Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2
Abstract
:1. Introduction
2. Results
2.1. Subjects
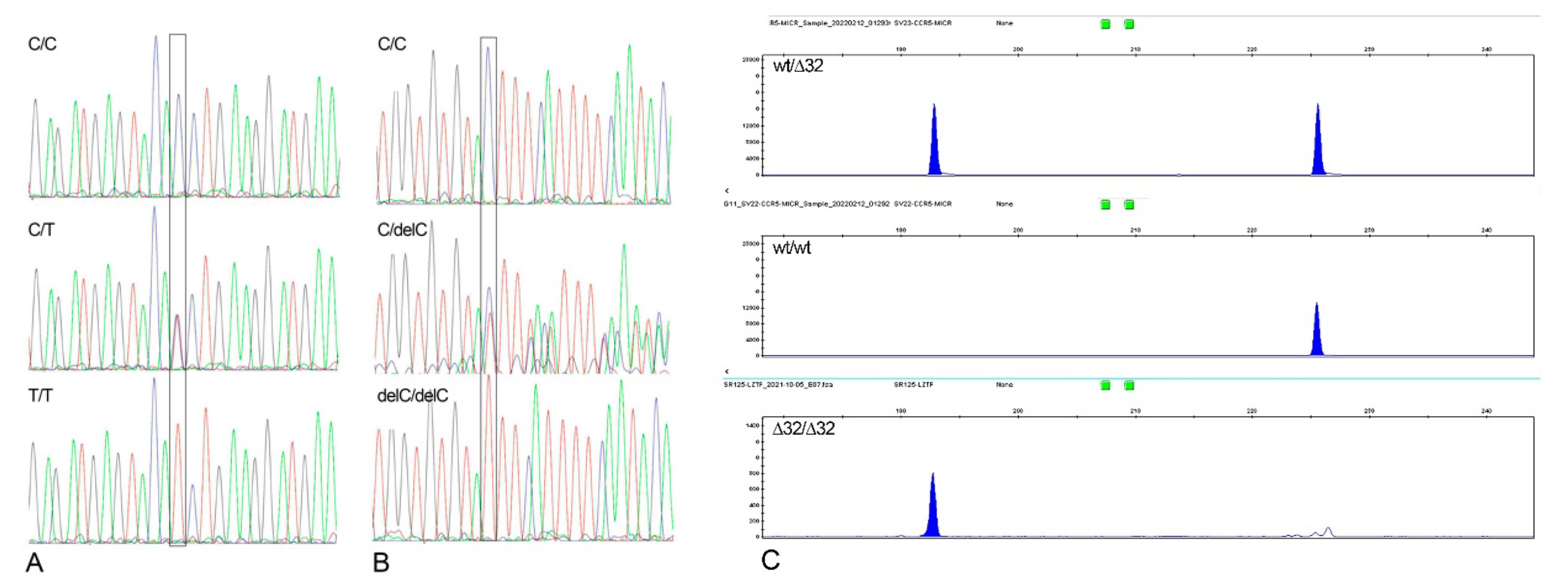
2.2. TMPRSS2 Gene
2.3. CCR5 Gene
2.4. TMPRSS2, CCR5 Gene Polymorphisms and COVID-19 Disease Severity
- Headache, rhinitis, pharyngitis, fatigue, mild cough, muscle aches.
- Fever < 37.5 °C.
- Peripheral SpO2 saturation >95.
- Convulsive cough.
- Fever < 37.5 °C.
- Mild dyspnea.
- Peripheral SpO2 saturation >90–<95.
- Severe dyspnea.
- Fever > 37.5 °C.
- Marked Desaturation SpO2 < 90; P/F < 300.
2.5. Genotype Concordance between Infected and Non Infected Members in Cohabiting Subjects
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Nasopharyngeal Swabs for SARS-CoV-2 Detection
4.3. DNA Preparation and Analysis
- TMPRSS2 gene, polymorphism rs12329760:
- for: 5′-TCTGCTGTCTGTTACTGTCACT-3′,
- rev 5′-ACTCATGGATAATCCTCCCTC-3′.
