.3390/ijms23169172
Olaya-Abril A, Luque-Almagro VM, Hidalgo-Carrillo J, Chicano-Gálvez E, Urbano FJ, Moreno-Vivián C, Richardson DJ, Roldán MD. The NtrYX Two-Component System of Paracoccus denitrificans Is Required for the Maintenance of Cellular Iron Homeostasis and for a Complete Denitrification under Iron-Limited Conditions. International Journal of Molecular Sciences. 2022; 23(16):9172. https://doi.org/10.3390/ijms23169172
Chicago/Turabian StyleOlaya-Abril, Alfonso, Víctor M. Luque-Almagro, Jesús Hidalgo-Carrillo, Eduardo Chicano-Gálvez, Francisco J. Urbano, Conrado Moreno-Vivián, David J. Richardson, and María Dolores Roldán. 2022. "The NtrYX Two-Component System of Paracoccus denitrificans Is Required for the Maintenance of Cellular Iron Homeostasis and for a Complete Denitrification under Iron-Limited Conditions" International Journal of Molecular Sciences 23, no. 16: 9172. https://doi.org/10.3390/ijms23169172
APA StyleOlaya-Abril, A., Luque-Almagro, V. M., Hidalgo-Carrillo, J., Chicano-Gálvez, E., Urbano, F. J., Moreno-Vivián, C., Richardson, D. J., & Roldán, M. D. (2022). The NtrYX Two-Component System of Paracoccus denitrificans Is Required for the Maintenance of Cellular Iron Homeostasis and for a Complete Denitrification under Iron-Limited Conditions. International Journal of Molecular Sciences, 23(16), 9172. https://doi.org/10.3390/ijms23169172