- TMPRSS2 gene, polymorphism rs35074065:
- for: 5′-GGGCCCCCAAAGTAACCAATGGA-3′,
- rev 5′-ATGGACCATTGAGCCAGTGCTTATGT-3′.
- CCR5 gene, polymorphism rs333:
- for: 5′-[6FAM] CTGTGTTTGCGTCTCTCCCA-3′,
- rev 5′-CCTCTTCTTCTCATTTCGACAC-3′.
4.4. Statistical Analysis
- (1)
- The differences between four COVID-19 categories (asymptomatic, pauci-symptomatic, symptomatic, and severe) and the genotypes of each of the three loci analyzed in 66 subjects with intellectual disability using a 4 × 3 contingency table.
- (2)
- The genotypes and allele frequencies between infected and non-infected patients using a 3 × 2 contingency table McNemar test. p values less than 0.05 were considered statistically significant. If the total N for a 2 × 2 chi-square table was less than about 40, the Yates continuity correction was used to compensate for deviations from the theoretical probability distribution. The calculation of the Pearson Correlation Coefficient was conducted using online website software: https://www.socscistatistics.com, accessed on 5 May 2022. Sample size: the statistical appropriateness of the chi-squared tests used in this study was assessed a posteriori by first calculating the effect size and then calculating the corresponding sample size required with α = 0.05 (www.statskingdom.com/34test_power_chi2.html, accessed on 5 May 2022).
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mulder, C.; Conti, E.; Saccone, S.; Federico, C. Beyond virology: Environmental constraints of the first wave of COVID-19 cases in Italy. Environ. Sci. Pollut. Res. Int. 2021, 28, 31996–32004. [Google Scholar] [CrossRef] [PubMed]
- Ciencewicki, J.; Jaspers, I. Air pollution and respiratory viral infection. Inhal. Toxicol. 2007, 19, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Sedlmaier, N.; Hoppenheidt, K.; Krist, H.; Lehmann, S.; Lang, H.; Büttner, M. Generation of avian influenza virus (AIV) contaminated fecal fine particulate matter (PM(2.5): Genome and infectivity detection and calculation of immission. Vet. Microbiol. 2009, 139, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Song, F.; Guo, W.; Tan, J.; Zhang, X.; Qiao, F.; Guo, J.; Zhang, L.; Jia, X. Potential Genes Associated with COVID-19 and Comorbidity. Int. J. Med. Sci. 2022, 19, 402–415. [Google Scholar] [CrossRef]
- Mjaess, G.; Karam, A.; Aoun, F.; Albisinni, S.; Roumeguère, T. COVID-19 and the male susceptibility: The role of ACE2, TMPRSS2 and the androgen receptor. Progrès Urol. 2020, 30, 484–487. [Google Scholar] [CrossRef]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Bhanushali, A.; Rao, P.; Raman, V.; Kokate, P.; Ambekar, A.; Mandva, S.; Bhatia, S.; Das, B.R. Status of TMPRSS2-ERG fusion in prostate cancer patients from India: Correlation with clinico-pathological details and TMPRSS2 Met160Val polymorphism. Prostate Int. 2018, 6, 145–150. [Google Scholar] [CrossRef]
- Onder, G.; Razza, G.; Brusaferro, S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. JAMA 2020, 323, 1775–1776. [Google Scholar] [CrossRef]
- Scortichini, M.; Schneider dos Santos, R.; De’ Donato, F.; De Sario, M.; Michelozzi, P.; Davoli, M.; Masselot, P.; Sera, F.; Gasparrini, A. Excess mortality during the COVID-19 outbreak in Italy: A two-stage interrupted time-series analysis. Int. J. Epidemiol. 2020, 49, 1909–1917. [Google Scholar] [CrossRef]
- Onozuka, D.; Tanoue, Y.; Nomura, S.; Kawashima, T.; Yoneoka, D.; Eguchi, A.; Ng, C.F.S.; Matsuura, K.; Shi, S.; Makiyama, K.; et al. Reduced mortality during the COVID-19 outbreak in Japan, 2020: A two-stage interrupted time-series design. Int. J. Epidemiol. 2022, 51, 75–84. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Simmons, G.; Zmora, P.; Gierer, S.; Heurich, A.; Pöhlmann, S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013, 100, 605–614. [Google Scholar] [CrossRef]
- Latini, A.; Agolini, E.; Novelli, A.; Borgiani, P.; Giannini, R.; Gravina, P.; Smarrazzo, A.; Dauri, M.; Andreoni, M.; Rogliani, P.; et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes 2020, 11, 1010. [Google Scholar] [CrossRef]
- Murray, M.F.; Kenny, E.E.; Ritchie, M.D.; Rader, D.J.; Bale, A.E.; Giovanni, M.A.; Abul-Husn, N.S. COVID-19 outcomes and the human genome. Genet. Med. 2020, 22, 1175–1177. [Google Scholar] [CrossRef]
- Huang, S.-W.; Wang, S.-F. SARS-CoV-2 entry related viral and host genetic variations: Implications on COVID-19 severity, immune escape, and infectivity. Int. J. Mol. Sci. 2021, 22, 3060. [Google Scholar] [CrossRef]
- Lodhi, N.; Singh, R.; Rajput, S.P.; Saquib, Q. SARS-CoV-2: Understanding the transcriptional regulation of ACE2 and TMPRSS2 and the role of single nucleotide polymorphism (SNP) at codon 72 of p53 in the innate immune response against virus infection. Int. J. Mol. Sci. 2021, 22, 8660. [Google Scholar] [CrossRef]
- Ravikantha, V.; Sasikalaa, M.; Naveenb, V.; Lathac, S.S.; Parsac, K.V.L.; Vijayasarathya, K.; Amanchyd, R.; Avanthia, S.; Govardhana, B.; Rakesha, K.; et al. A variant in TMPRSS2 is associated with decreased disease severity in COVID-19. Meta Gene 2021, 29, 100930. [Google Scholar] [CrossRef]
- Russo, R.; Andolfo, I.; Lasorsa, V.A.; Iolascon, A.; Capasso, M. Genetic analysis of the coronavirus SARS-CoV-2 host protease TMPRSS2 in different populations. Front. Genet. 2020, 11, 872. [Google Scholar] [CrossRef]
- Hubacek, J.A.; Dusek, L.; Majek, O.; Adamek, V.; Cervinkova, T.; Dlouha, D.; Pavel, J.; Adamkova, V. CCR5Delta32 deletion as a protective factor in Czech first-wave COVID-19 subjects. Physiol. Res. 2021, 70, 111–115. [Google Scholar] [CrossRef]
- Cuesta-Llavona, E.; Gómez, J.; Albaiceta, G.M.; Amado-Rodríguez, L.; García-Clemente, M.; Gutiérrez-Rodríguez, J.; López-Alonso, I.; Hermida, T.; Enríquez, A.I.; Hernández-González, C.; et al. Variant-genetic and transcript-expression analysis showed a role for the chemokine-receptor CCR5 in COVID-19 severity. Int. Immunopharmacol. 2021, 98, 107825. [Google Scholar] [CrossRef]
- Gómez, J.; Cuesta-Llavona, E.; Albaiceta, G.M.; García-Clemente, M.; López-Larrea, C.; Amado-Rodríguez, L.; López-Alonso, I.; Hermida, T.; Henríquez, A.I.; Gil, H.; et al. The CCR5-delta32 variant might explain part of the association between COVID-19 and the chemokine-receptor gene cluster. medRxiv 2020. [Google Scholar] [CrossRef]
- Després, V.R.; Huffman, J.A.; Burrows, S.M.; Hoose, C.; Safatov, A.S.; Buryak, G.; Fröhlich-Nowoisky, J.; Elbert, W.; Andreae, M.O.; Pöschl, U.; et al. Primary biological aerosol particles in the atmosphere: A review. Tellus B 2012, 64, 15598. [Google Scholar] [CrossRef]
- Panda, A.K.; Padhi, A.; Prusty, B.A.K. CCR5 Δ32 minor allele is associated with susceptibility to SARS-CoV-2 infection and death: An epidemiological investigation. Clin. Chim. Acta 2020, 510, 60–61. [Google Scholar] [CrossRef]
- Cizmarevic, S.N.; Tota, M.; Ristić, S. Does the CCR5-Δ32 mutation explain the variable coronavirus-2019 pandemic statistics in Europe? Croat. Med. J. 2020, 61, 525–526. [Google Scholar] [CrossRef]
- Bernas, S.N.; Baldauf, H.; Wendler, S.; Heidenreich, F.; Lange, V.; Hofmann, J.A.; Sauter, J.; Schmidt, A.H.; Schetelig, J. CCR5-Δ32 mutations do not determine COVID-19 disease course. Int. J. Infect. Dis. 2021, 105, 653–655. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Choudhari, R.; Nema, V.; Khan, A.A. ACE2 and TMPRSS2 polymorphisms in various diseases with special reference to its impact on COVID-19 disease. Microb. Pathog. 2021, 150, 104621. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Nascimento, I.J.; Ferreira da Silva-Júnior, E.; Mendonça de Aquino, T. Molecular modeling targeting Transmembrane Serine Protease 2 (TMPRSS2) as an alternative drug target against coronaviruses. Curr. Drug Targets 2022, 23, 240–259. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F. New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020, 18, 216. [Google Scholar] [CrossRef] [PubMed]
- Wulandari, L.; Hamidah, B.; Pakpahan, C.; Damayanti, N.S.; Kurniati, N.D.; Adiatmaja, C.O.; Wigianita, M.R.; Soedarsono, H.D.; Tinduh, D.; Prakoeswa, C.R.S.; et al. Initial study on TMPRSS2 p.Val160Met genetic variant in COVID-19 patients. Hum. Genom. 2021, 15, 29. [Google Scholar] [CrossRef]
- Torre-Fuentes, L.; Matías-Guiu, J.; Hernández-Lorenzo, L.; Montero-Escribano, P.; Pytel, V.; Porta-Etessam, J.; Gómez-Pinedo, U.; Matías-Guiu, J.A. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J. Med. Virol. 2021, 93, 863–869. [Google Scholar] [CrossRef]
- Schönfelder, K.; Breuckmann, K.; Elsner, C.; Dittmer, U.; Fistera, D.; Herbstreit, F.; Risse, J.; Schmidt, K.; Sutharsan, S.; Taube, C.; et al. Transmembrane serine protease 2 polymorphisms and susceptibility to Severe Acute Respiratory Syndrome Coronavirus Type 2 Infection: A german case-control study. Front. Genet. 2021, 12, 667231. [Google Scholar] [CrossRef]
- Musumeci, A.; Vinci, M.; L’Episcopo, F.; Ragalmuto, A.; Neri, V.; Roccella, M.; Quatrosi, G.; Vetri, L.; Calì, F. Implementation of sample pooling procedure using a rapid SARS-CoV-2 diagnostic Real-Time PCR test performed prior to hospital admission of people with intellectual disabilities. Int. J. Environ. Res. Public Health 2021, 18, 9317. [Google Scholar] [CrossRef]
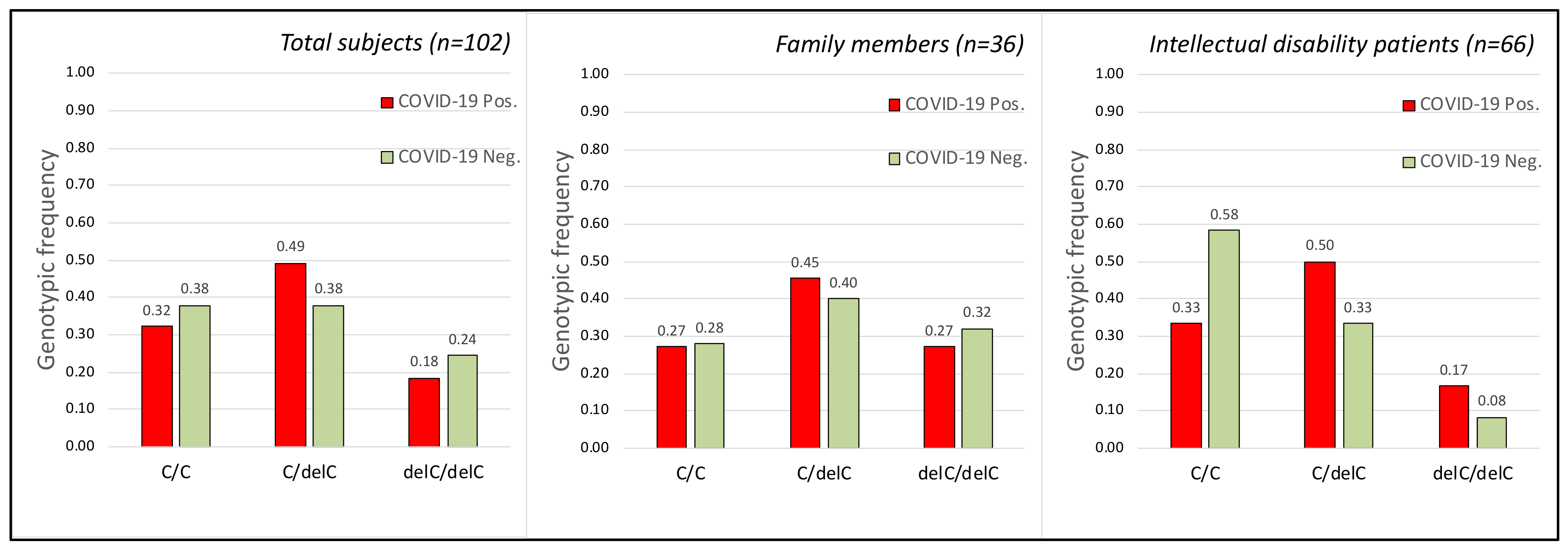
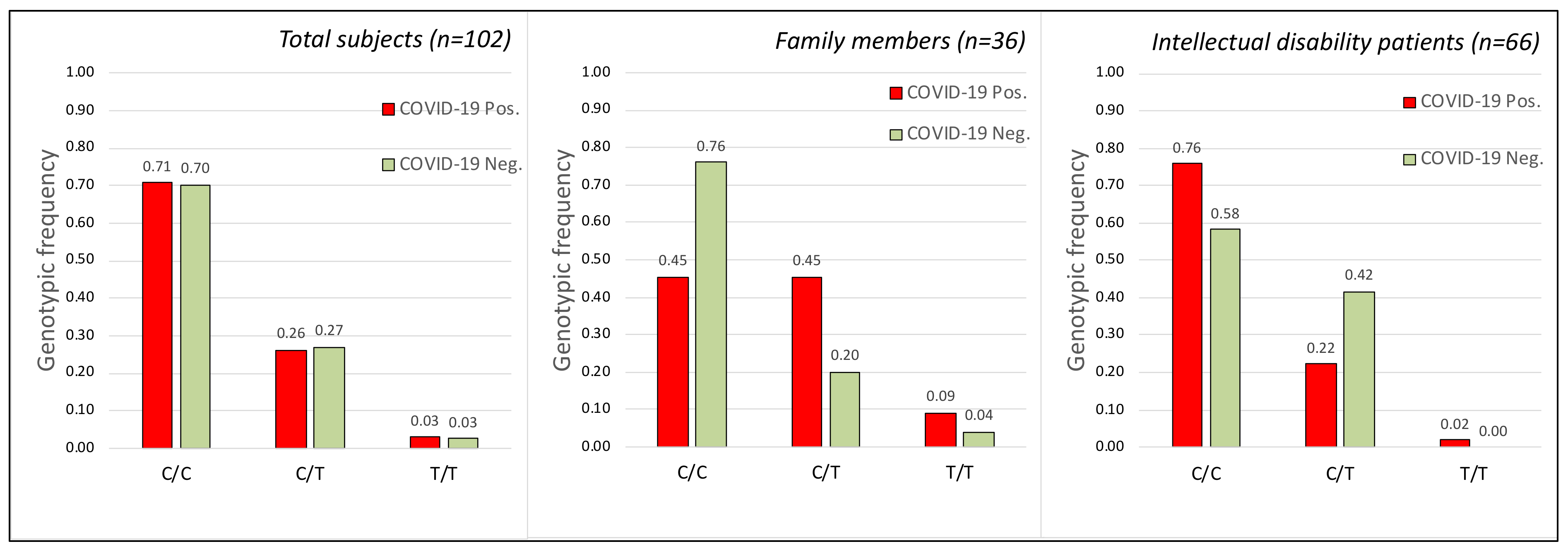
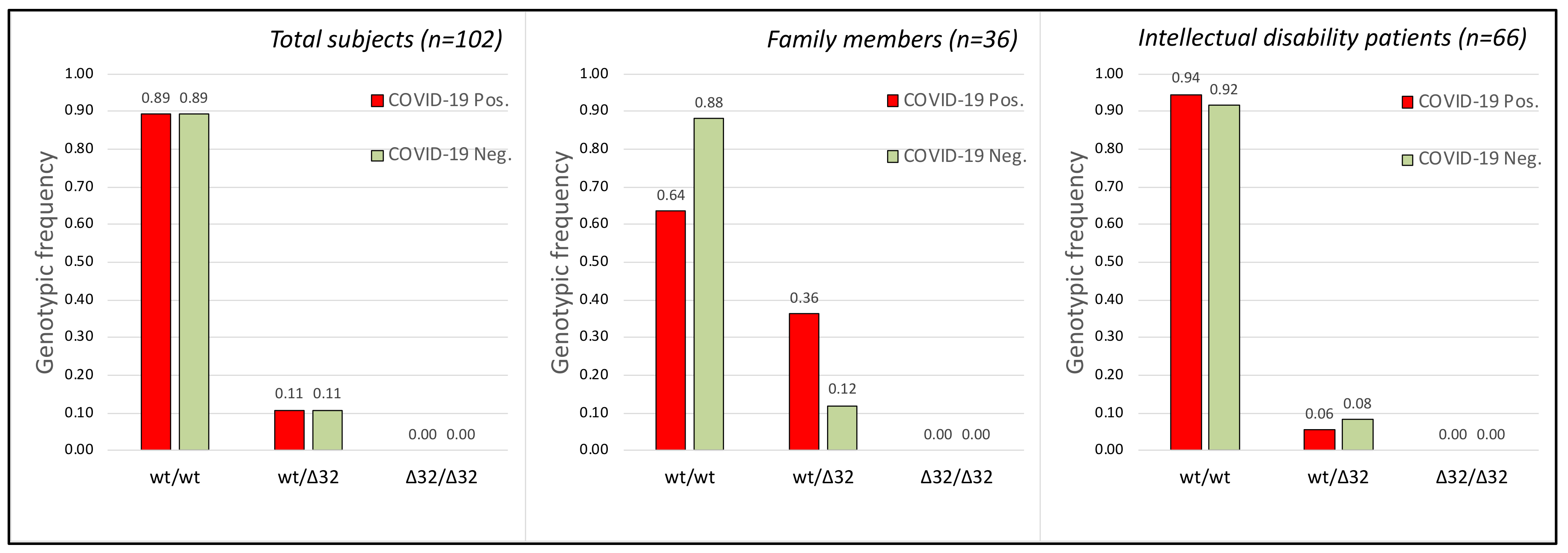
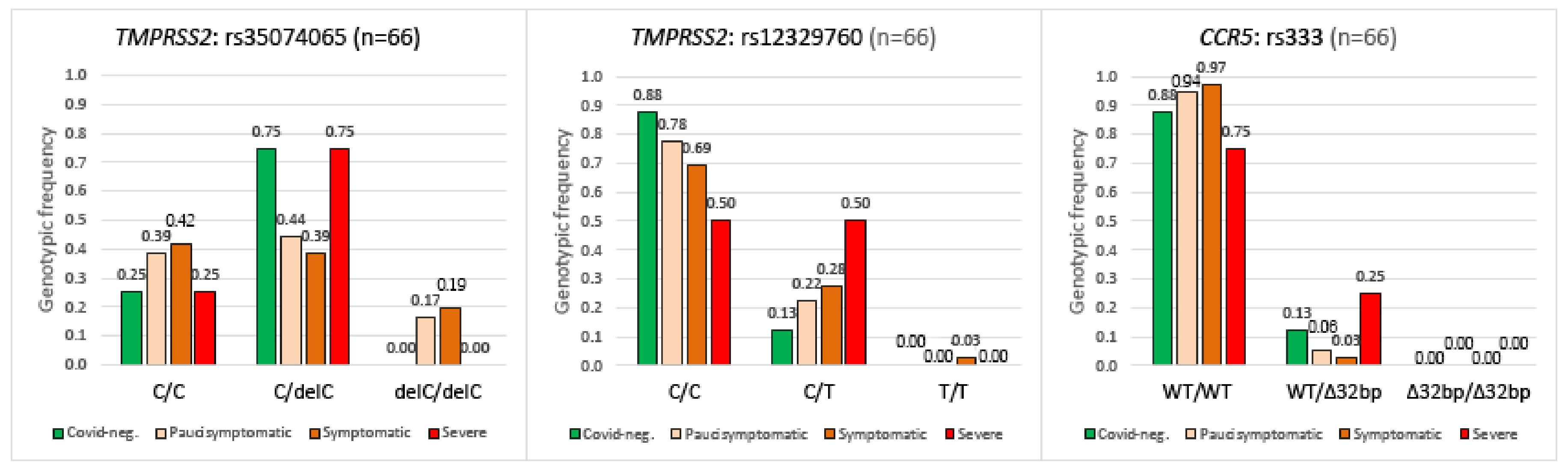
| With Intellectual Disability (1) | Family Members (2) | |||
|---|---|---|---|---|
| COVID-19 Infected | COVID-19 Non-Infected | COVID-19 Infected | COVID-19 Non-Infected | |
| N° | 54 | 12 | 11 | 25 |
| Mean age (±SD) | 42.42 (±11.38) | 40.49 (±17.05) | 36.78 (±18.11) | 32.64 (±17.11) |
| Female/Male | 49/5 | 10/2 | 5/6 | 15/10 |
| (Female %) | (90.7%) | (83.3%) | (45.4%) | (60.0%) |
| Infected vs. Non-Infected | TMPRSS2 rs35074065 | TMPRSS2 rs12329760 | CCR5 rs333 |
|---|---|---|---|
| Females | Chi = 2.157 p = 0.34 | Chi = 1.33 p = 0.51 | Chi = 2.22 p = 0.14 |
| Males | Chi = 0.71 p = 0.70 | Chi = 2.4 p = 0.12 | Chi = 0.15 p = 0.696 |
| SNP rs12329760 | SNP rs35074065 | SNP rs333 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | C/C | C/T | T/T | C/C | C/delC | delC/delC | wt/wt | wt/∆32 | ∆32/∆32 |
| Positive members | 5 | 5 | 1 | 3 | 5 | 3 | 7 | 4 * | 0 |
| Negative members | 19 | 5 | 1 | 7 | 10 | 8 | 22 | 3 | 0 |
| Family numbers with positive and negative members (1) | 5 | 3 | 0 | 1 | 3 | 2 | 7 | 2 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitello, G.A.; Federico, C.; Bruno, F.; Vinci, M.; Musumeci, A.; Ragalmuto, A.; Sturiale, V.; Brancato, D.; Calì, F.; Saccone, S. Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 9171. https://doi.org/10.3390/ijms23169171
Vitello GA, Federico C, Bruno F, Vinci M, Musumeci A, Ragalmuto A, Sturiale V, Brancato D, Calì F, Saccone S. Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2. International Journal of Molecular Sciences. 2022; 23(16):9171. https://doi.org/10.3390/ijms23169171
Chicago/Turabian StyleVitello, Girolamo Aurelio, Concetta Federico, Francesca Bruno, Mirella Vinci, Antonino Musumeci, Alda Ragalmuto, Valentina Sturiale, Desiree Brancato, Francesco Calì, and Salvatore Saccone. 2022. "Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2" International Journal of Molecular Sciences 23, no. 16: 9171. https://doi.org/10.3390/ijms23169171
APA StyleVitello, G. A., Federico, C., Bruno, F., Vinci, M., Musumeci, A., Ragalmuto, A., Sturiale, V., Brancato, D., Calì, F., & Saccone, S. (2022). Allelic Variations in the Human Genes TMPRSS2 and CCR5, and the Resistance to Viral Infection by SARS-CoV-2. International Journal of Molecular Sciences, 23(16), 9171. https://doi.org/10.3390/ijms23169171










